In Silico Study Probes Potential Inhibitors of Human Dihydrofolate Reductase for Cancer Therapeutics
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Dataset
2.2. Generation of Pharmacophore Model
2.3. Pharmacophore Validation
2.4. Virtual Screening and Drug-Likeness Prediction
2.5. Molecular Docking Simulation
2.6. Molecular Dynamics (MD) Simulation
2.7. Binding Free Energy Calculations
3. Results
3.1. Generation of Pharmacophore Model
3.2. Pharmacophore Validation
3.2.1. Fischer’s Randomization Test
3.2.2. Test Set Validation
3.2.3. Decoy Set Validation
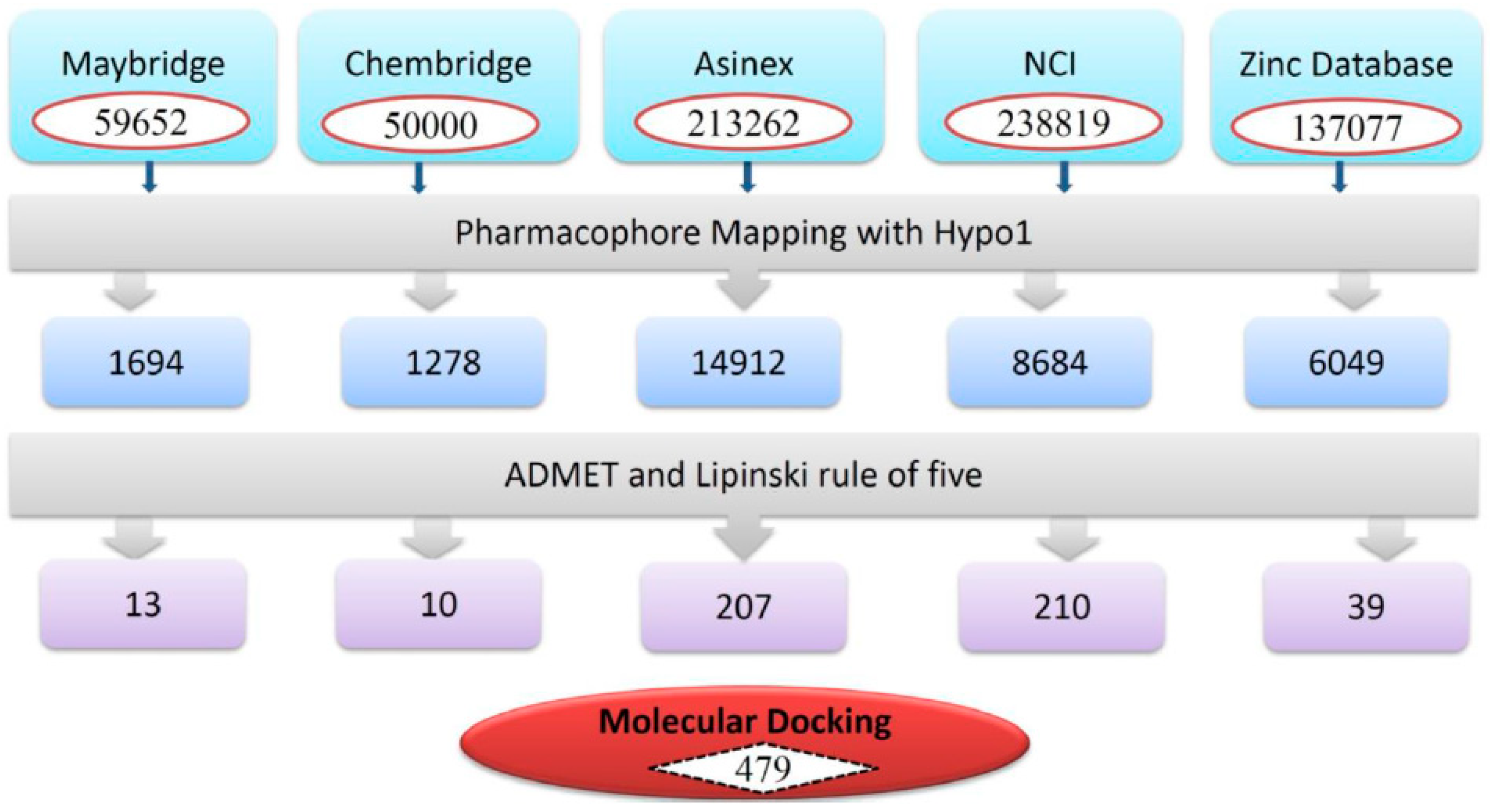
3.3. Virtual Screening of Chemical Databases
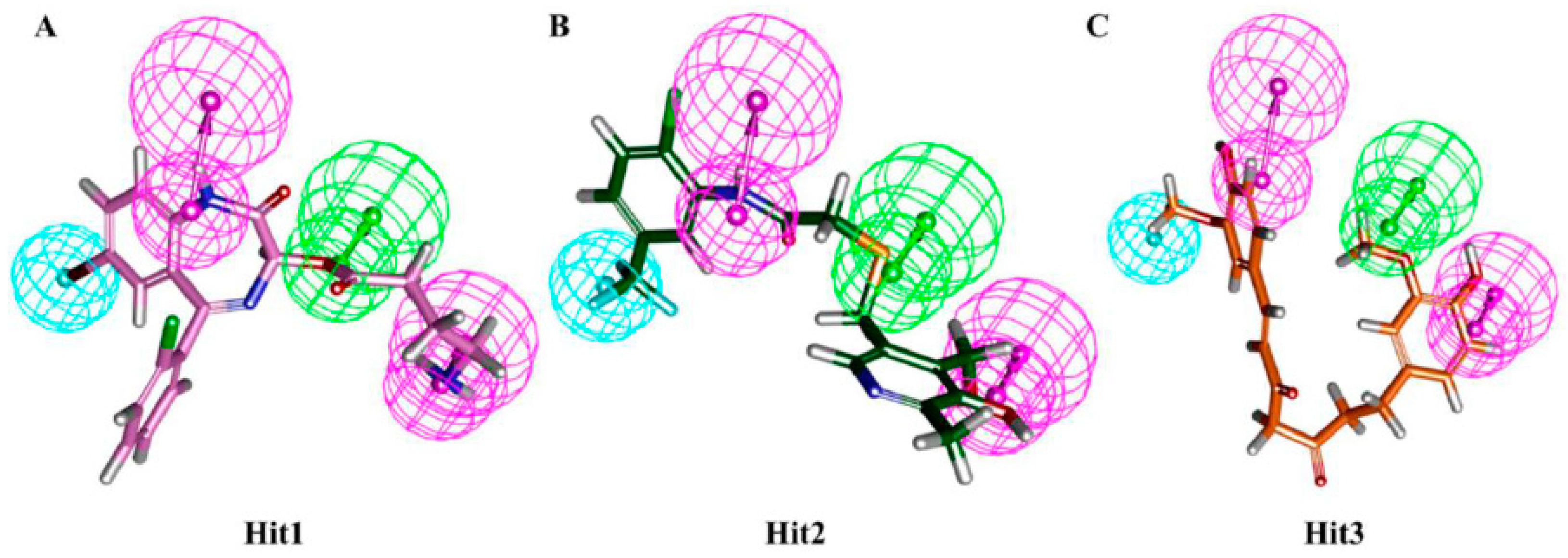
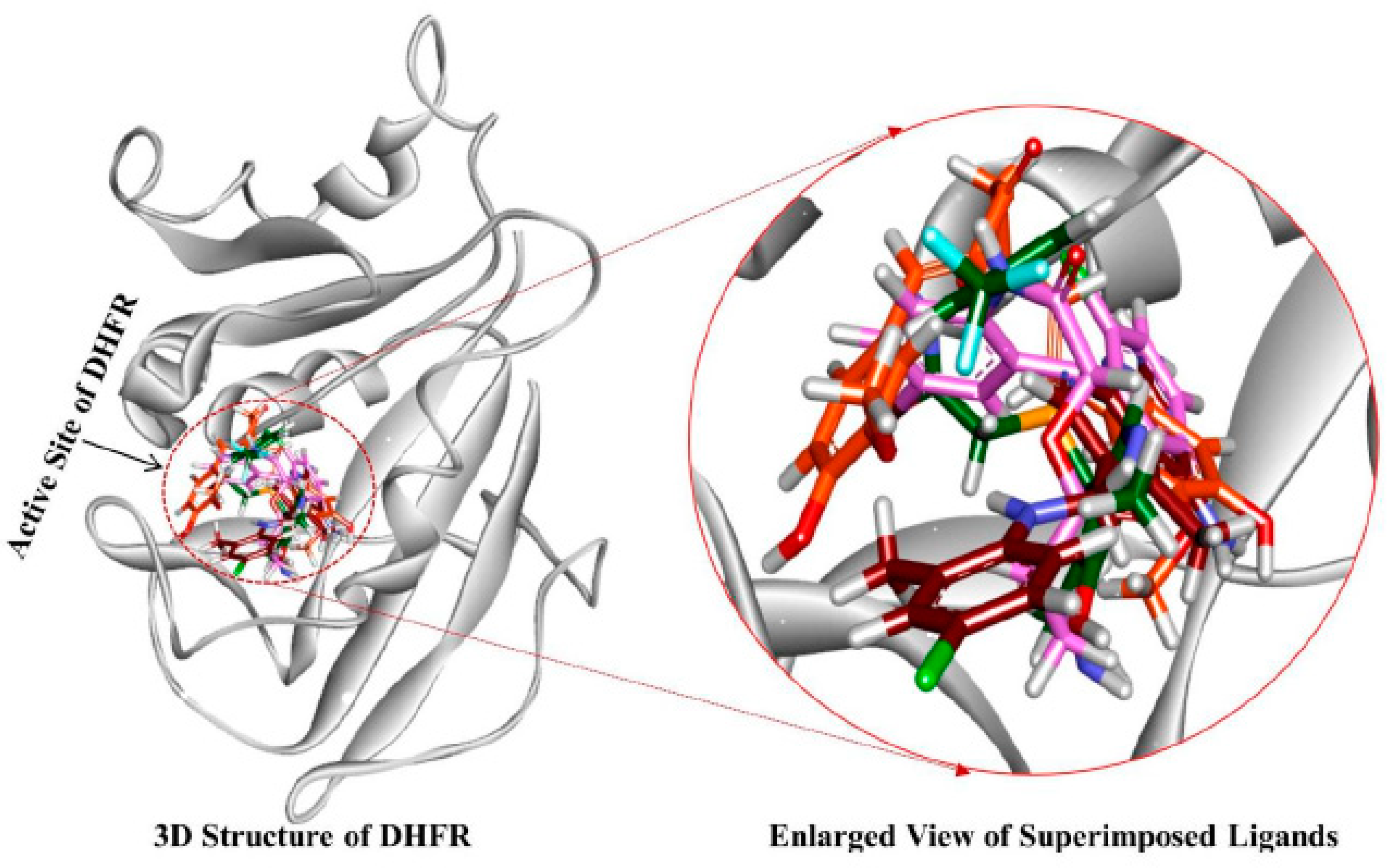
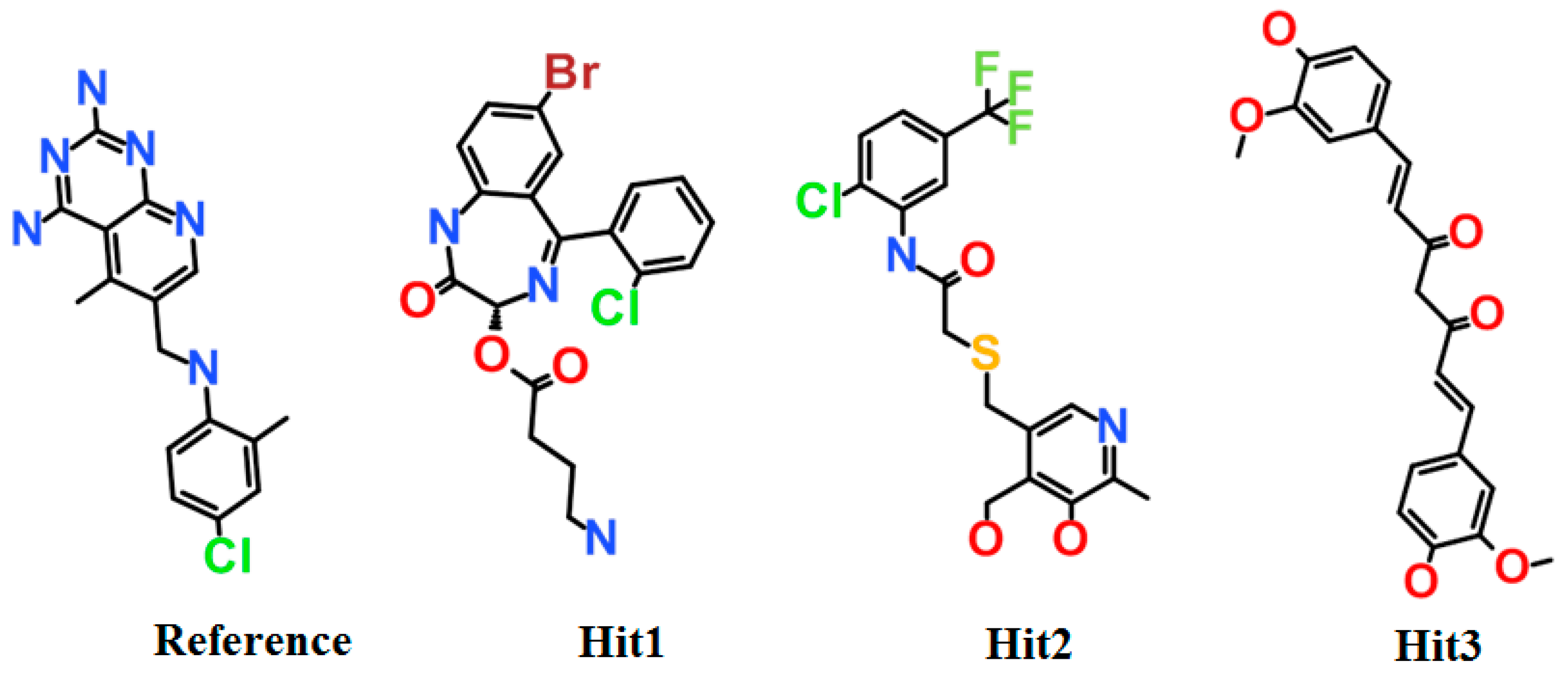
3.4. Molecular Docking Simulation
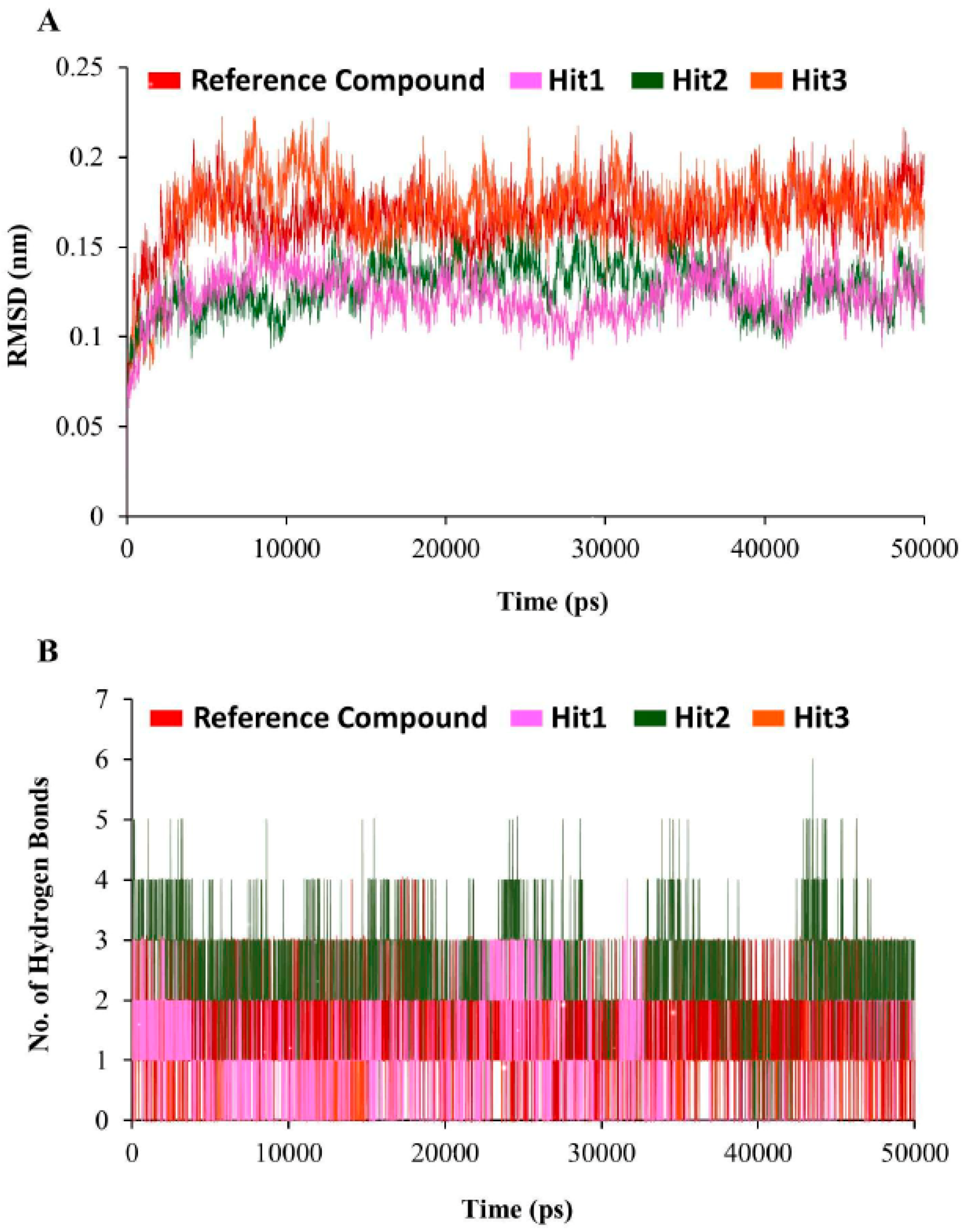
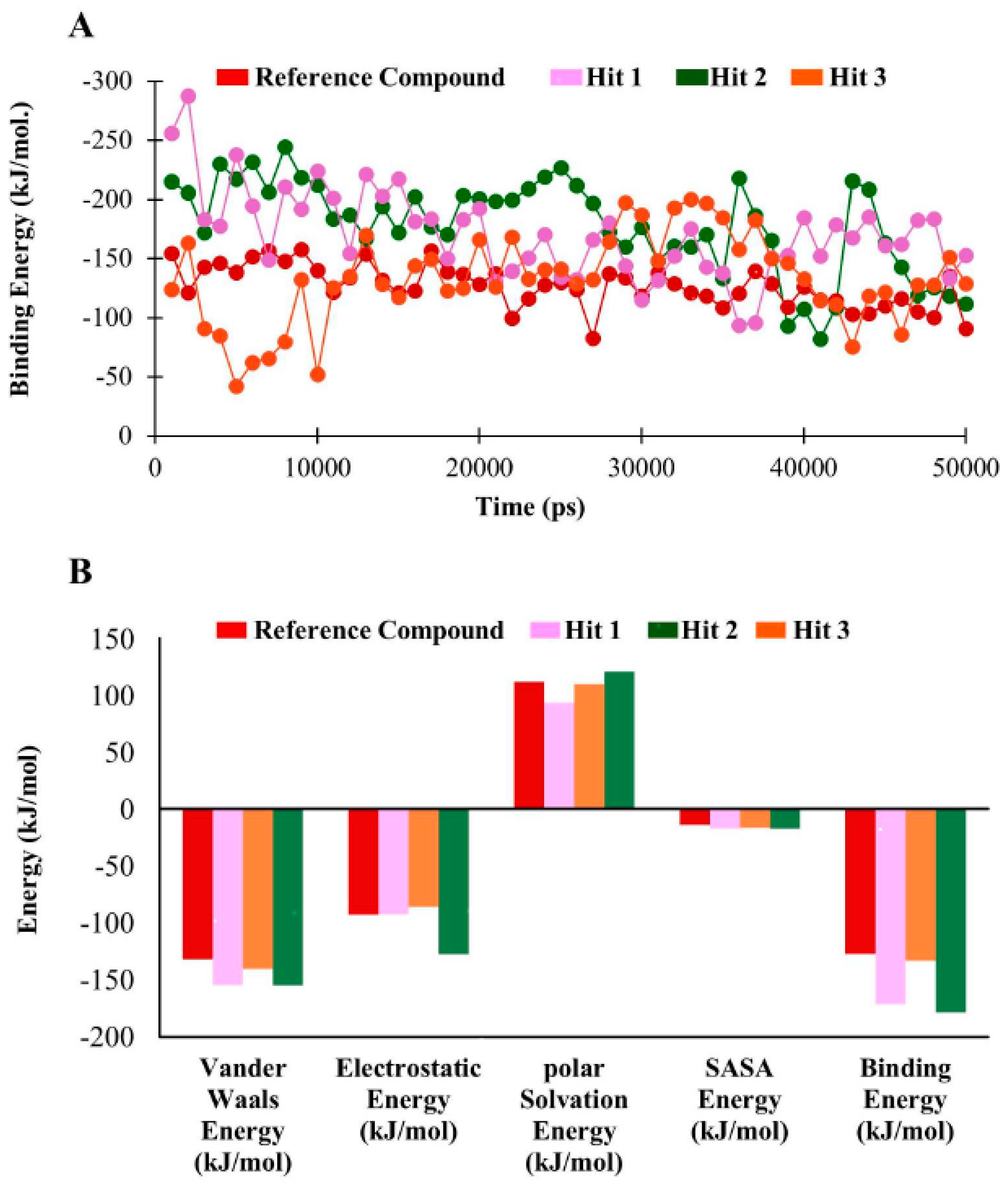
3.5. Molecular Dynamics Simulation
3.6. Binding Free Energy Analysis
3.7. Toxicity Evaluation by TOPKAT
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tobias, A.M.; Toska, D.; Lange, K.; Eck, T.; Bhat, R.; Janson, C.A.; Rotella, D.P.; Gubler, U.; Goodey, N.M. Expression, purification, and inhibition profile of dihydrofolate reductase from the filarial nematode Wuchereria bancrofti. PLoS ONE 2018. [Google Scholar] [CrossRef] [PubMed]
- Snapka, R.M.; Ge, S.; Trask, J.; Robertson, F. Unbalanced growth in mouse cells with amplified dhfr genes. Cell Prolif. 1997, 30, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, S.; Motmaen, S.R.; Afsharpad, M.; Djadid, N.D. Molecular characterization of antifolates resistance-associated genes, (dhfr and dhps) in Plasmodium vivax isolates from the Middle East. Malar. J. 2009, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.R.; Dyson, H.J.; Wright, P.E. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 119–159. [Google Scholar] [CrossRef] [PubMed]
- Osorio, E.; Aguilera, C.; Naranjo, N.; Marín, M.; Muskus, C. Biochemical characterization of the bifunctional enzyme dihydrofolate reductase-thymidylate synthase from Leishmania (Viannia) and its evaluation as a drug target. Biomedica 2013, 33, 393–401. [Google Scholar] [PubMed]
- Polshakov, V.I. Dihydrofolate reductase: Structural aspects of mechanisms of enzyme catalysis and inhibition. Russ. Chem. Bull. 2001, 50, 1733–1751. [Google Scholar] [CrossRef]
