Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcript Expression Analysis Using Oncomine Platform
2.2. Transcript Expression Analysis Using GENT
2.3. Transcript Expression Analysis Using GEPIA
2.4. Methylation Status Analysis Using TCGA Wanderer
2.5. PPI Analysis Using STRING
2.6. Analysis of Gene Expression and Mutation Alterations Using cBioPortal
2.7. Survival Analysis Using Kaplan-Meier Plotter
2.8. Survival Analysis Using PROGgeneV2
2.9. Prognosis Analysis Using PrognoScan
2.10. Prognosis Analysis Using SurvExpress
2.11. Survival and Correlation Analysis Using R2
2.12. Statistical Analysis
3. Results
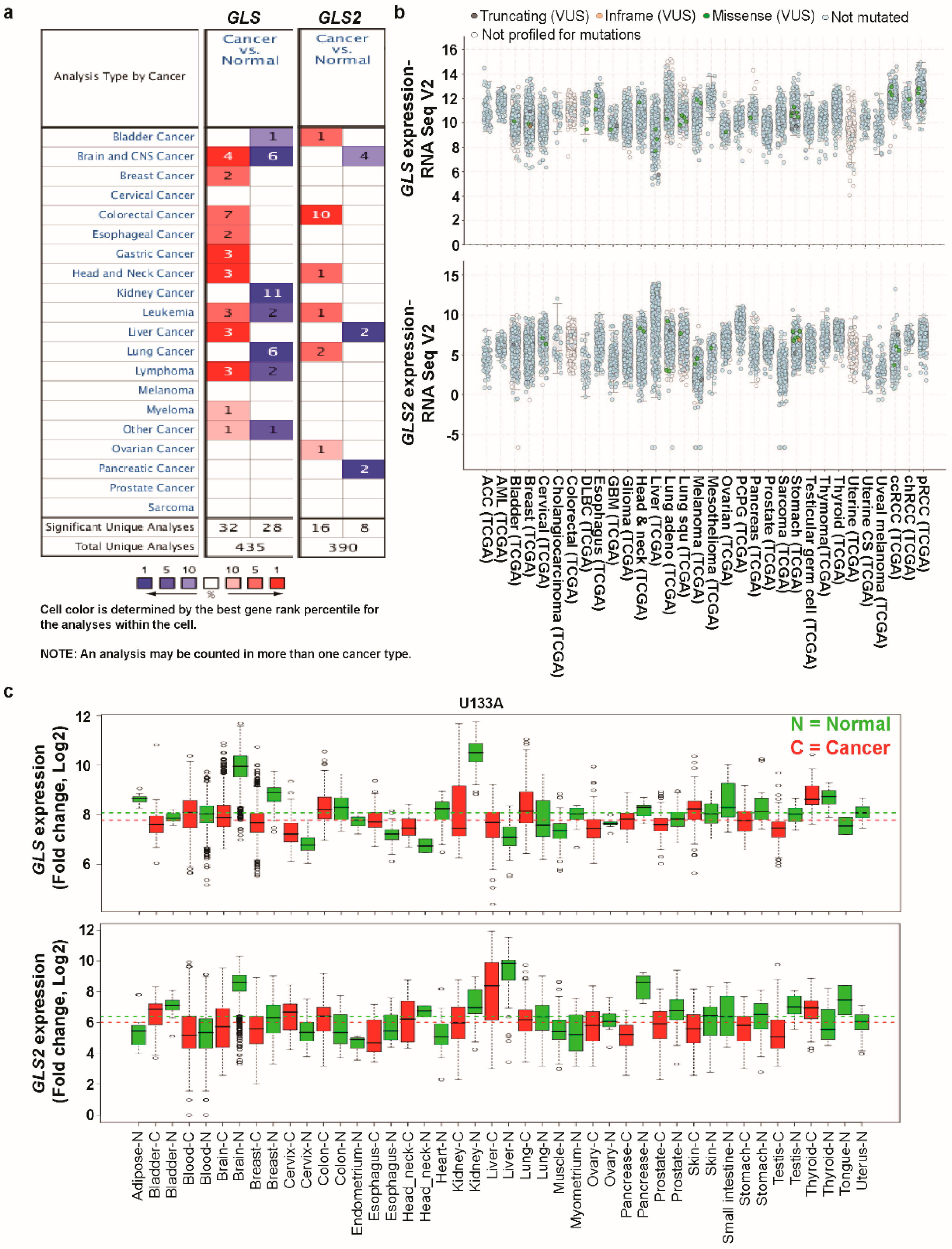
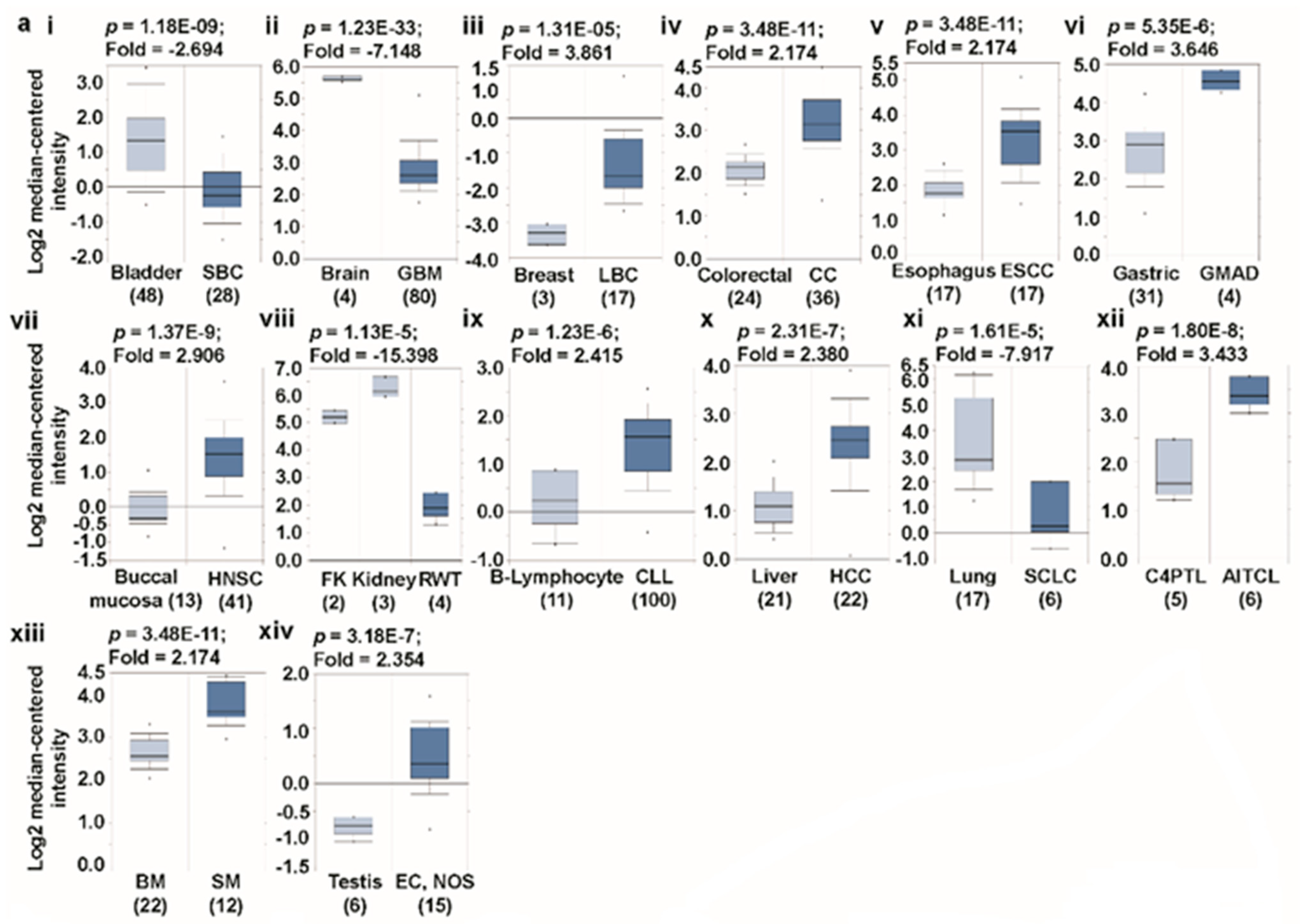
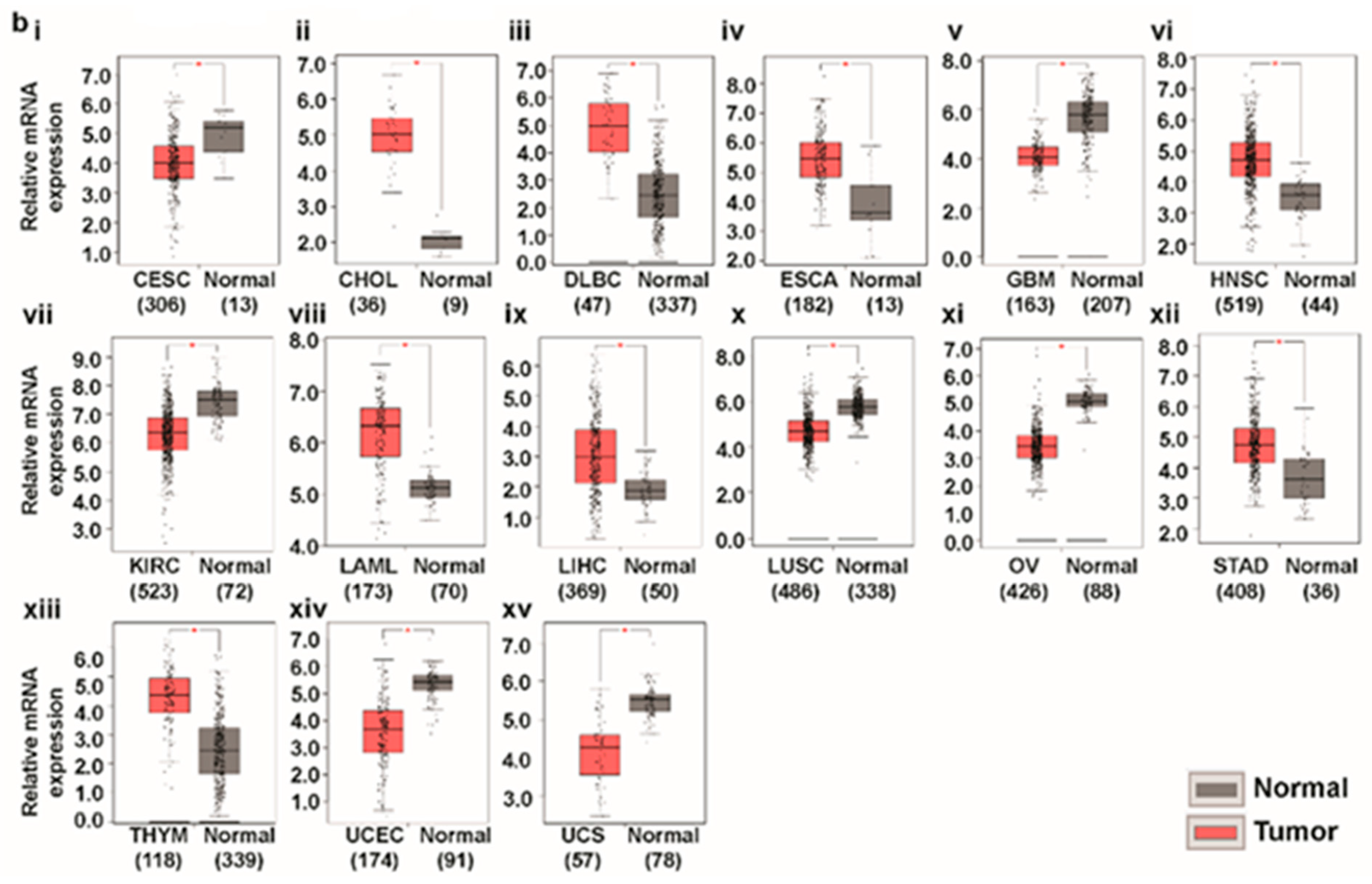
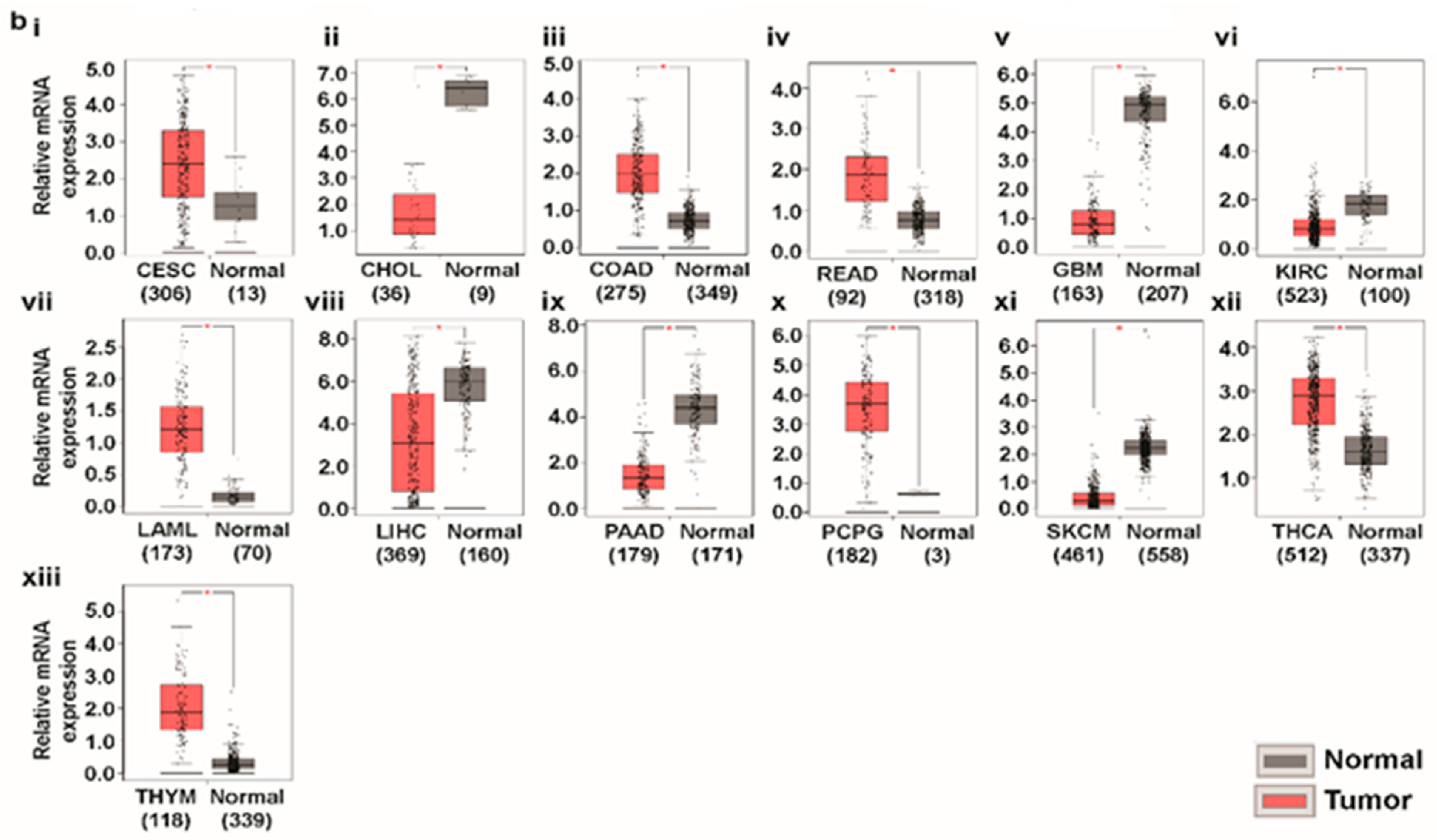
3.1. Transcript Expression Analysis of Glutaminases
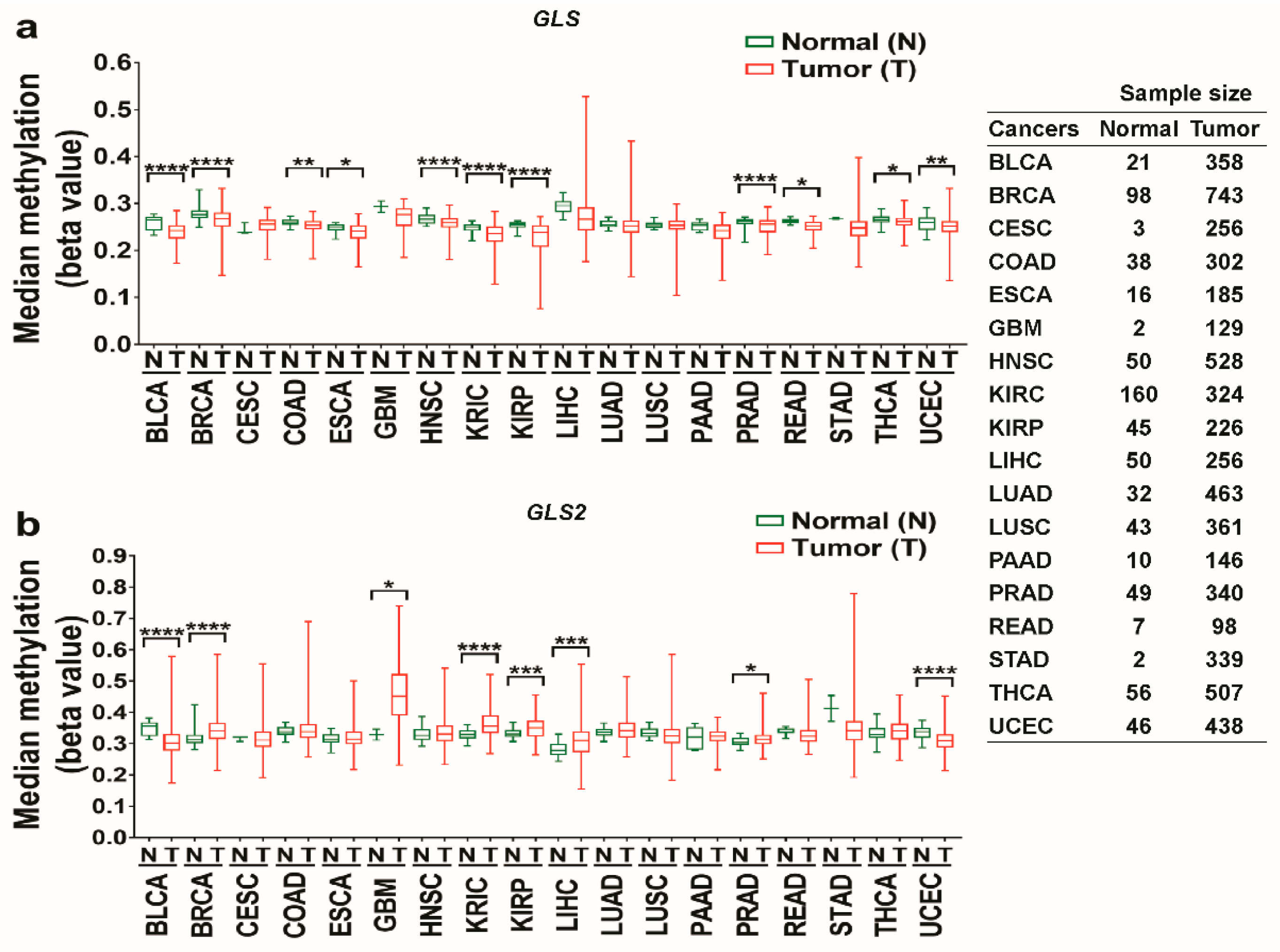
3.2. The Promoter Methylation Status of Glutaminases in Different Types of Cancer
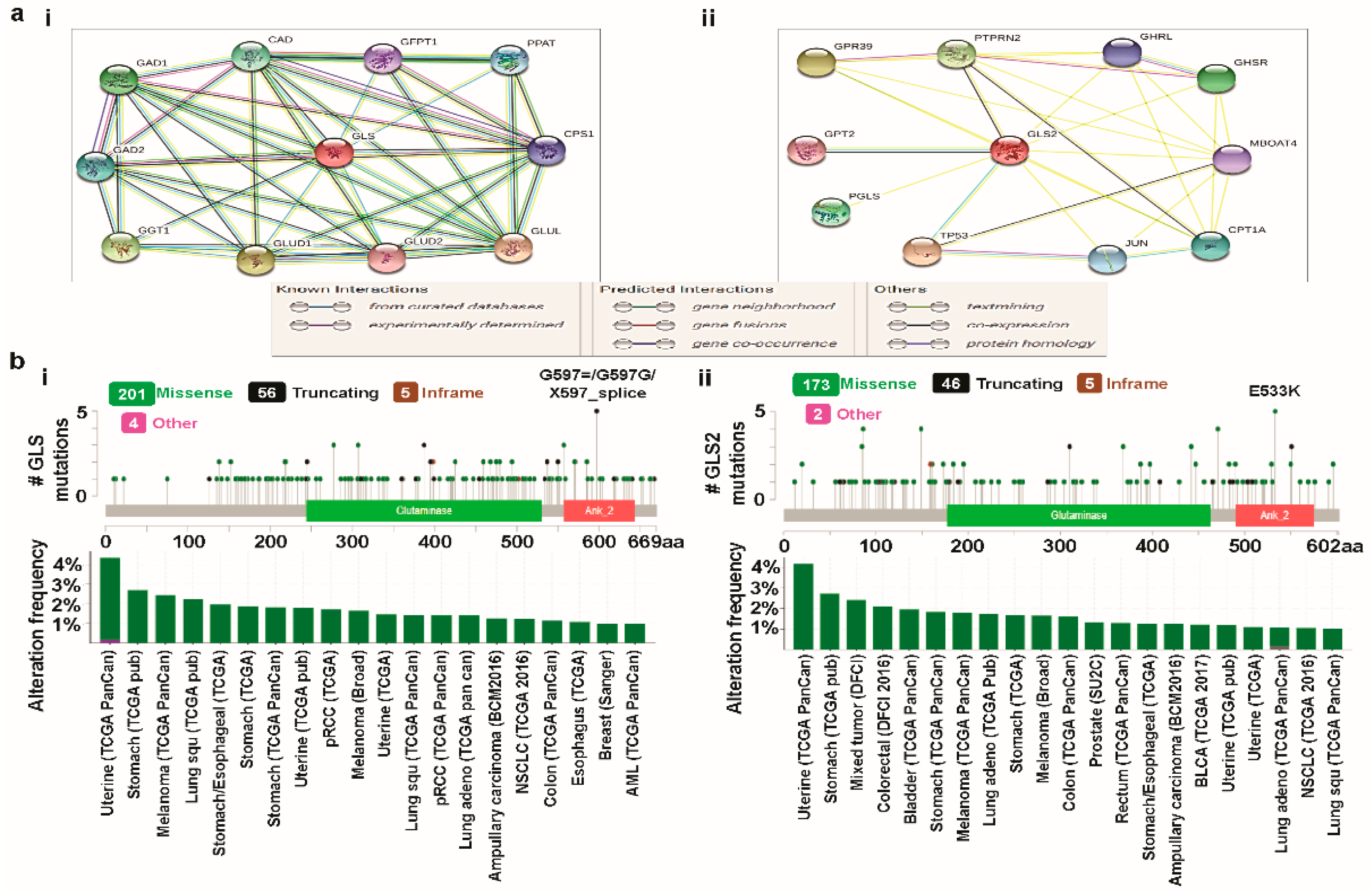
3.3. Predicting Protein-Protein Interactions (PPIs) of Glutaminases
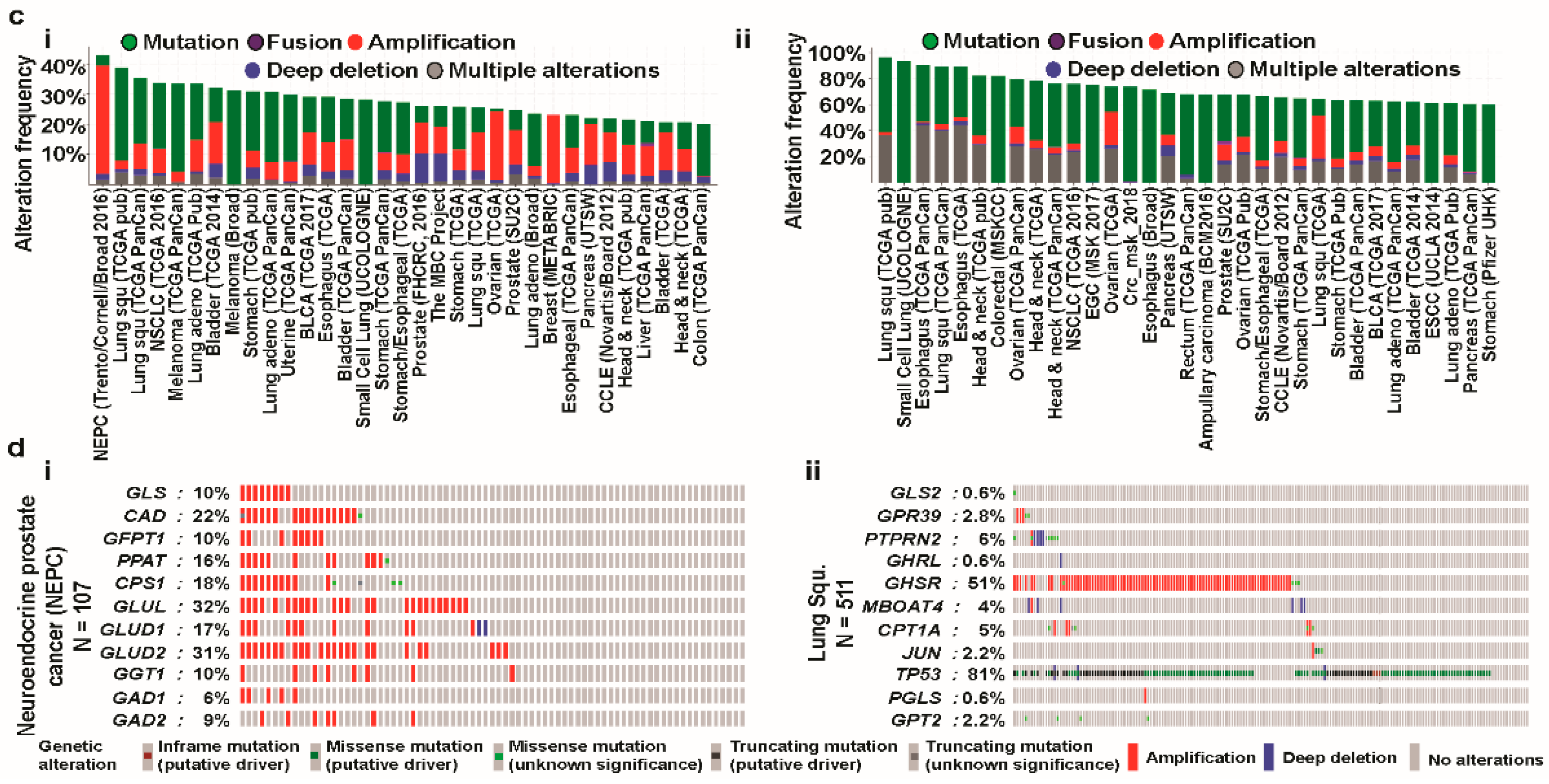
3.4. Cross-Cancer Relationship of Glutaminases with Mutations and Copy Number Alterations (CNAs)
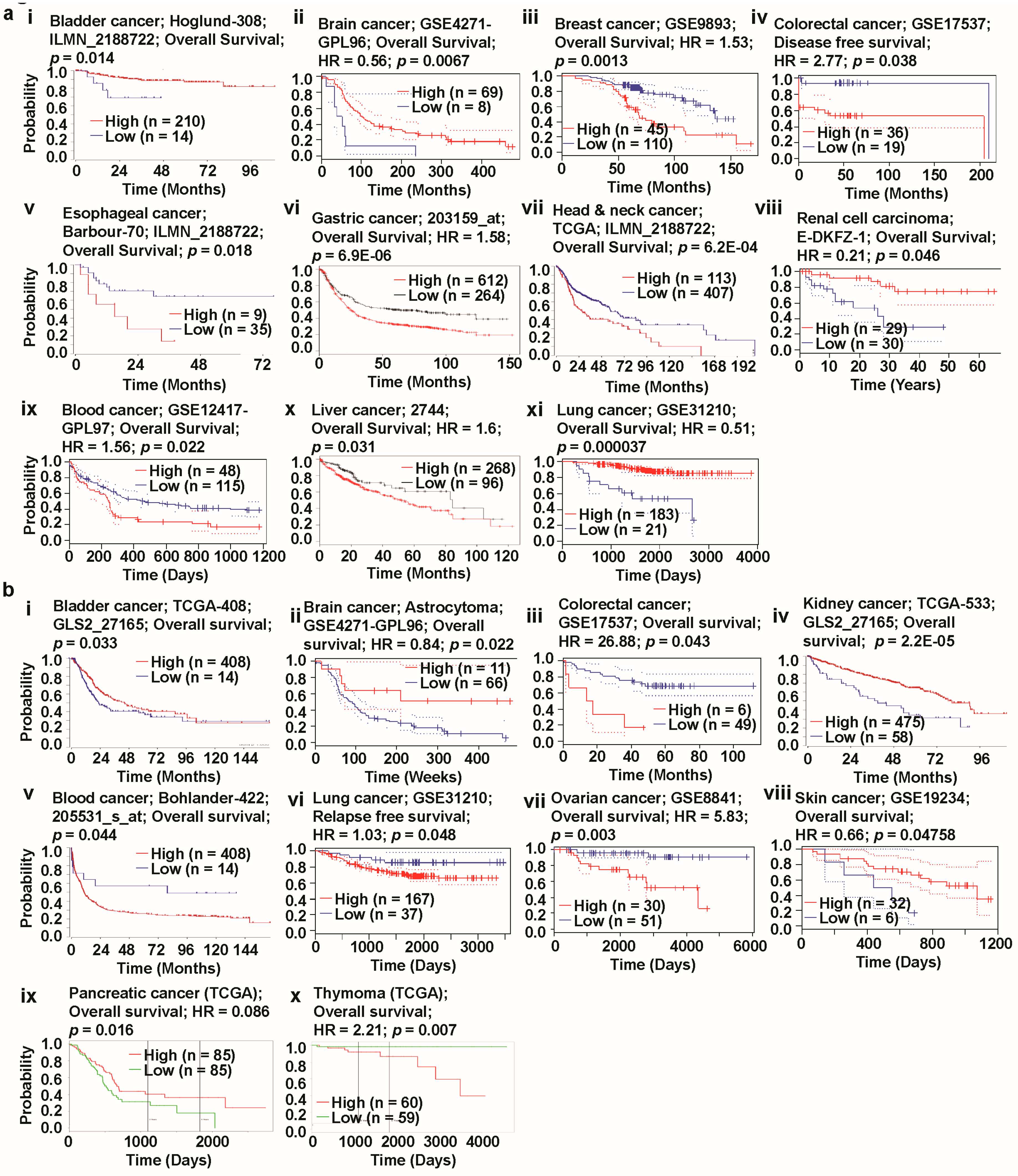
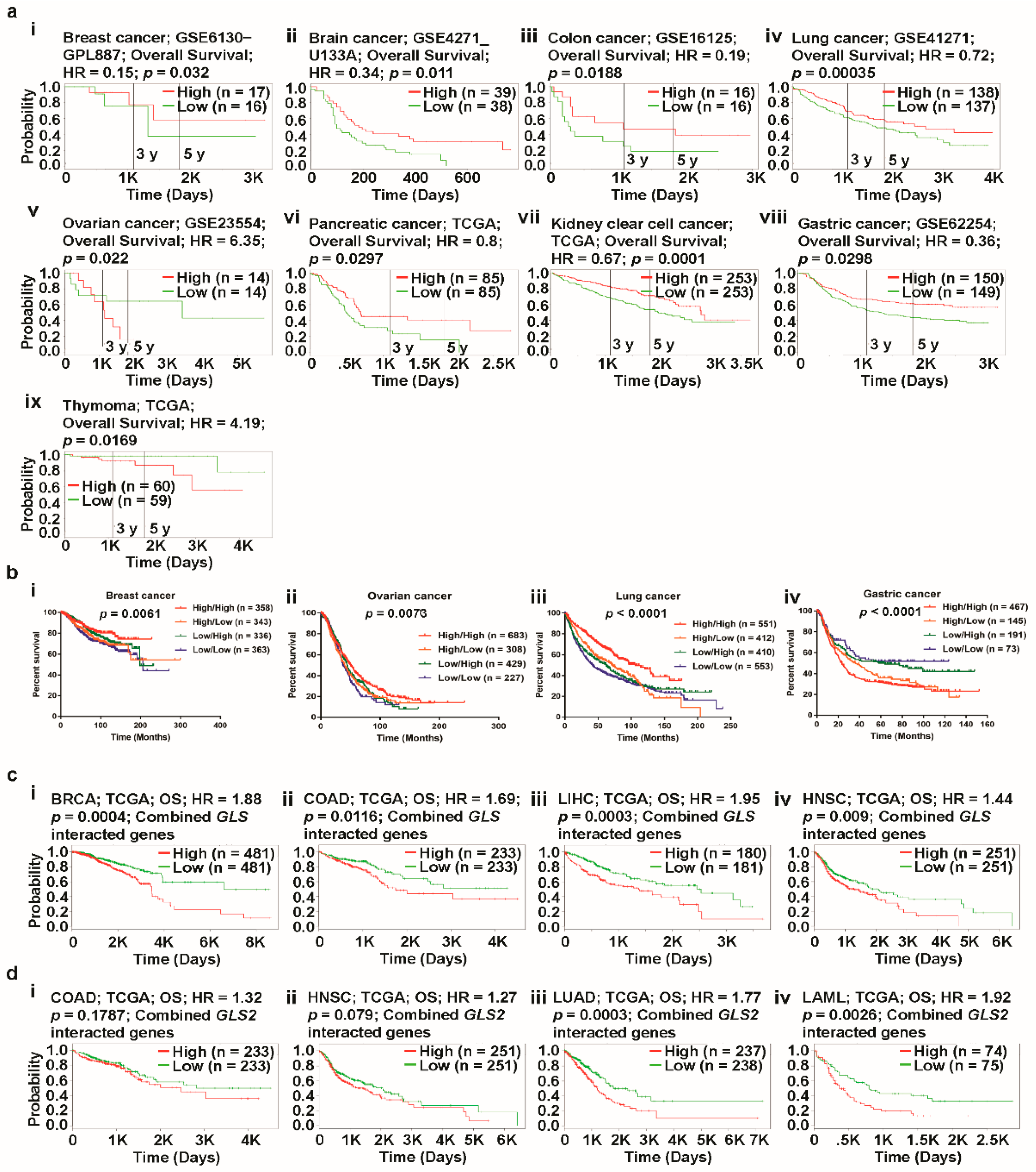
3.5. Prognosis Estimation of Glutaminases
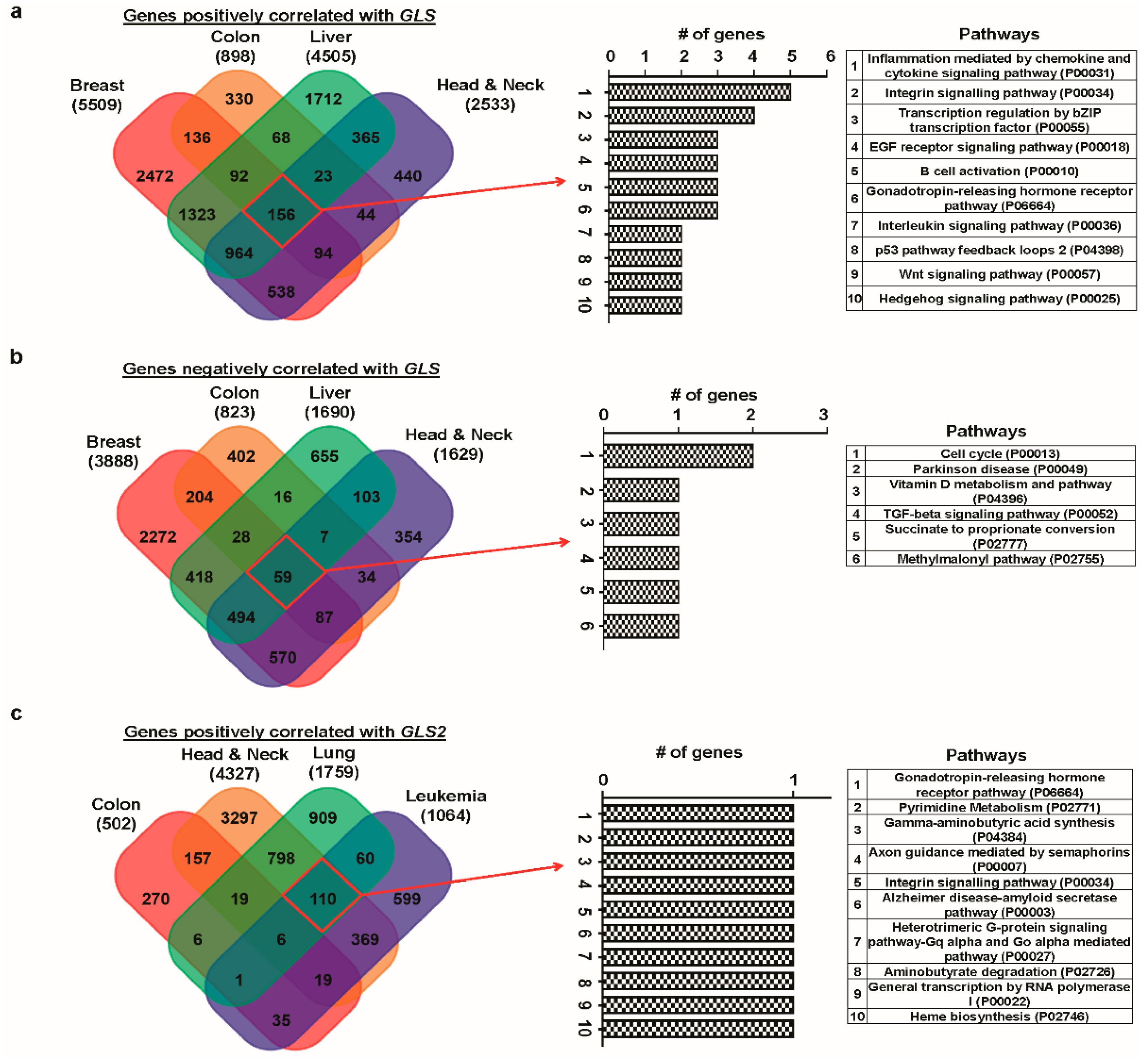
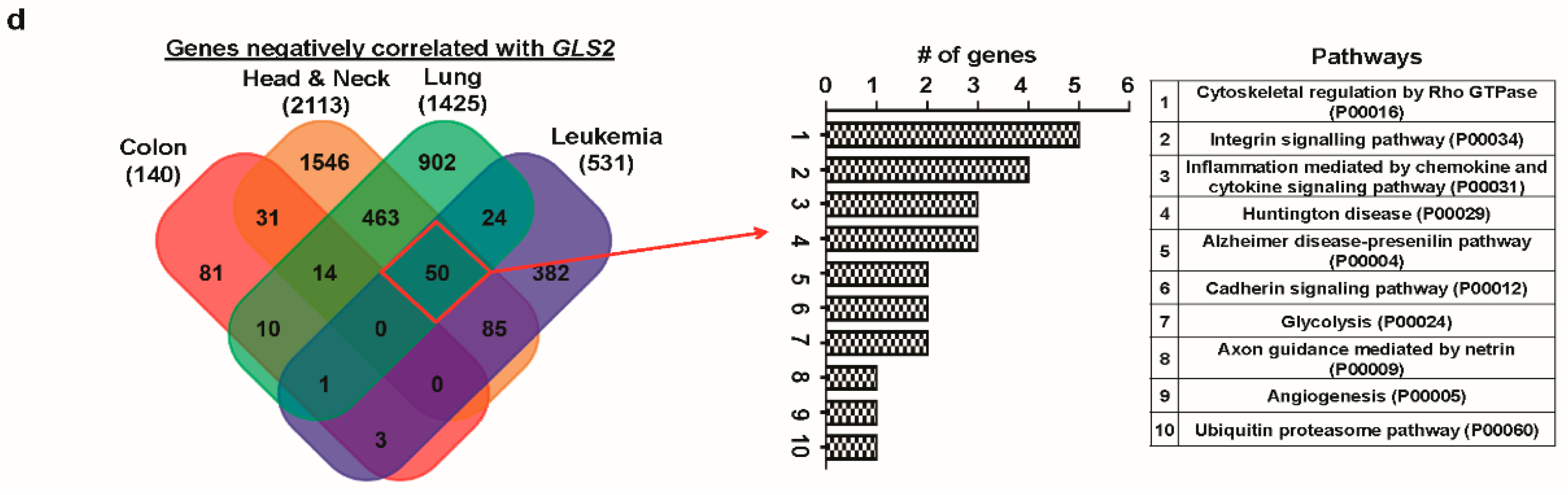
3.6. Correlated Genes of Glutaminases and Their Functional Gene Ontology and Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. Cancer Metabolism: The Known, Unknowns. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 1. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Gonçalves, V.; Lameirinhas, A.; Henrique, R.; Jeronimo, C. Metabolism and epigenetic interplay in cancer: Regulation and putative therapeutic targets. Front. Genet. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Luengo, A.; Gui, D.Y.; Vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; Márquez, J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Mates, J.M.; Segura, J.A.; Martin-Rufian, M.; Campos-Sandoval, J.A.; Alonso, F.J.; Marquez, J. Glutaminase Isoenzymes as Key Regulators in Metabolic and Oxidative Stress Against Cancer. Curr. Mol. Med. 2013, 13, 514–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Erickson, J.W.; Fuji, R.; Ramachandran, S.; Gao, P.; Dinavahi, R.; Wilson, K.F.; Ambrosio, A.L.; Dias, S.M.; Dang, C.V.; et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 2010, 18, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; Lokshin, M.; Hosokawa, H.; Nakayama, T.; Suzuki, Y.