Investigating the Role of Everolimus in mTOR Inhibition and Autophagy Promotion as a Potential Host-Directed Therapeutic Target in Mycobacterium tuberculosis Infection
Abstract
1. Introduction
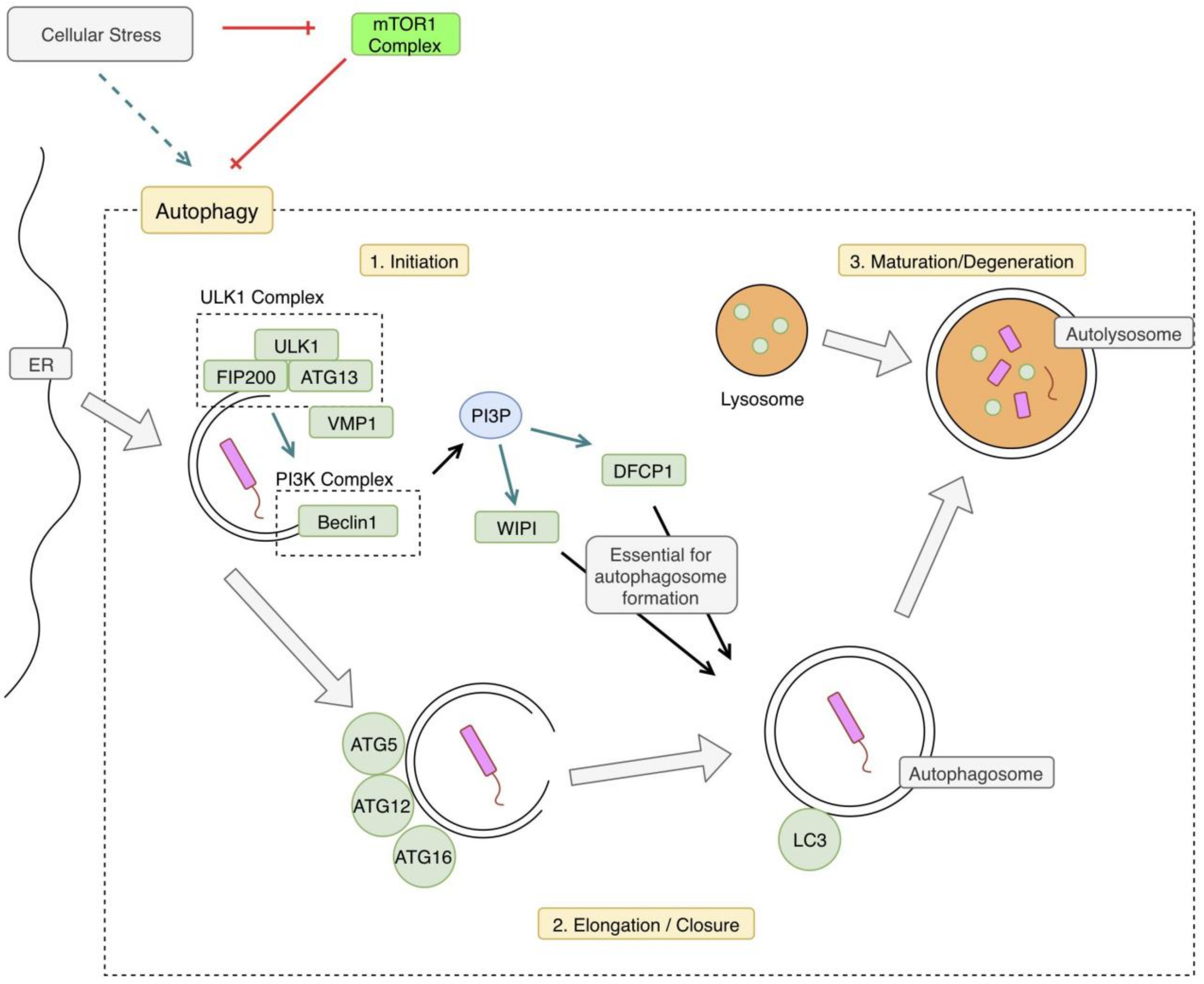
2. Autophagy Overview
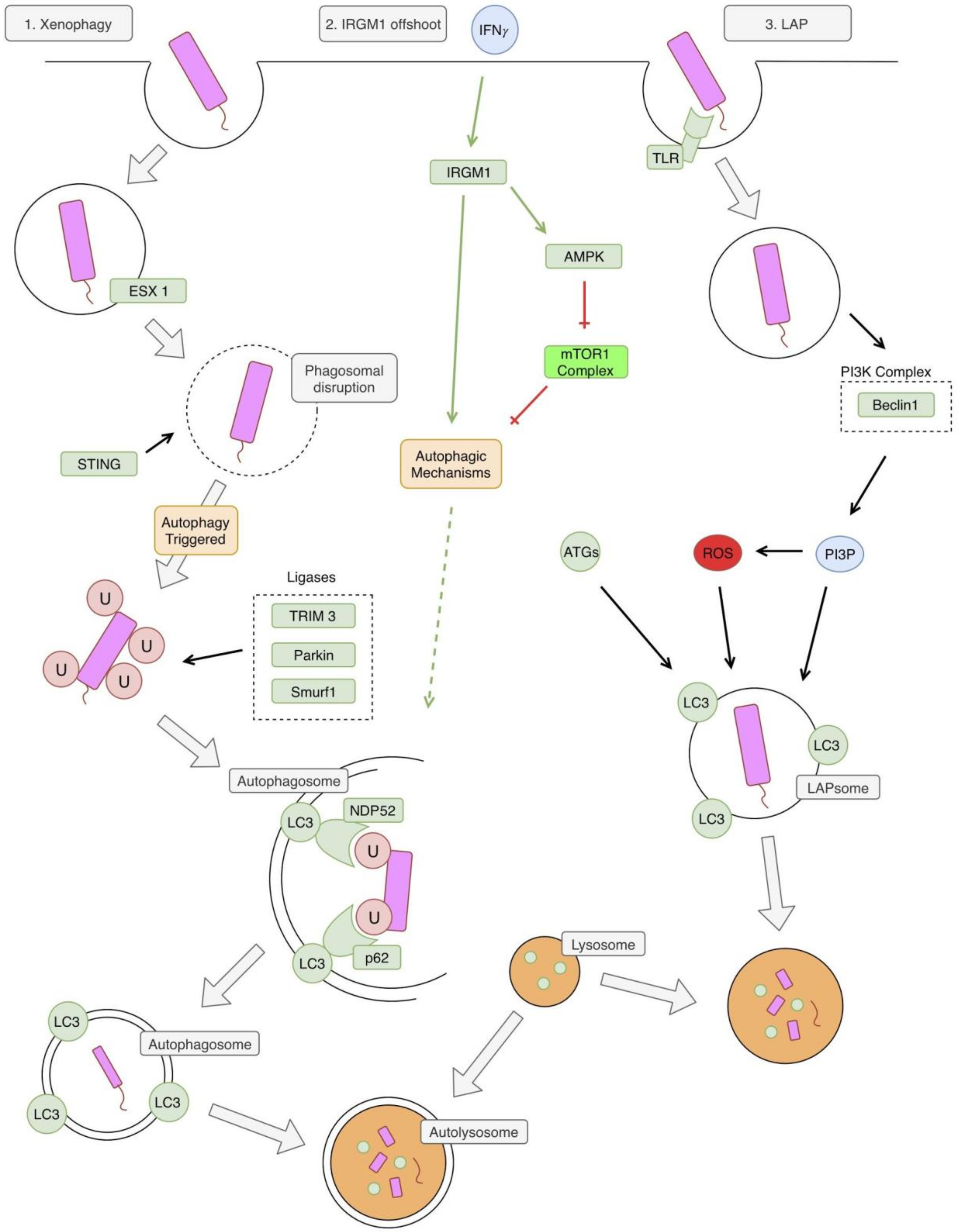
3. Autophagy and TB in the Innate Immune System
4. Autophagy and TB in the Adaptive Immune System
4.1. Type 1 T helper (TH1)
4.2. Granuloma Formation
4.3. Autophagy and Adaptive Immunity
5. mTOR
6. mTOR in TB
7. Treatment
8. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Barberis, I.; Bragazzi, N.L.; Galluzzo, L.; Martini, M. The history of tuberculosis: From the first historical records to the isolation of Koch’s bacillus. J. Prev. Med. Hyg. 2017, 58, E9–E12. [Google Scholar]
- Mittal, C.; Gupta, S. Noncompliance to DOTS: How it can be Decreased. Indian J. Community Med. Off. Publ. Indian Assoc. Prev. Soc. Med. 2011, 36, 27–30. [Google Scholar] [CrossRef]
- Kolloli, A.; Subbian, S. Host-Directed Therapeutic Strategies for Tuberculosis. Front. Med. 2017, 4, 171. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef]
- Sharma, V.; Verma, S.; Seranova, E.; Sarkar, S.; Kumar, D. Selective Autophagy and Xenophagy in Infection and Disease. Front. Cell Dev. Biol. 2018, 6, 147. [Google Scholar] [CrossRef]
- Singh, P.; Subbian, S. Harnessing the mTOR Pathway for Tuberculosis Treatment. Front. Microbiol. 2018, 9, 70. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef]
- Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef]
- Watson, R.O.; Manzanillo, P.S.; Cox, J.S. Extracellular M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell 2012, 150, 803–815. [Google Scholar] [CrossRef]
- Wong, K.-W. The Role of ESX-1 in Mycobacterium tuberculosis Pathogenesis. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Paik, S.; Kim, J.K.; Chung, C.; Jo, E.-K. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence 2018, 1–12. [Google Scholar] [CrossRef]
- Manzanillo, P.S.; Ayres, J.S.; Watson, R.O.; Collins, A.C.; Souza, G.; Rae, C.S.; Schneider, D.S.; Nakamura, K.; Shiloh, M.U.; Cox, J.S. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013, 501, 512–516. [Google Scholar] [CrossRef]
- Franco, L.H.; Nair, V.R.; Scharn, C.R.; Xavier, R.J.; Torrealba, J.R.; Shiloh, M.U.; Levine, B. The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe 2017, 21, 59–72. [Google Scholar] [CrossRef]