- Oefner, C.; D’Arcy, A.; Winkler, F.K. Crystal structure of human dihydrofolate reductase complexed with folate. Eur. J. Biochem. 1988, 174, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Tosso, R.D.; Andujar, S.A.; Gutierrez, L.; Angelina, E.; Rodríguez, R.; Nogueras, M.; Baldoni, H.; Suvire, F.D.; Cobo, J.; Enriz, R.D. Molecular modeling study of dihydrofolate reductase inhibitors. Molecular dynamics simulations, quantum mechanical calculations, and experimental corroboration. J. Chem. Inf. Model. 2013, 53, 2018–2032. [Google Scholar] [CrossRef]
- Zhang, Q.; Nguyen, T.; Mcmichael, M.; Velu, S.; Zou, J.; Zhou, X.; Wu, H. New small-molecule inhibitors of dihydrofolate reductase inhibit Streptococcus mutans HHS Public Access. Int. J. Antimicrob. Agents 2015, 46, 174–182. [Google Scholar] [CrossRef]
- Yuthavong, Y.; Tarnchompoo, B.; Vilaivan, T.; Chitnumsub, P.; Kamchonwongpaisan, S.; Charman, S.A.; Mclennan, D.N.; White, K.L.; Vivas, L.; Bongard, E.; et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. PNAS 2012, 109, 16823–16828. [Google Scholar] [CrossRef]
- Then, R.L. Antimicrobial dihydrofolate reductase inhibitors-achievements and future options. Rev. J. Chemother. 2004, 16, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Scocchera, E.; Reeve, S.M.; Keshipeddy, S.; Lombardo, M.N.; Hajian, B.; Sochia, A.E.; Alverson, J.B.; Priestley, N.D.; Anderson, A.C.; Wright, D.L. Charged nonclassical antifolates with activity against gram-positive and gram-negative pathogens. ACS Med. Chem. Lett. 2016, 692–696. [Google Scholar] [CrossRef]
- Kubbies, M.; Stockinger, H. Cell cycle-dependent DHFR and t-PA production in cotransfected, MTX-amplified CHO cells revealed by dual-laser flow cytometry. Exp. Cell Res. 1990, 188, 267–271. [Google Scholar] [CrossRef]
- Neradil, J.; Pavlasova, G.; Veselska, R. New mechanisms for an old drug; DHFR- and non-DHFR-mediated effects of methotrexate in cancer cells. Klin. Onkol. 2012, 25, 2S87–2S92. [Google Scholar] [PubMed]
- Rodríguez, M.; Coma, S.; Noé, V.; Ciudad, C.J. Development and effects of immunoliposomes carrying an antisense oligonucleotide against DHFR RNA and directed toward human breast cancer cells overexpressing HER2. Antisense Nucleic Acid Drug Dev. 2002, 12, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Giletti, A.; Esperon, P. Genetic markers in methotrexate treatments. Pharmacogenomics J. 2018, 18, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kaur, M.; Sachdeva, S. Mechanism inspired development of rationally designed dihydrofolate reductase inhibitors as anticancer agents. J. Med. Chem. 2012, 55, 6381–6390. [Google Scholar] [CrossRef]
- Rao, A.S.; Road, K. A study on dihydrofolate reductase and its inhibitors: A review. Int. J. 2013, 4, 2535–2547. [Google Scholar]