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 2010, 107, 7461–7466. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, C.; Wu, R.; Sun, Y.; Levine, A.; Feng, Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA 2010, 107, 7455–7460. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef]
- Jacque, N.; Ronchetti, A.M.; Larrue, C.; Meunier, G.; Birsen, R.; Willems, L.; Saland, E.; Decroocq, J.; Maciel, T.T.; Lambert, M.; et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015, 126, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Shin, G.; Kang, T.W.; Yang, S.; Baek, S.J.; Jeong, Y.S.; Kim, S.Y. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011, 10, 149–157. [Google Scholar] [CrossRef]
- Tang, Z.F.; Li, C.W.; Kang, B.X.; Gao, G.; Li, C.; Zhang, Z.M. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015, 8, 22. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data (vol 2, pg 401, 2012). Cancer Discov. 2012, 2, 960. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.C.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Lanczky, A.; Nagy, A.; Bottai, G.; Munkacsy, G.; Szabo, A.; Santarpia, L.; Gyorffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Tr. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Goswami, C.P.; Nakshatri, H. PROGgeneV2: Enhancements on the existing database. BMC Cancer 2014, 14, 970. [Google Scholar] [CrossRef]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2009, 2, 18. [Google Scholar] [CrossRef]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martinez-Ledesma, E.; Martinez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Trevino, V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Koster, J.; Molenaar, J.J.; Versteeg, R. R2: Accessible web-based genomics analysis and visualization platform for biomedical researchers. Cancer Res 2015, 75. [Google Scholar] [CrossRef]
- R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl http://r2platform.com (accessed on 25 January 2019).
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Mi, H.Y.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Sanchez-Carbayo, M.; Socci, N.D.; Lozano, J.; Saint, F.; Cordon-Cardo, C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006, 24, 778–789. [Google Scholar] [CrossRef]
- Murat, A.; Migliavacca, E.; Gorlia, T.; Lambiv, W.L.; Shay, T.; Hamou, M.F.; de Tribolet, N.; Regli, L.; Wick, W.; Kouwenhoven, M.C.M.; et al. Stem cell-related “Self-Renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008, 26, 3015–3024. [Google Scholar] [CrossRef]
