Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study
Abstract
1. Introduction
2. Materials and Methods
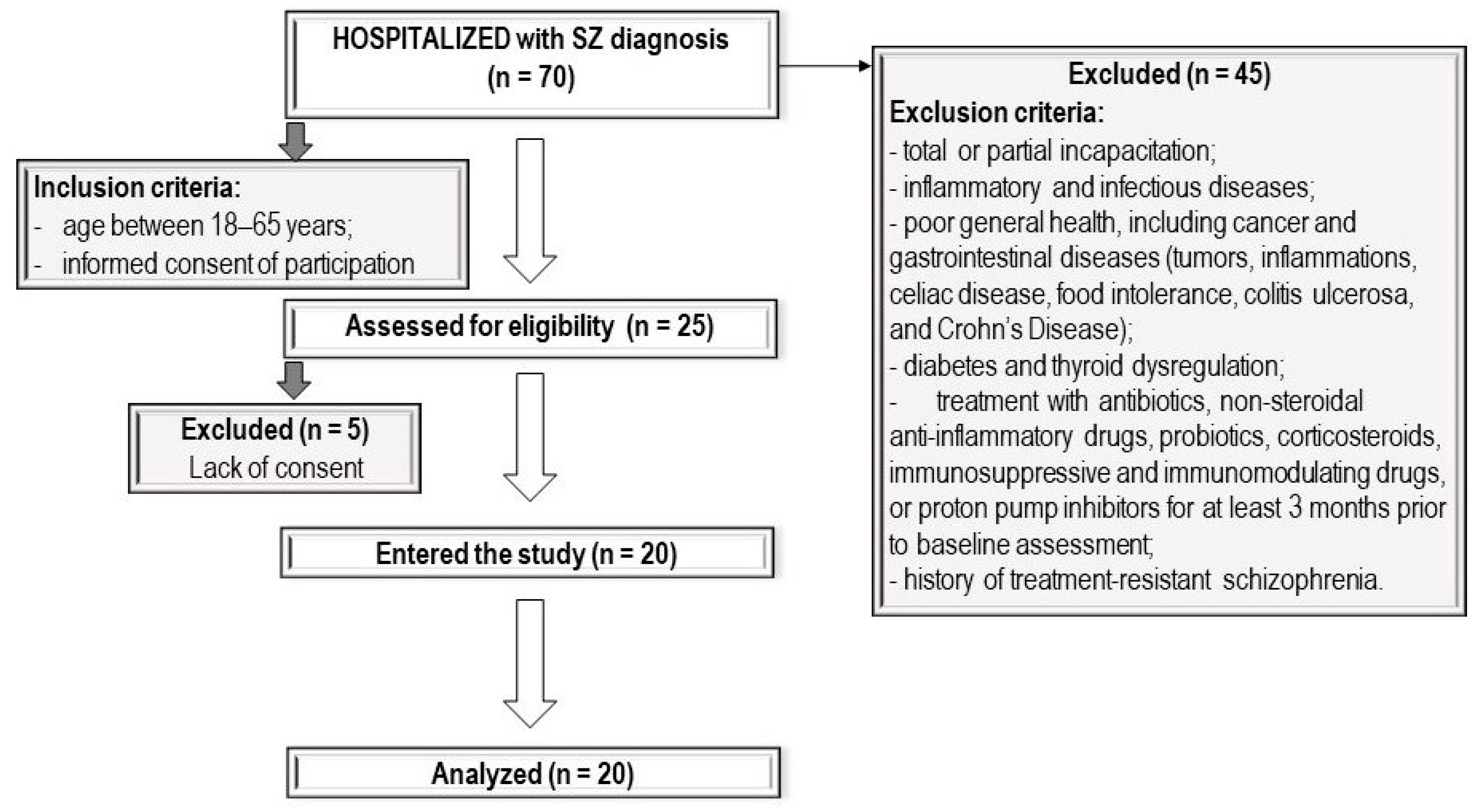
2.1. Patients
2.2. Study Protocol