- Lin, J.T.; Mbewe, B.; Taylor, S.M.; Luntamo, M.; Meshnick, S.R.; Ashorn, P. Increased prevalence of dhfr and dhps mutants at delivery in Malawian pregnant women receiving intermittent preventive treatment for malaria. Trop. Med. Int. Health 2013, 18, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Askari, B.S.; Krajinovic, M. Dihydrofolate reductase gene variations in susceptibility to disease and treatment outcomes. Curr. Genomics 2010, 11, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Dias-Polak, D.; Bergman, R.; Avitan-Hersh, E. Mycophenolate mofetil therapy in adult patients with recalcitrant atopic dermatitis. J. Dermatolog. Treat. 2018, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xiang, G.; Yap, C.-W.; Chui, W.-K. 3D-QSAR Study on dihydro-1,3,5-triazines and their spiro derivatives as DHFR inhibitors by comparative molecular field analysis (CoMFA). Bioorg. Med. Chem. Lett. 2012, 22, 3194–3197. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lin, Y.; Wen, X.; Jorissen, R.N.; Gilson, M.K. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Suzek, T.; Zhang, J.; Wang, J.; He, S.; Cheng, T.; Shoemaker, B.A.; Gindulyte, A.; Bryant, S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014, 42, D1075–D1082. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.K. Pharmacophore Mapping of a Series of 2,4-Diamino-5-deazapteridine inhibitors of Mycobacterium avium complex dihydrofolate reductase. J. Med. Chem. 2001, 45, 41–53. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Bioinforma. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Rampogu, S.; Son, M.; Baek, A.; Park, C.; Rana, R.M.; Zeb, A.; Parameswaran, S.; Lee, K.W. Targeting natural compounds against HER2 kinase domain as potential anticancer drugs applying pharmacophore based molecular modelling approaches. Comput. Biol. Chem. 2018, 74, 327–338. [Google Scholar] [CrossRef]
- Rosta, E.; Buchete, N.-V.; Hummer, G. Thermostat artifacts in replica exchange molecular dynamics simulations. J. Chem. Theory Comput. 2009, 5, 1393–1399. [Google Scholar] [CrossRef]
- Haigis, V.; Belkhodja, Y.; Vuilleumier, R.; Boutin, A. Challenges in first-principles NPT molecular dynamics of Soft Porous Crystals: A case study on MIL-53(Ga). J. Chem. Phys. 2014, 141, 064703. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Chin, S.P.; Sukumaran, S.D.; Buckle, M.J.C.; Kiew, L.V.; Chung, L.Y. In Silico and in vitro analysis of bacoside A aglycones and its derivatives as the constituents responsible for the cognitive effects of Bacopa monnieri. PLoS ONE 2015, 10, e0126565. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Sakkiah, S.; Lee, K.W. Pharmacophore-based virtual screening and density functional theory approach to identifying novel butyrylcholinesterase inhibitors. Acta Pharmacol. Sin. 2012, 33, 964–978. [Google Scholar] [CrossRef] [PubMed]