- Zhao, H.J.; Langerod, A.; Ji, Y.; Nowels, K.W.; Nesland, J.M.; Tibshirani, R.; Bukholm, I.K.; Karesen, R.; Botstein, D.; Borresen-Dale, A.L.; et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol. Biol. Cell 2004, 15, 2523–2536. [Google Scholar] [CrossRef]
- Skrzypczak, M.; Goryca, K.; Rubel, T.; Paziewska, A.; Mikula, M.; Jarosz, D.; Pachlewski, J.; Oledzki, J.; Ostrowsk, J. Modeling Oncogenic Signaling in Colon Tumors by Multidirectional Analyses of Microarray Data Directed for Maximization of Analytical Reliability. PLoS ONE 2010, 5, e13091. [Google Scholar] [CrossRef]
- Hu, N.; Clifford, R.J.; Yang, H.H.; Wang, C.Y.; Goldstein, A.M.; Ding, T.; Taylor, P.R.; Lee, M.P. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genom. 2010, 11, 576. [Google Scholar] [CrossRef]
- D’Errico, M.; de Rinaldis, E.; Blasi, M.F.; Viti, V.; Falchetti, M.; Calcagnile, A.; Sera, F.; Saieua, C.; Ottini, L.; Palli, D.; et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur. J. Cancer 2009, 45, 461–469. [Google Scholar] [CrossRef]
- Ginos, M.A.; Page, G.P.; Michalowicz, B.S.; Patel, K.J.; Volker, S.E.; Pambuccian, S.E.; Ondrey, F.G.; Adams, G.L.; Gaffney, P.M. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004, 64, 55–63. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Kuiper, R.P.; Boethe, T.; Ljungberg, B.; van Kessel, A.G.; Kovacs, G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 2009, 9, 152. [Google Scholar] [CrossRef]
- Haslinger, C.; Schweifer, N.; Stilgenbauer, S.; Dohner, H.; Lichter, P.; Kraut, N.; Stratowa, C.; Abseher, R. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J. Clin. Oncol. 2004, 22, 3937–3949. [Google Scholar] [CrossRef]
- Roessler, S.; Jia, H.L.; Budhu, A.; Forgues, M.; Ye, Q.H.; Lee, J.S.; Thorgeirsson, S.S.; Sun, Z.T.; Tang, Z.Y.; Qin, L.X.; et al. A Unique Metastasis Gene Signature Enables Prediction of Tumor Relapse in Early-Stage Hepatocellular Carcinoma Patients. Cancer Res. 2010, 70, 10202–10212. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Richards, W.G.; Staunton, J.; Li, C.; Monti, S.; Vasa, P.; Ladd, C.; Beheshti, J.; Bueno, R.; Gillette, M.; et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA 2001, 98, 13790–13795. [Google Scholar] [CrossRef]
- Piccaluga, P.P.; Agostinelli, C.; Califano, A.; Rossi, M.; Basso, K.; Zupo, S.; Went, P.; Klein, U.; Zinzani, P.L.; Baccarani, M.; et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J. Clin. Investig. 2007, 117, 823–834. [Google Scholar] [CrossRef]