- Chauhan, S.; Kumar, S.; Jain, A.; Ponpuak, M.; Mudd, M.H.; Kimura, T.; Choi, S.W.; Peters, R.; Mandell, M.; Bruun, J.-A.; et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell 2016, 39, 13–27. [Google Scholar] [CrossRef]
- Chauhan, S.; Mandell, M.A.; Deretic, V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell 2015, 58, 507–521. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Sanjuan, M.A.; Dillon, C.P.; Tait, S.W.G.; Moshiach, S.; Dorsey, F.; Connell, S.; Komatsu, M.; Tanaka, K.; Cleveland, J.L.; Withoff, S.; et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007, 450, 1253–1257. [Google Scholar] [CrossRef]
- Martinez, J.; Malireddi, R.S.; Lu, Q.; Cunha, L.D.; Pelletier, S.; Gingras, S.; Orchard, R.; Guan, J.-L.; Tan, H.; Peng, J.; et al. Molecular characterization of LC3-associated phagocytosis (LAP) reveals distinct roles for Rubicon, NOX2, and autophagy proteins. Nat. Cell Biol. 2015, 17, 893–906. [Google Scholar] [CrossRef]
- Bandyopadhyay, U.; Overholtzer, M. LAP: The protector against autoimmunity. Cell Res. 2016, 26, 865–866. [Google Scholar] [CrossRef]
- Saleh, M.; Longhi, G. Macrophage Infection by Mycobacteria. Mycobact. Dis. 2016, 6. [Google Scholar] [CrossRef]
- Reiner, N.E. Altered cell signaling and mononuclear phagocyte deactivation during intracellular infection. Immunol. Today 1994, 15, 374–381. [Google Scholar] [CrossRef]
- Vergne, I.; Fratti, R.A.; Hill, P.J.; Chua, J.; Belisle, J.; Deretic, V. Mycobacterium tuberculosis Phagosome Maturation Arrest: Mycobacterial Phosphatidylinositol Analog Phosphatidylinositol Mannoside Stimulates Early Endosomal Fusion. Mol. Biol. Cell 2004, 15, 751–760. [Google Scholar] [CrossRef]
- Deretic, V.; Singh, S.; Master, S.; Harris, J.; Roberts, E.; Kyei, G.; Davis, A.; Haro, S.D.; Naylor, J.; Lee, H.-H.; et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell. Microbiol. 2006, 8, 719–727. [Google Scholar] [CrossRef]
- Vandal, O.H.; Nathan, C.F.; Ehrt, S. Acid Resistance in Mycobacterium tuberculosis. J. Bacteriol. 2009, 191, 4714–4721. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy, an immunologic magic bullet: Mycobacterium tuberculosis phagosome maturation block and how to bypass it. Future Microbiol. 2008, 3, 517–524. [Google Scholar] [CrossRef]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef]