- Gangjee, A.; Vidwans, A.P.; Vasudevan, A.; Queener, S.F.; Kisliuk, R.L.; Cody, V.; Li, R.; Galitsky, N.; Luft, J.R.; Pangborn, W. Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents. J. Med. Chem. 1998, 41, 3426–3434. [Google Scholar] [CrossRef] [PubMed]
- Algul, O.; Paulsen, J.L.; Anderson, A.C. 2,4-Diamino-5-(2′-arylpropargyl)pyrimidine derivatives as new nonclassical antifolates for human dihydrofolate reductase inhibition. J. Mol. Graph. Model 2012, 29, 608–613. [Google Scholar]
- Luo, W.; Yu, Q.; Kulkarni, S.S.; Parrish, D.A.; Holloway, H.W.; Tweedie, D.; Shafferman, A.; Lahiri, D.K.; Brossi, A.; Greig, N.H. Inhibition of human acetyl- and butyrylcholinesterase by novel carbamates of (−)- and (+)-tetrahydrofurobenzofuran and methanobenzodioxepine. J. Med. Chem. 2006, 49, 2174–2185. [Google Scholar] [CrossRef]
- Lu, X.; Yang, H.; Chen, Y.; Li, Q.; He, S.; Jiang, X.; Feng, F.; Qu, W.; Sun, H. The development of pharmacophore modeling: Generation and recent applications in drug discovery. Curr. Pharm. Des. 2018, 24, 3424–3439. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, H.; Lian, Z.; Han, D.; Jin, X. Pharmacophore modeling and virtual screening for the discovery of new fatty acid amide hydrolase inhibitors. Acta Pharm. Sin. B 2011, 1, 27–35. [Google Scholar] [CrossRef]
- Tripathi, A.; Bankaitis, V.A. Molecular docking: From lock and key to combination lock. J. Mol. Med. Clin. Appl. 2017, 2, 1–19. [Google Scholar]
- Fradera, X.; Babaoglu, K. Overview of methods and strategies for conducting virtual small molecule screening. In Current Protocols in Chemical Biology; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2017; Volume 9, pp. 196–212. [Google Scholar]
- Nowak, W.; Cody, V.; Wojtczak, A. Computer modeling studies of the structural role of NADPH binding to active site mutants of human dihydrofolate reductase in complex with piritrexim. Acta Biochim. Pol. 2001, 48, 903–916. [Google Scholar] [PubMed]
- El-Hamamsy, M.H.R.I.; Smith, A.W.; Thompson, A.S.; Threadgill, M.D. Structure-based design, synthesis and preliminary evaluation of selective inhibitors of dihydrofolate reductase from Mycobacterium tuberculosis. Bioorg. Med. Chem. 2007, 15, 4552–4576. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Discovery Studio (D.S.). Predictive Toxicology in Discovery Studio. Available online: http://accelrys.com/products/datasheets/ds_topkat.pdf (accessed on 14 August 2018).