- Zhan, F.H.; Barlogie, B.; Arzoumanian, V.; Huang, Y.S.; Hollmig, K.; Pineda-Roman, M.; Tricot, G.; van Rhee, F.; Zangari, M.; Dhodapkar, M.; et al. A gene expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 2007, 109, 1692–1700. [Google Scholar] [CrossRef] [PubMed]
- Korkola, J.E.; Houldsworth, J.; Chadalavada, R.S.V.; Olshen, A.B.; Dobrzynski, D.; Reuter, V.E.; Bosl, G.J.; Chaganti, R.S.K. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006, 66, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; den Boon, J.A.; Chen, I.H.; Newton, M.A.; Dahl, D.B.; Chen, M.; Cheng, Y.J.; Westra, W.H.; Chen, C.J.; Hildesheim, A.; et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006, 66, 7999–8006. [Google Scholar] [CrossRef] [PubMed]
- Stearman, R.S.; Dwyer-Nield, L.; Zerbe, L.; Blaine, S.A.; Chan, Z.; Bunn, P.A.; Johnson, G.L.; Hirsch, F.R.; Merrick, D.T.; Franklin, W.A.; et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005, 167, 1763–1775. [Google Scholar] [CrossRef]
- Yoshihara, K.; Tajima, A.; Komata, D.; Yamamoto, T.; Kodama, S.; Fujiwara, H.; Suzuki, M.; Onishi, Y.; Hatae, M.; Sueyoshi, K.; et al. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009, 100, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Iacobuzio-Donahue, C.A.; Maitra, A.; Olsen, M.; Lowe, A.W.; Van Heek, N.T.; Rosty, C.; Walter, K.; Sato, N.; Parker, A.; Ashfaq, R.; et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003, 162, 1151–1162. [Google Scholar] [CrossRef]
- Gaedcke, J.; Grade, M.; Jung, K.; Camps, J.; Jo, P.; Emons, G.; Gehoff, A.; Sax, U.; Schirmer, M.; Becker, H.; et al. Mutated KRAS Results in Overexpression of DUSP4, a MAP-Kinase Phosphatase, and SMYD3, a Histone Methyltransferase, in Rectal Carcinomas. Gene Chromosome Cancer 2010, 49, 1024–1034. [Google Scholar] [CrossRef]
- Lampa, M.; Arlt, H.; He, T.; Ospina, B.; Reeves, J.; Zhang, B.L.; Murtie, J.; Deng, G.J.; Barberis, C.; Hoffmann, D.; et al. Glutaminase is essential for the growth of triple-negative breast cancer cells with a deregulated glutamine metabolism pathway and its suppression synergizes with mTOR inhibition. PLoS ONE 2017, 12, e0185092. [Google Scholar] [CrossRef]
- Yu, D.C.; Shi, X.B.; Meng, G.; Chen, J.; Yan, C.; Jiang, Y.; Wei, J.W.; Ding, Y.T. Kidney-type glutaminase (GLS1) is a biomarker for pathologic diagnosis and prognosis of hepatocellular carcinoma. Oncotarget 2015, 6, 7619–7631. [Google Scholar] [CrossRef]