- Chizzolini, C.; Chicheportiche, R.; Burger, D.; Dayer, J.M. Human Th1 cells preferentially induce interleukin (IL)-1beta while Th2 cells induce IL-1 receptor antagonist production upon cell/cell contact with monocytes. Eur. J. Immunol. 1997, 27, 171–177. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium tuberculosis Survival in Infected Macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef]
- Taylor, G.A.; Feng, C.G.; Sher, A. p47 GTPases: Regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 2004, 4, 100–109. [Google Scholar] [CrossRef]
- Harris, J. Autophagy and cytokines. Cytokine 2011, 56, 140–144. [Google Scholar] [CrossRef]
- Caruso, A.M.; Serbina, N.; Klein, E.; Triebold, K.; Bloom, B.R.; Flynn, J.L. Mice Deficient in CD4 T Cells Have Only Transiently Diminished Levels of IFN-γ, Yet Succumb to Tuberculosis. J. Immunol. 1999, 162, 5407–5416. [Google Scholar]
- Boom, W.H.; Canaday, D.H.; Fulton, S.A.; Gehring, A.J.; Rojas, R.E.; Torres, M. Human immunity to M. tuberculosis: T cell subsets and antigen processing. Tuberculosis 2003, 83, 98–106. [Google Scholar] [CrossRef]
- Cooper, A.M.; Magram, J.; Ferrante, J.; Orme, I.M. Interleukin 12 (IL-12) Is Crucial to the Development of Protective Immunity in Mice Intravenously Infected with Mycobacterium tuberculosis. J. Exp. Med. 1997, 186, 39–45. [Google Scholar] [CrossRef]
- Cooper, A.M.; Dalton, D.K.; Stewart, T.A.; Griffin, J.P.; Russell, D.G.; Orme, I.M. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 1993, 178, 2243–2247. [Google Scholar] [CrossRef]
- North, R.J.; Jung, Y.-J. Immunity to Tuberculosis. Annu. Rev. Immunol. 2004, 22, 599–623. [Google Scholar] [CrossRef]
- O’Garra, A.; Britton, W.J. Cytokines in Tuberculosis. In Handbook of Tuberculosis; John Wiley & Sons, Ltd: Hoboken, NJ, USA, 2017; pp. 185–225. [Google Scholar]
- Algood, H.M.S.; Chan, J.; Flynn, J.L. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003, 14, 467–477. [Google Scholar] [CrossRef]
- Flynn, J.L.; Goldstein, M.M.; Chan, J.; Triebold, K.J.; Pfeffer, K.; Lowenstein, C.J.; Schrelber, R.; Mak, T.W.; Bloom, B.R. Tumor necrosis factor-α is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 1995, 2, 561–572. [Google Scholar] [CrossRef]
- Bean, A.G.D.; Roach, D.R.; Briscoe, H.; France, M.P.; Korner, H.; Sedgwick, J.D.; Britton, W.J. Structural Deficiencies in Granuloma Formation in TNF Gene-Targeted Mice Underlie the Heightened Susceptibility to Aerosol Mycobacterium tuberculosis Infection, Which Is Not Compensated for by Lymphotoxin. J. Immunol. 1999, 162, 3504–3511. [Google Scholar]