- Prival, M.J. Evaluation of the TOPKAT system for predicting the carcinogenicity of chemicals. Environ. Mol. Mutagen. 2001, 37, 55–69. [Google Scholar] [CrossRef]
- Golbamaki, A.; Cassano, A.; Lombardo, A.; Moggio, Y.; Colafranceschi, M.; Benfenati, E. Comparison of in silico models for prediction of Daphnia magna acute toxicity. SAR QSAR Environ. Res. 2014, 25, 673–694. [Google Scholar] [CrossRef] [PubMed]
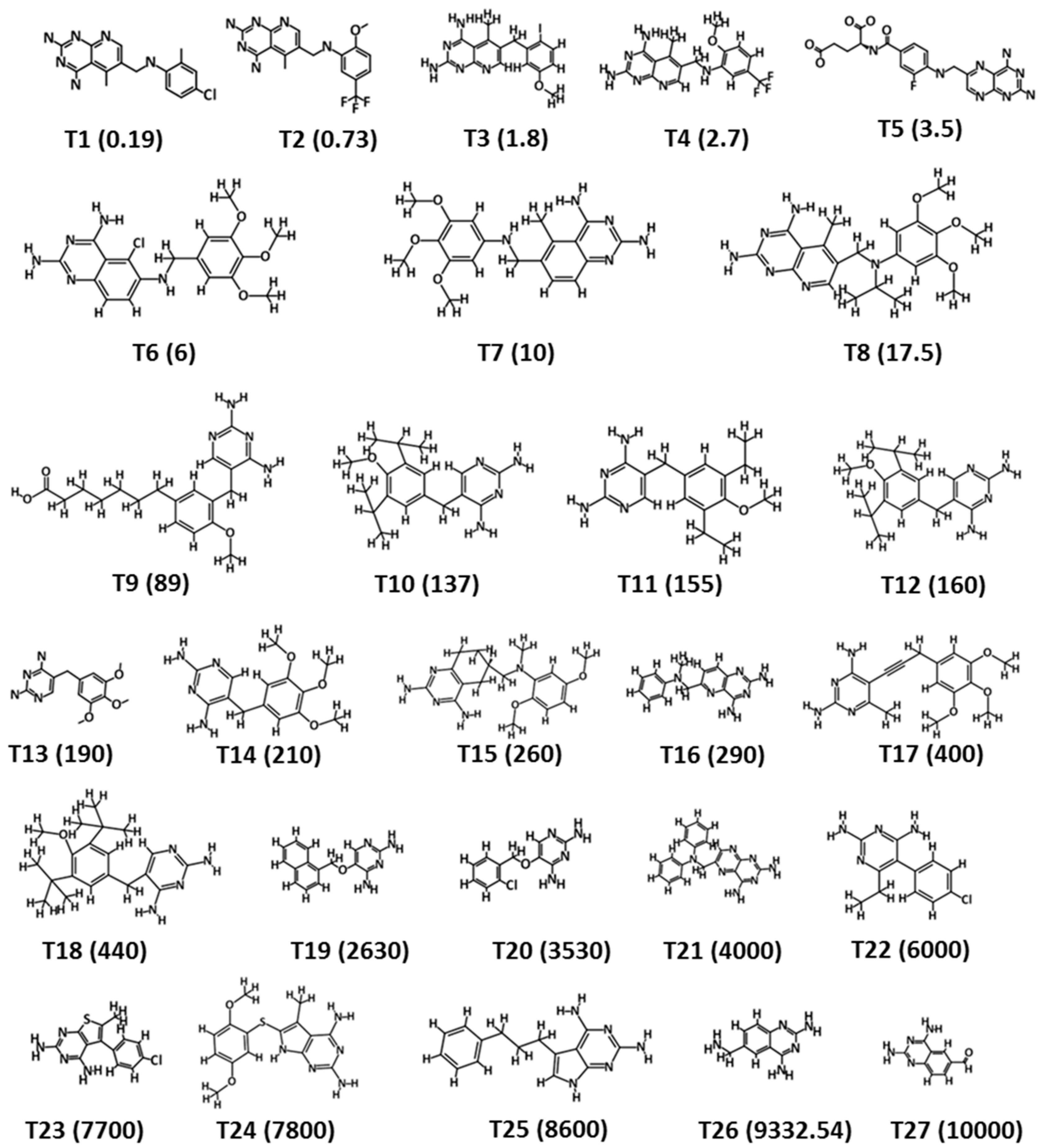
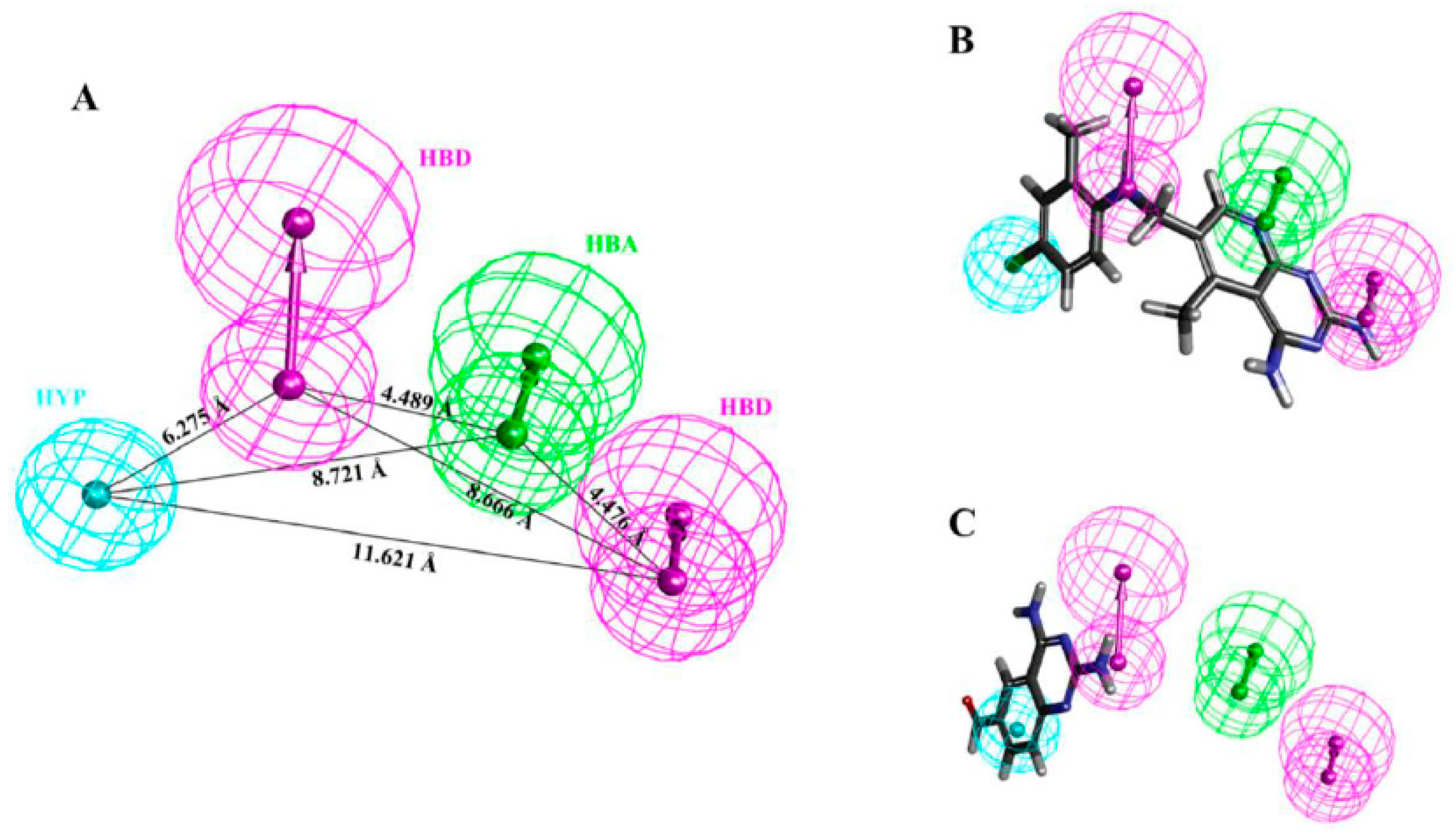
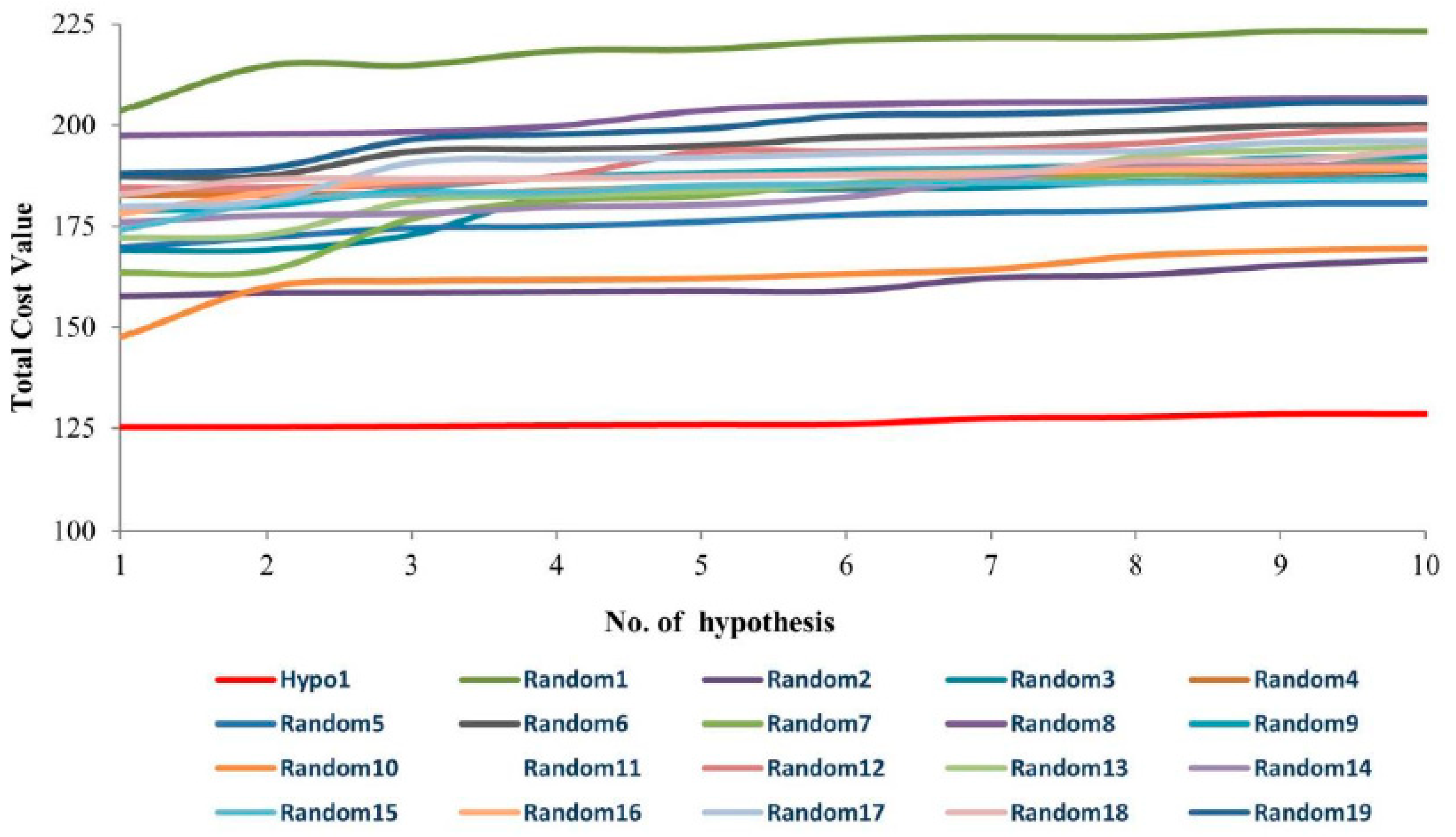
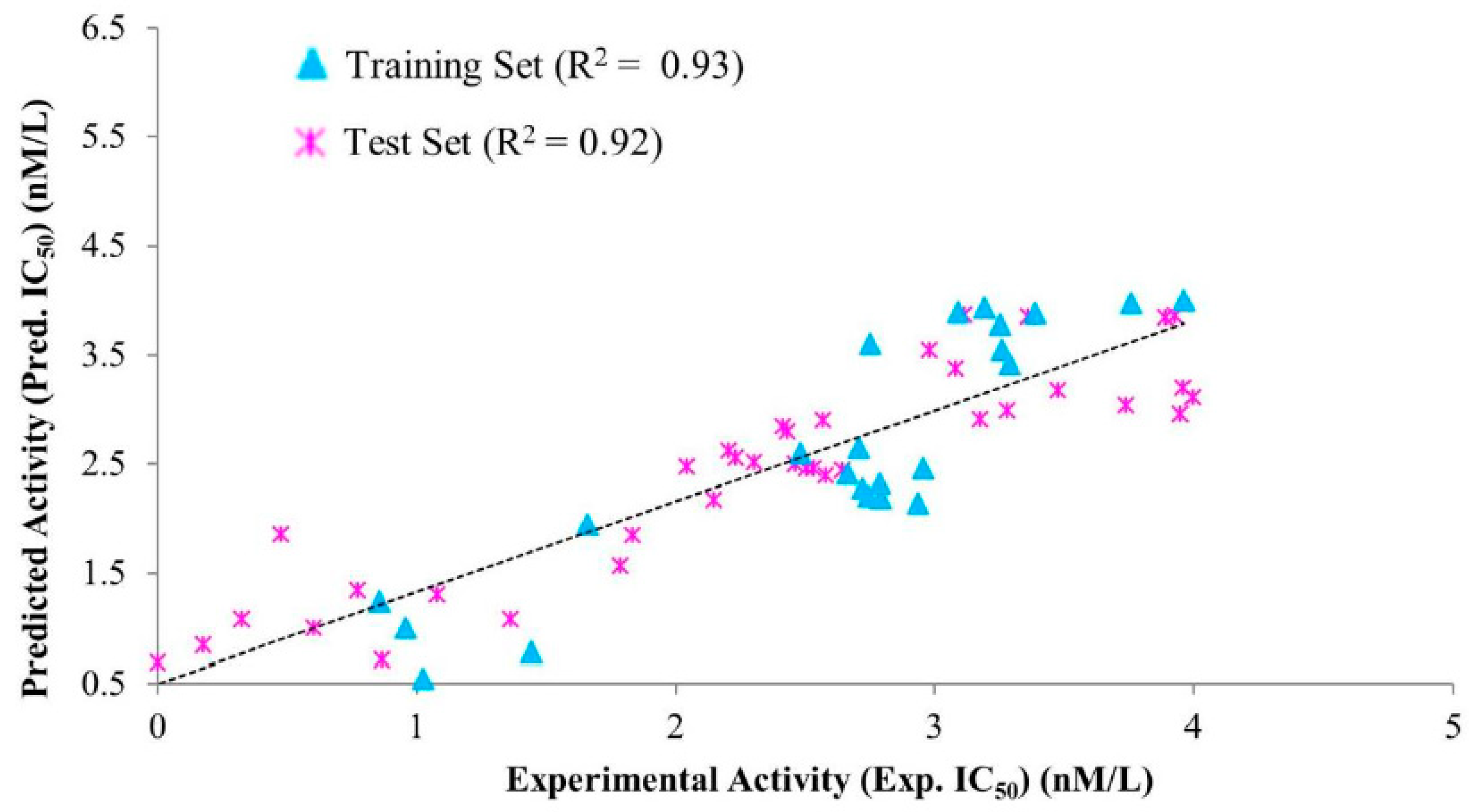
| Hypo. No. | Total Cost | Cost Difference a | Root Means Square Deviation (RMSD) b | Correlation (R2) | Max Fit | Features c |
|---|---|---|---|---|---|---|
| Hypo1 | 125.276 | 78.308 | 0.99 | 0.94 | 11.15 | HBA, HBD, HBD, HYP |
| Hypo2 | 125.362 | 78.304 | 1.05 | 0.93 | 9.94 | HBA, HBD, HBD, HYP |
| Hypo3 | 125.509 | 78.157 | 1.05 | 0.93 | 10.04 | HBA, HBD, HBD, HYP |
| Hypo4 | 125.358 | 77.701 | 1.05 | 0.93 | 10.04 | HBA, HBD, HBD, HYP |
| Hypo5 | 125.867 | 77.799 | 1.06 | 0.93 | 10.21 | HBA, HBD, HBD, HYP |
| Hypo6 | 125.965 | 77.946 | 1.03 | 0.93 | 10.74 | HBA, HBD, HBD, HYP |
| Hypo7 | 127.533 | 76.133 | 1.13 | 0.92 | 9.97 | HBA, HBD, HBD, HYP |
| Hypo8 | 125.781 | 75.885 | 1.14 | 0.92 | 9.75 | HBA, HBD, HBD, HYP |
| Hypo9 | 128.568 | 75.098 | 1.15 | 0.92 | 10.24 | HBA, HBD, HBD, HYP |
| Hypo10 | 128.568 | 75.098 | 1.15 | 0.92 | 10.26 | HBA, HBD, HBD, HYP |
| Compound No. | Fit Value | Experimental IC50 nM/L | Predicted IC50 nM/L | a Error | b Exp. Scale | b Pred. Scale |
|---|---|---|---|---|---|---|
| 1 | 9.63 | 0.19 | 0.21 | 1.10 | +++ | +++ |
| 2 | 7.42 | 0.73 | 7.55 | 10.34 | +++ | +++ |
| 3 | 6.87 | 1.8 | 27.09 | 15.05 | +++ | +++ |
| 4 | 8.24 | 2.7 | 1.15 | −2.36 | +++ | +++ |