- Huang, F.; Zhang, Q.Y.; Ma, H.; Lv, Q.; Zhang, T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int. J. Clin. Exp. Pathol. 2014, 7, 1093–1100. [Google Scholar]
- Wang, J.B.; Erickson, J.W.; Fuji, R.; Ramachandran, S.; Gao, P.; Dinavahi, R.; Wilson, K.F.; Ambrosio, A.L.B.; Dias, S.M.G.; Dang, C.V.; et al. Targeting Mitochondrial Glutaminase Activity Inhibits Oncogenic Transformation (vol 18, pg 207, 2010). Cancer Cell 2010, 18, 397. [Google Scholar] [CrossRef]
- Zhang, J.F.; Wang, L.S.; Mao, S.Y.; Liu, M.N.; Zhang, W.T.; Zhang, Z.W.; Guo, Y.D.; Huang, B.S.; Yan, Y.; Huang, Y.; et al. miR-1-3p Contributes to Cell Proliferation and Invasion by Targeting Glutaminase in Bladder Cancer Cells. Cell Physiol. Biochem. 2018, 51, 513–527. [Google Scholar] [CrossRef]
- Pieter, A.; van den Heuvel, J.; Jing, J.P.; Wooster, R.F.; Bachman, K.E. Analysis of glutamine dependency in non-small cell lung cancer GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol. Ther. 2012, 13, 1185–1194. [Google Scholar]
- Li, H.J.; Li, X.; Pang, H.; Pan, J.J.; Xie, X.J.; Chen, W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 2015, 45, 1055–1063. [Google Scholar] [CrossRef]
- Katt, W.P.; Lukey, M.J.; Cerione, R.A. A tale of two glutaminases: Homologous enzymes with distinct roles in tumorigenesis. Future Med. Chem. 2017, 9, 223–243. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Lin, M.H.; Zhu, W.; Liang, Y.J.; Hong, X.H.; Zhao, Y.H.; Young, K.H.; Hu, W.W.; Feng, Z.H. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 2014, 5, 2635–2647. [Google Scholar] [CrossRef]
- Udali, S.; Guarini, P.; Ruzzenente, A.; Ferrarini, A.; Guglielmi, A.; Lotto, V.; Tononi, P.; Pattini, P.; Moruzzi, S.; Campagnaro, T.; et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin. Epigenetics 2015, 7, 43. [Google Scholar] [CrossRef]
- Flanagan, J.M.; Wilson, A.; Koo, C.; Masrour, N.; Gallon, J.; Loomis, E.; Flower, K.; Wilhelm-Benartzi, C.; Hergovich, A.; Cunnea, P.; et al. Platinum-Based Chemotherapy Induces Methylation Changes in Blood DNA Associated with Overall Survival in Patients with Ovarian Cancer. Clin. Cancer Res. 2017, 23, 2213–2222. [Google Scholar] [CrossRef]
- Shu, R.; He, J.; Wu, C.; Gao, J. The association between RARβ and FHIT promoter methylation and the carcinogenesis of patients with cervical carcinoma: A meta-analysis. Tumor Biol. 2017, 39, 1010428317709126. [Google Scholar] [CrossRef]