- Valdivia, A.; Ly, J.; Gonzalez, L.; Hussain, P.; Saing, T.; Islamoglu, H.; Pearce, D.; Ochoa, C.; Venketaraman, V. Restoring Cytokine Balance in HIV-Positive Individuals with Low CD4 T Cell Counts. AIDS Res. Hum. Retroviruses 2017, 33, 905–918. [Google Scholar] [CrossRef]
- Lagman, M.; Ly, J.; Saing, T.; Kaur Singh, M.; Vera Tudela, E.; Morris, D.; Chi, P.-T.; Ochoa, C.; Sathananthan, A.; Venketaraman, V. Investigating the causes for decreased levels of glutathione in individuals with type II diabetes. PLoS ONE 2015, 10, e0118436. [Google Scholar] [CrossRef]
- Teskey, G.; Cao, R.; Islamoglu, H.; Medina, A.; Prasad, C.; Prasad, R.; Sathananthan, A.; Fraix, M.; Subbian, S.; Zhong, L.; et al. The Synergistic Effects of the Glutathione Precursor, NAC and First-Line Antibiotics in the Granulomatous Response against Mycobacterium tuberculosis. Front. Immunol. 2018, 9, 2069. [Google Scholar] [CrossRef]
- Teskey, G.; Abrahem, R.; Cao, R.; Gyurjian, K.; Islamoglu, H.; Lucero, M.; Martinez, A.; Paredes, E.; Salaiz, O.; Robinson, B.; et al. Chapter Five—Glutathione as a Marker for Human Disease. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 87, pp. 141–159. [Google Scholar]
- Crotzer, V.L.; Blum, J.S. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. Baltim. Md 1950 2009, 182, 3335–3341. [Google Scholar] [CrossRef]
- Dörfel, D.; Appel, S.; Grünebach, F.; Weck, M.M.; Müller, M.R.; Heine, A.; Brossart, P. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood 2005, 105, 3199–3205. [Google Scholar] [CrossRef]
- Brazil, M.I.; Weiss, S.; Stockinger, B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur. J. Immunol. 1997, 27, 1506–1514. [Google Scholar] [CrossRef]
- Jagannath, C.; Lindsey, D.R.; Dhandayuthapani, S.; Xu, Y.; Hunter, R.L.; Eissa, N.T. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009, 15, 267–276. [Google Scholar] [CrossRef]
- Kim, Y.C.; Guan, K.-L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef]
- Jung, C.H.; Ro, S.-H.; Cao, J.; Otto, N.M.; Kim, D.-H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef]
- Zeng, H.; Cohen, S.; Guy, C.; Shrestha, S.; Neale, G.; Brown, S.A.; Cloer, C.; Kishton, R.J.; Gao, X.; Youngblood, B.; et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity 2016, 45, 540–554. [Google Scholar] [CrossRef]