2.3. Processing of Raw Data and Statistical Analysis
3. Results
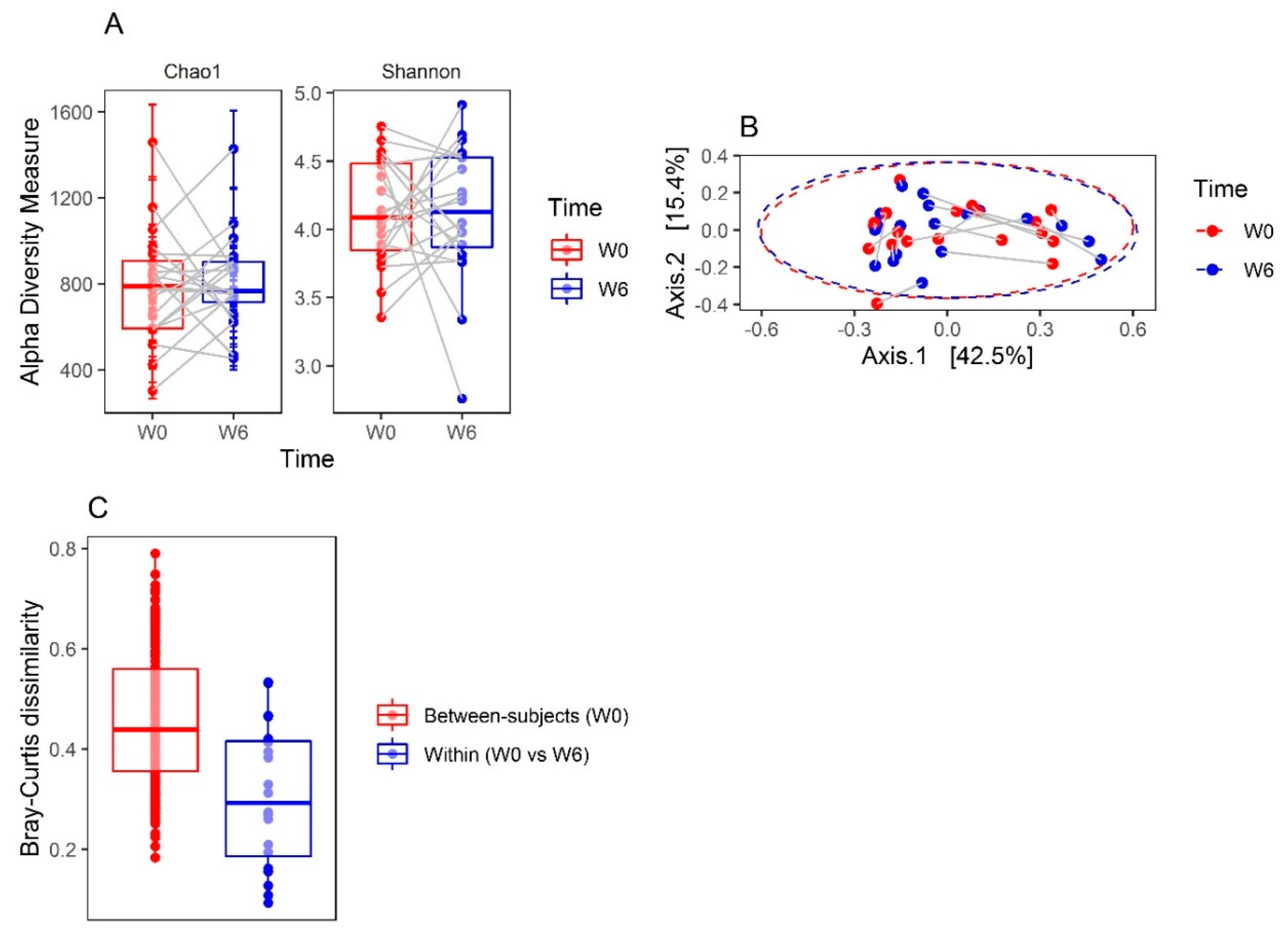
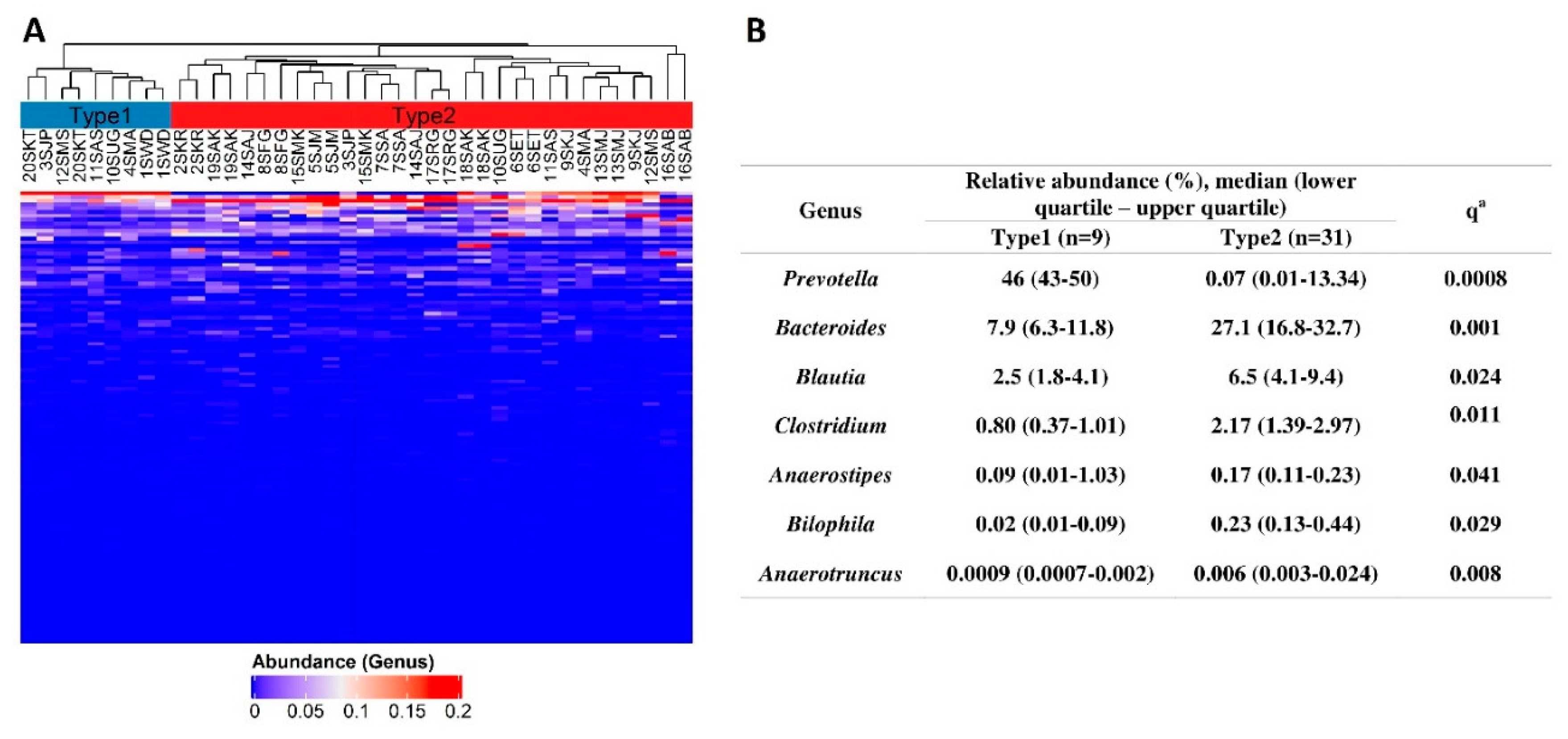
3.1. Microbiota Compositions
3.2. Clinical Improvement and BMI Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO Schizophrenia. Available online: https://www.who.int/news-room/fact-sheets/detail/schizophrenia (accessed on 2 June 2019).