- Zhang, J.B.; Wang, C.; Chen, M.Q.; Cao, J.P.; Zhong, Y.; Chen, L.T.; Shen, H.M.; Xia, D.J. Epigenetic silencing of glutaminase 2 in human liver and colon cancers. BMC Cancer 2013, 13, 601. [Google Scholar] [CrossRef]
- Nagase, T.; Ishikawa, K.-I.; Suyama, M.; Kikuno, R.; Hirosawa, M.; Miyajima, N.; Tanaka, A.; Kotani, H.; Nomura, N.; Ohara, O. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998, 5, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Elgadi, K.; Meguid, R.; Qian, M.; Souba, W.; Abcouwer, S. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genom. 1999, 1, 51. [Google Scholar] [CrossRef] [PubMed]
- Porter, L.D.; Ibrahim, H.; Taylor, L.; Curthoys, N.P. Complexity and species variation of the kidney-type glutaminase gene. Physiol. Genom. 2002, 9, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Cassago, A.; Ferreira, A.P.S.; Ferreira, I.M.; Fornezari, C.; Gomes, E.R.M.; Greene, K.S.; Pereira, H.M.; Garratt, R.C.; Dias, S.M.G.; Ambrosio, A.L.B. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. USA 2012, 109, 1092–1097. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Erickson, J.W.; Stalnecker, C.A.; Katt, W.P.; Huang, Q.Q.; Cerione, R.A.; Ramachandran, S. Mechanistic Basis of Glutaminase Activation: A Key Enzyme that Promotes Glutamine Metabolism in Cancer Cells. J. Biol. Chem. 2016, 291, 20900–20910. [Google Scholar] [CrossRef] [PubMed]
- McGivan, J.; Lacey, J.; Joseph, S. Localization and some properties of phosphate-dependent glutaminase in disrupted liver mitochondria. Biochem. J. 1980, 192, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Park, K.G. Targeting Glutamine Metabolism for Cancer Treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Momcilovic, M.; Bailey, S.T.; Lee, J.T.; Fishbein, M.C.; Magyar, C.; Braas, D.; Graeber, T.; Jackson, N.J.; Czernin, J.; Emberley, E.; et al. Targeted Inhibition of EGFR and Glutaminase Induces Metabolic Crisis in EGFR Mutant Lung Cancer. Cell Rep. 2017, 18, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, T.N.; Patwardhan, P.P.; Cremers, S.; Schwartz, G.K. Targeted inhibition of glutaminase as a potential new approach for the treatment of NF1 associated soft tissue malignancies. Oncotarget 2017, 8, 94054–94068. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, H.; Opalinska, J.; Zhou, L.; Sohal, D.; Fazzari, M.J.; Yu, Y.T.; Montagna, C.; Montgomery, E.A.; Canto, M.; Dunbar, K.B.; et al. Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis. PLoS Genet. 2011, 7, e1001356. [Google Scholar] [CrossRef]