- Kim, J.E.; Chen, J. Regulation of Peroxisome Proliferator–Activated Receptor-γ Activity by Mammalian Target of Rapamycin and Amino Acids in Adipogenesis. Diabetes 2004, 53, 2748–2756. [Google Scholar] [CrossRef]
- Lachmandas, E.; Beigier-Bompadre, M.; Cheng, S.; Kumar, V.; van Laarhoven, A.; Wang, X.; Ammerdorffer, A.; Boutens, L.; de Jong, D.; Kanneganti, T.; et al. Rewiring cellular metabolism via the AKT/mTOR pathway contributes to host defense against Mycobacterium tuberculosis in human and murine cells. Eur. J. Immunol. 2016, 46, 2574–2586. [Google Scholar] [CrossRef]
- Venketaraman, V. Understanding the Host Immune Response Against Mycobacterium Tuberculosis Infection; Springer: Berlin, Germany, 2018. [Google Scholar]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef]
- Huang, H.Y.; Wang, W.C.; Lin, P.Y.; Huang, C.P.; Chen, C.Y.; Chen, Y.K. The roles of autophagy and hypoxia in human inflammatory periapical lesions. Int. Endod. J. 2018, 51, e125–e145. [Google Scholar] [CrossRef]
- Liu, Q.; Thoreen, C.; Wang, J.; Sabatini, D.; Gray, N.S. mTOR Mediated Anti-Cancer Drug Discovery. Drug Discov. Today Ther. Strateg. 2009, 6, 47–55. [Google Scholar] [CrossRef]
- Restrepo, B.I. Metformin: Candidate host-directed therapy for tuberculosis in diabetes and non-diabetes patients. Tuberculosis 2016, 101, S69–S72. [Google Scholar] [CrossRef]
- Williams, A.; Sarkar, S.; Cuddon, P.; Ttofi, E.K.; Saiki, S.; Siddiqi, F.H.; Jahreiss, L.; Fleming, A.; Pask, D.; Goldsmith, P.; et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008, 4, 295–305. [Google Scholar] [CrossRef]
- Sarkar, S.; Floto, R.A.; Berger, Z.; Imarisio, S.; Cordenier, A.; Pasco, M.; Cook, L.J.; Rubinsztein, D.C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005, 170, 1101–1111. [Google Scholar] [CrossRef]
- Sohn, H.; Kim, K.; Lee, K.-S.; Choi, H.-G.; Lee, K.-I.; Shin, A.-R.; Kim, J.-S.; Shin, S.J.; Song, C.-H.; Park, J.-K.; et al. Lithium inhibits growth of intracellular Mycobacterium kansasii through enhancement of macrophage apoptosis. J. Microbiol. 2014, 52, 299–306. [Google Scholar] [CrossRef]
- Klümpen, H.-J.; Beijnen, J.H.; Gurney, H.; Schellens, J.H.M. Inhibitors of mTOR. Oncologist 2010, 15, 1262–1269. [Google Scholar] [CrossRef]
- Klawitter, J.; Nashan, B.; Christians, U. Everolimus and Sirolimus in Transplantation-Related but Different. Expert Opin. Drug Saf. 2015, 14, 1055–1070. [Google Scholar] [CrossRef]