- Müller, N. Inflammation in Schizophrenia: Pathogenetic Aspects and Therapeutic Considerations. Schizophr. Bull. 2018, 44, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Purves-Tyson, T.D.; Owens, S.J.; Rothmond, D.A.; Halliday, G.M.; Double, K.L.; Stevens, J.; McCrossin, T.; Weickert, C.S. Putative presynaptic dopamine dysregulation in schizophrenia is supported by molecular evidence from post-mortem human midbrain. Transl. Psychiatry 2017, 7, e1003. [Google Scholar] [CrossRef] [PubMed]
- Guest, F.L.; Martins-de-Souza, D.; Rahmoune, H.; Bahn, S.; Guest, P.C. The effects of stress on hypothalamic-pituitary-adrenal (HPA) axis function in subjects with schizophrenia. Arch. Clin. Psychiatry (São Paulo) 2013, 40, 20–27. [Google Scholar] [CrossRef]
- Guest, P.C.; Martins-De-Souza, D.; Vanattou-Saifoudine, N.; Harris, L.W.; Bahn, S. Abnormalities in Metabolism and Hypothalamic–Pituitary–Adrenal Axis Function in Schizophrenia. Int. Rev. Neurobiol. 2011, 101, 145–168. [Google Scholar]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The role of the gut microbiome in systemic inflammatory disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.-N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Waclawiková, B.; El Aidy, S. Role of Microbiota and Tryptophan Metabolites in the Remote Effect of Intestinal Inflammation on Brain and Depression. Pharmaceuticals (Basel) 2018, 11, 63. [Google Scholar] [CrossRef]
- Rogers, G.B.; Keating, D.J.; Young, R.L.; Wong, M.-L.; Licinio, J.; Wesselingh, S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry 2016, 21, 738–748. [Google Scholar] [CrossRef]
- Schwarz, E.; Maukonen, J.; Hyytiäinen, T.; Kieseppä, T.; Orešič, M.; Sabunciyan, S.; Mantere, O.; Saarela, M.; Yolken, R.; Suvisaari, J. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr. Res. 2018, 192, 398–403. [Google Scholar] [CrossRef]
- Zheng, P.; Zeng, B.; Liu, M.; Chen, J.; Pan, J.; Han, Y.; Liu, Y.; Cheng, K.; Zhou, C.; Wang, H.; et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci. Adv. 2019, 5, eaau8317. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.; Clarke, G.; Scully, P.; Dinan, T.G. Kynurenine pathway in psychosis: Evidence of increased tryptophan degradation. J. Psychopharmacol. 2009, 23, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Llenos, I.C.; Dulay, J.R.; Weis, S. Upregulation of the initiating step of the kynurenine pathway in postmortem anterior cingulate cortex from individuals with schizophrenia and bipolar disorder. Brain Res. 2006, 1073, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Hálfdánarson, Ó.; Zoëga, H.; Aagaard, L.; Bernardo, M.; Brandt, L.; Fusté, A.C.; Furu, K.; Garuoliené, K.; Hoffmann, F.; Huybrechts, K.F.; et al. International trends in antipsychotic use: A study in 16 countries, 2005–2014. Eur. Neuropsychopharmacol. 2017, 27, 1064–1076. [Google Scholar]
- Vancampfort, D.; Correll, C.U.; Galling, B.; Probst, M.; De Hert, M.; Ward, P.B.; Rosenbaum, S.; Gaughran, F.; Lally, J.; Stubbs, B. Diabetes mellitus in people with schizophrenia, bipolar disorder and major depressive disorder: A systematic review and large scale meta-analysis. World Psychiatry 2016, 15, 166–174. [Google Scholar] [CrossRef]
- Galling, B.; Roldán, A.; Nielsen, R.E.; Nielsen, J.; Gerhard, T.; Carbon, M.; Stubbs, B.; Vancampfort, D.; De Hert, M.; Olfson, M.; et al. Type 2 Diabetes Mellitus in Youth Exposed to Antipsychotics. JAMA Psychiatry 2016, 73, 247–259. [Google Scholar] [CrossRef]