| 5 | 7.33 | 3.5 | 9.40 | 2.69 | +++ | +++ |
| 6 | 7.66 | 6 | 4.41 | −1.36 | +++ | +++ |
| 7 | 7.32 | 10 | 9.68 | −1.03 | +++ | +++ |
| 8 | 7.36 | 17.5 | 8.69 | −2.01 | +++ | +++ |
| 9 | 6.26 | 89 | 110.31 | 1.24 | +++ | ++ |
| 10 | 5.74 | 137 | 364.60 | 2.66 | ++ | ++ |
| 11 | 5.75 | 155 | 357.03 | 2.30 | ++ | ++ |
| 12 | 5.77 | 160 | 339.73 | 2.12 | ++ | ++ |
| 13 | 5.52 | 190 | 606.05 | 3.19 | ++ | + |
| 14 | 5.78 | 210 | 331.91 | 1.58 | ++ | ++ |
| 15 | 5.62 | 260 | 482.36 | 1.86 | ++ | ++ |
| 16 | 5.38 | 290 | 827.48 | 2.85 | ++ | + |
| 17 | 5.75 | 400 | 352.12 | −1.14 | ++ | ++ |
| 18 | 5.80 | 440 | 315.88 | −1.39 | ++ | ++ |
| 19 | 5.11 | 2630 | 1566.48 | −1.68 | + | + |
| 20 | 4.84 | 3530 | 2915.70 | −1.21 | + | + |
| 21 | 5.18 | 4000 | 1327.90 | −3.01 | + | + |
| 22 | 4.78 | 6000 | 3296.55 | −1.82 | + | + |
| 23 | 5.01 | 7700 | 1946.96 | −3.95 | + | + |
| 24 | 5.06 | 7800 | 1749.88 | −4.46 | + | + |
| 25 | 5.53 | 8600 | 597.43 | −14.40 | + | + |
| 26 | 4.52 | 9332 | 6062.97 | −1.54 | + | + |
| 27 | 4.34 | 10,000 | 9094.55 | −1.10 | + | + |
| Parameters | Values |
|---|---|
| Total no. of molecules in database (D) | 75 |
| Total no. of actives in database (A) | 8 |
| Total no. of hit molecules from the database (Ht) | 8 |
| Total no. of active molecules in hit list (Ha) | 7 |
| Percentage Yield of actives ((Ha/Ht) × 100) | 87.5% |
| Percentage Ratio of actives ((Ha/A) × 100) | 88% |
| Enrichment Factor (EF = (Ha/Ht)/(A/D)) | 8.23 |
| False negatives (A − Ha) | 1 |
| False positive (Ht − Ha) | 1 |
| Goodness of fit score (GF) | 0.86 |
| System | Goldfitness Score | Chemscore | Estimated IC50 (nM/L) |
|---|---|---|---|
| DHFR + a Reference | 44.67 | −23.35 | 0.21 |
| DHFR + Hit1 | 70.05 | −34.51 | 0.12 |