- Vincent, A.; Omura, N.; Hong, S.M.; Jaffe, A.; Eshleman, J.; Goggins, M. Genome-Wide Analysis of Promoter Methylation Associated with Gene Expression Profile in Pancreatic Adenocarcinoma. Clin. Cancer Res. 2011, 17, 4341–4354. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.J.; Rubbi, L.; Dawson, D.W.; Donahue, T.R.; Pellegrini, M. Pancreatic Cancer Patient Survival Correlates with DNA Methylation of Pancreas Development Genes. PLoS ONE 2015, 10, e0128814. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.-L.; Zhong, W.-Z.; An, S.-J.; Yang, J.-J.; Su, J.; Chen, Z.-H.; Yan, H.-H.; Chen, Z.-Y.; Huang, Z.-M.; Zhang, X.-C. KRAS mutation in patients with lung cancer: A predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann. Surg. Oncol. 2013, 20, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Marabese, M.; Ganzinelli, M.; Garassino, M.C.; Shepherd, F.A.; Piva, S.; Caiola, E.; Macerelli, M.; Bettini, A.; Lauricella, C.; Floriani, I. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy. Oncotarget 2015, 6, 34014. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, C.; Kagara, N.; Naoi, Y.; Shimoda, M.; Shimomura, A.; Maruyama, N.; Shimazu, K.; Kim, S.J.; Noguchi, S. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat. 2015, 150, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C.; et al. Prognostic Role of KRAS and BRAF in Stage II and III Resected Colon Cancer: Results of the Translational Study on the PETACC-3, EORTC 40993, SAKK 60-00 Trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Merkel, D.E.; McGuire, W.L. Ploidy, proliferative activity and prognosis. DNA flow cytometry of solid tumors. Cancer 1990, 65, 1194–1205. [Google Scholar] [CrossRef]
- Kallioniemi, O.P.; Hietanen, T.; Mattila, J.; Lehtinen, M.; Lauslahti, K.; Koivula, T. Aneuploid DNA content and high S-phase fraction of tumour cells are related to poor prognosis in patients with primary breast cancer. Eur. J. Cancer Clin. Oncol. 1987, 23, 277–282. [Google Scholar] [CrossRef]
- Zimmerman, P.; Bint, M.; Hawson, G.; Parsons, P. Ploidy as a prognostic determinant in surgically treated lung cancer. Lancet 1987, 330, 530–533. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.M.; Walsh, L.A.; Desrichard, A.; Huse, J.T.; Wu, W.; Gao, J.J.; Bose, P.; Lee, W.; Chan, T.A. Integrated Genomics for Pinpointing Survival Loci within Arm-Level Somatic Copy Number Alterations. Cancer Cell 2016, 29, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yao, D.M.; Liu, W.; Wang, N.; Lv, H.J.; Zhang, G.J.; Ji, M.J.; Xu, L.; He, N.Y.; Shi, B.Y.; et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer 2012, 12, 50. [Google Scholar] [CrossRef]