- Galling, B.; Correll, C.U. Do antipsychotics increase diabetes risk in children and adolescents? Expert Opin. Drug Saf. 2015, 14, 219–241. [Google Scholar] [CrossRef]
- De Hert, M.; Vancampfort, D.; Correll, C.U.; Mercken, V.; Peuskens, J.; Sweers, K.; Van Winkel, R.; Mitchell, A.J. Guidelines for screening and monitoring of cardiometabolic risk in schizophrenia: Systematic evaluation. Br. J. Psychiatry 2011, 199, 99–105. [Google Scholar] [CrossRef]
- Bak, M.; Fransen, A.; Janssen, J.; Van Os, J.; Drukker, M. Almost All Antipsychotics Result in Weight Gain: A Meta-Analysis. PLoS ONE 2014, 9, e94112. [Google Scholar] [CrossRef]
- Musil, R.; Obermeier, M.; Russ, P.; Hamerle, M. Weight gain and antipsychotics: A drug safety review. Expert Opin. Drug Saf. 2015, 14, 73–96. [Google Scholar] [CrossRef]
- Spertus, J.; Horvitz-Lennon, M.; Abing, H.; Normand, S.-L. Risk of weight gain for specific antipsychotic drugs: A meta-analysis. NPJ Schizophr. 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Iglesias, R.; Martínez-García, O.; Pardo-Garcia, G.; Amado, J.A.; Garcia-Unzueta, M.T.; Tabares-Seisdedos, R.; Crespo-Facorro, B. Course of weight gain and metabolic abnormalities in first treated episode of psychosis: The first year is a critical period for development of cardiovascular risk factors. Int. J. Neuropsychopharmacol. 2014, 17, 41–51. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association; American Psychiatric Association; American Association of Clinical Endocrinologists; North American Association for the Study of Obesity Consensus development conference on antipsychotic drugs and obesity and diabetes. Diabetes Care 2004, 27, 596–601.
- De Hert, M.; Dekker, J.M.; Wood, D.; Kahl, K.G.; Holt, R.I.G.; Möller, H.-J. Cardiovascular disease and diabetes in people with severe mental illness position statement from the European Psychiatric Association (EPA), supported by the European Association for the Study of Diabetes (EASD) and the European Society of Cardiology (ESC). Eur. Psychiatry 2009, 24, 412–424. [Google Scholar] [CrossRef]
- Albaugh, V.L.; Henry, C.R.; Bello, N.T.; Hajnal, A.; Lynch, S.L.; Halle, B.; Lynch, C.J. Hormonal and metabolic effects of olanzapine and clozapine related to body weight in rodents. Obesity (Silver Spring) 2006, 14, 36–51. [Google Scholar] [CrossRef]
- Basson, B.R.; Kinon, B.J.; Taylor, C.C.; Szymanski, K.A.; Gilmore, J.A.; Tollefson, G.D. Factors Influencing Acute Weight Change in Patients with Schizophrenia Treated with Olanzapine, Haloperidol, or Risperidone. J. Clin. Psychiatry 2001, 62, 231–238. [Google Scholar] [CrossRef]
- Ou, J.-J.; Xu, Y.; Chen, H.-H.; Fan, X.; Gao, K.; Wang, J.; Guo, X.-F.; Wu, R.-R.; Zhao, J.-P. Comparison of metabolic effects of ziprasidone versus olanzapine treatment in patients with first-episode schizophrenia. Psychopharmacology 2013, 225, 627–635. [Google Scholar] [CrossRef]
- Davey, K.J.; Cotter, P.D.; O’Sullivan, O.; Crispie, F.; Dinan, T.G.; Cryan, J.F.; O’Mahony, S.M. Antipsychotics and the gut microbiome: Olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Transl. Psychiatry 2013, 3, e309. [Google Scholar] [CrossRef]
- Davey, K.J.; O’Mahony, S.M.; Schellekens, H.; O’Sullivan, O.; Bienenstock, J.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gender-dependent consequences of chronic olanzapine in the rat: Effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology 2012, 221, 155–169. [Google Scholar] [CrossRef]