- First Generic Drug Approvals. Available online: https://www.fda.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/DrugandBiologicApprovalReports/ANDAGenericDrugApprovals/default.htm (accessed on 10 January 2019).
- Mannick, J.B.; Del Giudice, G.; Lattanzi, M.; Valiante, N.M.; Praestgaard, J.; Huang, B.; Lonetto, M.A.; Maecker, H.T.; Kovarik, J.; Carson, S.; et al. mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 2014, 6, 268ra179. [Google Scholar] [CrossRef]
- AFINITOR (Everolimus) Tablets [Package Insert] 2016. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/022334s036lbl.pdf (accessed on 13 January 2019).
- Bento, C.F.; Empadinhas, N.; Mendes, V. Autophagy in the Fight against Tuberculosis. DNA Cell Biol. 2015, 34, 228–242. [Google Scholar] [CrossRef]
- Gupta, A.; Pant, G.; Mitra, K.; Madan, J.; Chourasia, M.K.; Misra, A. Inhalable Particles Containing Rapamycin for Induction of Autophagy in Macrophages Infected with Mycobacterium tuberculosis. Mol. Pharm. 2014, 11, 1201–1207. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerni, S.; Shafer, D.; To, K.; Venketaraman, V. Investigating the Role of Everolimus in mTOR Inhibition and Autophagy Promotion as a Potential Host-Directed Therapeutic Target in Mycobacterium tuberculosis Infection. J. Clin. Med. 2019, 8, 232. https://doi.org/10.3390/jcm8020232
Cerni S, Shafer D, To K, Venketaraman V. Investigating the Role of Everolimus in mTOR Inhibition and Autophagy Promotion as a Potential Host-Directed Therapeutic Target in Mycobacterium tuberculosis Infection. Journal of Clinical Medicine. 2019; 8(2):232. https://doi.org/10.3390/jcm8020232
Chicago/Turabian StyleCerni, Stephen, Dylan Shafer, Kimberly To, and Vishwanath Venketaraman. 2019. "Investigating the Role of Everolimus in mTOR Inhibition and Autophagy Promotion as a Potential Host-Directed Therapeutic Target in Mycobacterium tuberculosis Infection" Journal of Clinical Medicine 8, no. 2: 232. https://doi.org/10.3390/jcm8020232
APA StyleCerni, S., Shafer, D., To, K., & Venketaraman, V. (2019). Investigating the Role of Everolimus in mTOR Inhibition and Autophagy Promotion as a Potential Host-Directed Therapeutic Target in Mycobacterium tuberculosis Infection. Journal of Clinical Medicine, 8(2), 232. https://doi.org/10.3390/jcm8020232