- Srividya, M.R.; Thota, B.; Shailaja, B.C.; Arivazhagan, A.; Thennarasu, K.; Chandramouli, B.A.; Hegde, A.S.; Santosh, V. Homozygous 10q23/PTEN deletion and its impact on outcome in glioblastoma: A prospective translational study on a uniformly treated cohort of adult patients. Neuropathology 2011, 31, 376–383. [Google Scholar] [CrossRef]
- Deming, S.L.; Nass, S.J.; Dickson, R.B.; Trock, B.J. C-myc amplification in breast cancer: A meta-analysis of its occurrence and prognostic relevance. Brit. J. Cancer 2000, 83, 1688–1695. [Google Scholar] [CrossRef]
- Lee, Y.Z.; Yang, C.W.; Chang, H.Y.; Hsu, H.Y.; Chen, I.S.; Chang, H.S.; Lee, C.H.; Lee, J.C.; Kumar, C.R.; Qiu, Y.Q.; et al. Discovery of selective inhibitors of Glutaminase-2, which inhibit mTORC1, activate autophagy and inhibit proliferation in cancer cells. Oncotarget 2014, 5, 6087–6101. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, M.; Ling, Q.; Li, N.; Zheng, J.; Yu, K.; Bao, S.; Huang, C.; Cheng, Q.; Chen, M. Glutaminase 2-a valuable predictor for hepatocellular carcinoma patients’ survival. Int. J. Clin. Exp. Med. 2016, 9, 17322–17333. [Google Scholar]
- Su, L.; Li, J.; Yang, J.; Shi, J.; Yu, D.; Ding, Y. Expression of glutaminase 1 gene in breast cancer and its clinical significance in neoadjuvant chemotherapy. Int. J. Clin. Exp. Pathol. 2016, 9, 9311–9317. [Google Scholar]
- Chen, L.; Cui, H.M. Targeting Glutamine Induces Apoptosis: A Cancer Therapy Approach. Int. J. Mol. Sci. 2015, 16, 22830–22855. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, S.K.; Islam, S.M.R.; Abdullah-AL-Wadud, M.; Islam, S.; Ali, F.; Park, K.S. Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer. J. Clin. Med. 2019, 8, 355. https://doi.org/10.3390/jcm8030355
Saha SK, Islam SMR, Abdullah-AL-Wadud M, Islam S, Ali F, Park KS. Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer. Journal of Clinical Medicine. 2019; 8(3):355. https://doi.org/10.3390/jcm8030355
Chicago/Turabian StyleSaha, Subbroto Kumar, S.M. Riazul Islam, M. Abdullah-AL-Wadud, Saiful Islam, Farman Ali, and Kyoung Sik Park. 2019. "Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer" Journal of Clinical Medicine 8, no. 3: 355. https://doi.org/10.3390/jcm8030355
APA StyleSaha, S. K., Islam, S. M. R., Abdullah-AL-Wadud, M., Islam, S., Ali, F., & Park, K. S. (2019). Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer. Journal of Clinical Medicine, 8(3), 355. https://doi.org/10.3390/jcm8030355