| DHFR + Hit2 | 58.95 | −33.66 | 0.043 |
| DHFR + Hit3 | 57.31 | −37.27 | 0.17 |
| System | No. of TIP3P Water Molecules | No. of Na+ Ions | System Size (nm) |
|---|---|---|---|
| DHFR+NADPH + a Reference | 11,306 | 4 | 7.193 × 7.193 × 7.193 |
| DHFR+NADPH + Hit1 | 11,306 | 4 | 7.193 × 7.193 × 7.193 |
| DHFR+NADPH + Hit2 | 11,306 | 4 | 7.193 × 7.193 × 7.193 |
| DHFR+NADPH + Hit3 | 11,306 | 4 | 7.193 × 7.193 × 7.193 |
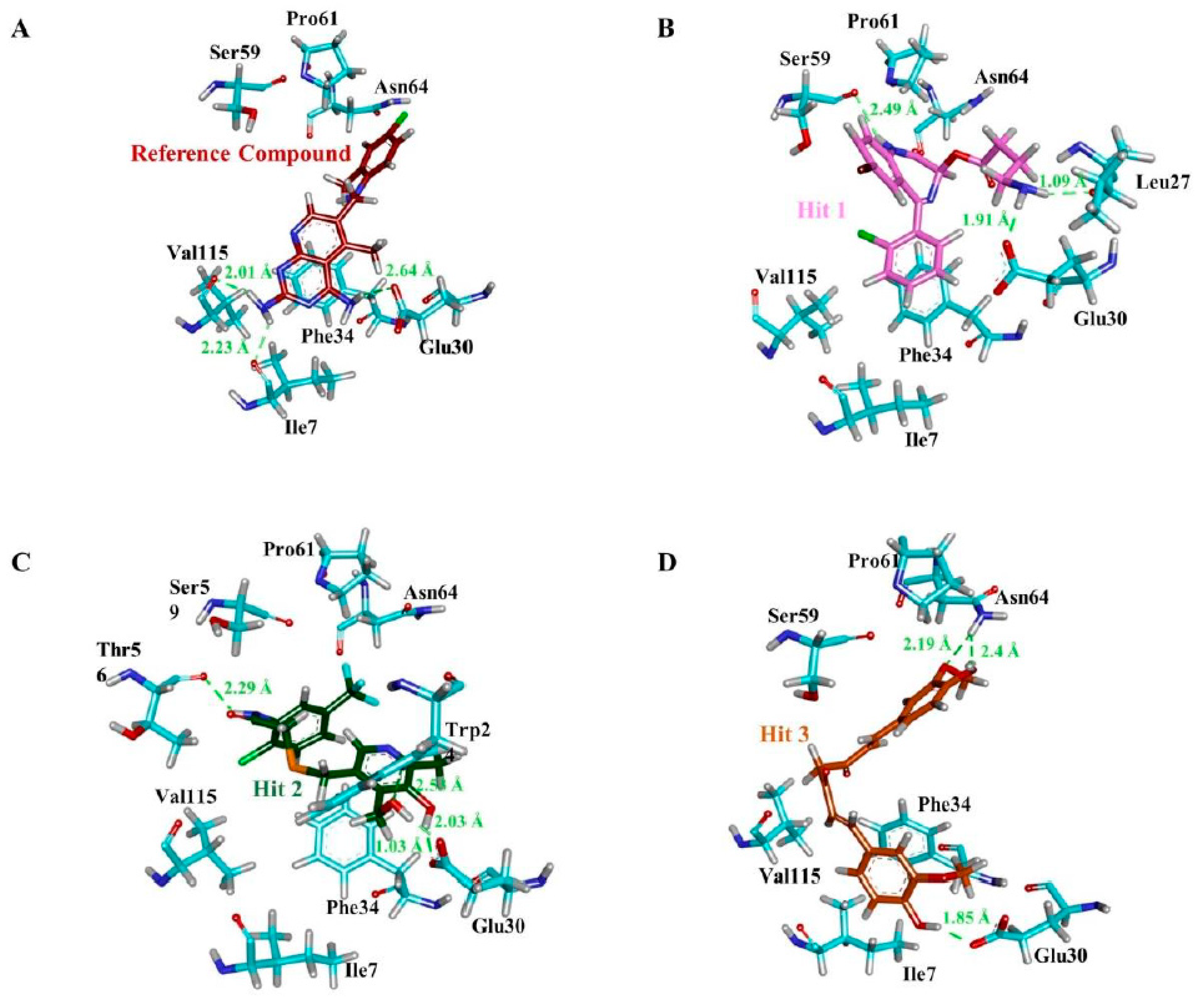
| Compound | Hydrogen Bond (<3 Å) | Van der Waals Interactions and Carbon Hydrogen Bond | π-Interaction |
|---|---|---|---|
| a Reference | Ile7, Glu30, Val115 | Val8, Gly31, Asn64, Gly116, Tyr121, NADPH | Ile7, Phe34, Ile60, Pro61, Leu67, Val115 |
| Hit1 | Leu27, Glu30, Ser59 | Val8, Ile16, Gly17, Asp21, Asp21, Pro26, Gly31, Gln35, Thr136, Pro61, Ile60 | Ala9, Leu22, Pro23, Phe34, Val115, NADPH |
| Hit2 | Trp24, Glu30 (2), Thr56, NADPH | Ala9, Gly31, Gln35, Ser59, Ile60, Pro61 | Phe34, Met52, Ile60, Leu67, Val115 |
| Hit3 | Glu30, Asn64 (2) | Val8, Leu22, Gly31, Gln35, Met52, Thr56, Pro61, Leu67, Val115, Tyr121, Thr136, NADPH | Ile7, Ala9, Ile60, Phe34 |
| Compounds | Van der Waals Energy (kJ/mol) | Electrostatic Energy (kJ/mol) | Polar Solvation Energy (kJ/mol) | SASA b Energy (kJ/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|---|
| a Reference | −131.78 | −92.84 | 111.73 | −13.9 | −127.05 |
| Hit1 | −154.58 | −92.63 | 93.27 | −17.218 | −171.12 |
| Hit2 | −154.75 | −127.45 | 120.79 | −17.31 | −178.47 |
| Hit3 | −140.36 | −86.23 | 109.75 | −16.47 | −133.16 |
| Name | ADMET Analysis | Lipinski’s Rule of Five | TOPKAT Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Solubility | BBB a | Hepato-Toxicity | Absorption | HBA b | HBD c | M.W (Da) d | Rat (Carcinogenicity) | Skin Irritancy | ||
| Female | Male | |||||||||
| MTX | 2 | 4 | TRUE | 3 | 12 | 5 | 454.447 | Single | Non | Non |
| Pralatexet | 2 | 4 | TRUE | 3 | 11 | 5 | 477.481 | Non | Non | Non |
| Pemetrexed | 3 | 4 | TRUE | 3 | 6 | 6 | 427.417 | Single | Non | Non |
| Hit1 | 3 | 3 | FALSE | 0 | 6 | 3 | 450.714 | Non | Non | Non |
| Hit2 | 3 | 3 | FALSE | 0 | 5 | 3 | 420.833 | Non | Non | Non |
| Hit3 | 3 | 3 | FALSE | 0 | 6 | 2 | 368.385 | Non | Non | Non |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rana, R.M.; Rampogu, S.; Zeb, A.; Son, M.; Park, C.; Lee, G.; Yoon, S.; Baek, A.; Parameswaran, S.; Park, S.J.; et al. In Silico Study Probes Potential Inhibitors of Human Dihydrofolate Reductase for Cancer Therapeutics. J. Clin. Med. 2019, 8, 233. https://doi.org/10.3390/jcm8020233
Rana RM, Rampogu S, Zeb A, Son M, Park C, Lee G, Yoon S, Baek A, Parameswaran S, Park SJ, et al. In Silico Study Probes Potential Inhibitors of Human Dihydrofolate Reductase for Cancer Therapeutics. Journal of Clinical Medicine. 2019; 8(2):233. https://doi.org/10.3390/jcm8020233
Chicago/Turabian StyleRana, Rabia Mukhtar, Shailima Rampogu, Amir Zeb, Minky Son, Chanin Park, Gihwan Lee, Sanghwa Yoon, Ayoung Baek, Sarvanan Parameswaran, Seok Ju Park, and et al. 2019. "In Silico Study Probes Potential Inhibitors of Human Dihydrofolate Reductase for Cancer Therapeutics" Journal of Clinical Medicine 8, no. 2: 233. https://doi.org/10.3390/jcm8020233
APA StyleRana, R. M., Rampogu, S., Zeb, A., Son, M., Park, C., Lee, G., Yoon, S., Baek, A., Parameswaran, S., Park, S. J., & Lee, K. W. (2019). In Silico Study Probes Potential Inhibitors of Human Dihydrofolate Reductase for Cancer Therapeutics. Journal of Clinical Medicine, 8(2), 233. https://doi.org/10.3390/jcm8020233