- Morgan, A.P.; Crowley, J.J.; Nonneman, R.J.; Quackenbush, C.R.; Miller, C.N.; Ryan, A.K.; Bogue, M.A.; Paredes, S.H.; Yourstone, S.; Carroll, I.M.; et al. The Antipsychotic Olanzapine Interacts with the Gut Microbiome to Cause Weight Gain in Mouse. PLoS ONE 2014, 9, e115225. [Google Scholar] [CrossRef]
- Flowers, S.A.; Evans, S.J.; Ward, K.M.; McInnis, M.G.; Ellingrod, V.L. Interaction Between Atypical Antipsychotics and the Gut Microbiome in a Bipolar Disease Cohort. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, M.; Rychlik, E.; Stoś, K.; Wierzejska, R.; Wojtasik, A.; Charzewska, J.; Mojska, H.; Szponar, L.; Sajór, I.; Kłosiewicz-Latoszek, L.; et al. Normy Żywienia dla Populacji Polski; Instytut Żywności i Żywienia: Warszawa, Poland, 2017; ISBN 978-83-86060-89-4. [Google Scholar]
- Ascher-Svanum, H.; Zhao, F.; Detke, H.C.; Nyhuis, A.W.; Lawson, A.H.; Stauffer, V.L.; Montgomery, W.; Witte, M.M.; McDonnell, D.P. Early response predicts subsequent response to olanzapine long-acting injection in a randomized, double-blind clinical trial of treatment for schizophrenia. BMC Psychiatry 2011, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 80. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B.; et al. Metabolic Reconstruction for Metagenomic Data and Its Application to the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Kao, A.C.-C.; Spitzer, S.; Anthony, D.C.; Lennox, B.; Burnet, P.W.J. Prebiotic attenuation of olanzapine-induced weight gain in rats: Analysis of central and peripheral biomarkers and gut microbiota. Transl. Psychiatry 2018, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xu, J.; Li, Z.; Huang, Y.; Yuan, Y.; Wang, J.; Zhang, M.; Hu, S.; Liang, Y. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophr. Res. 2018, 197, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Firth, J.; Rosenbaum, S.; Ward, P.B.; Curtis, J.; Teasdale, S.B.; Yung, A.R.; Sarris, J. Adjunctive nutrients in first-episode psychosis: A systematic review of efficacy, tolerability and neurobiological mechanisms. Early Interv. Psychiatry 2018, 12, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Leppik, L.; Kriisa, K.; Koido, K.; Koch, K.; Kajalaid, K.; Haring, L.; Vasar, E.; Zilmer, M. Profiling of Amino Acids and Their Derivatives Biogenic Amines Before and After Antipsychotic Treatment in First-Episode Psychosis. Front. Psychol. 2018, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, C.P.; Allott, K.A.; Murphy, B.P.; Yuen, H.P.; Proffitt, T.-M.; Papas, A.; Moral, J.; Pham, T.; O’Regan, M.K.; Phassouliotis, C.; et al. Adjunctive Taurine in First-Episode Psychosis: A Phase 2, Double-Blind, Randomized, Placebo-Controlled Study. J. Clin. Psychiatry 2016, 77, e1610–e1617. [Google Scholar] [CrossRef] [PubMed]
- Hatano, T.; Ohnuma, T.; Sakai, Y.; Shibata, N.; Maeshima, H.; Hanzawa, R.; Suzuki, T.; Arai, H. Plasma alanine levels increase in patients with schizophrenia as their clinical symptoms improve—Results from the Juntendo University Schizophrenia Projects (JUSP). Psychiatry Res. Neuroimaging 2010, 177, 27–31. [Google Scholar] [CrossRef]
- Errico, F.; Nuzzo, T.; Carella, M.; Bertolino, A.; Usiello, A. The Emerging Role of Altered d-Aspartate Metabolism in Schizophrenia: New Insights from Preclinical Models and Human Studies. Front. Psychol. 2018, 9, 559. [Google Scholar] [CrossRef]
- Cao, B.; Wang, D.-F.; Xu, M.-Y.; Liu, Y.-Q.; Yan, L.-L.; Wang, J.-Y.; Lu, Q.-B. Lower folate levels in schizophrenia: A meta-analysis. Psychiatry Res. 2016, 245, 1–7. [Google Scholar] [CrossRef]
- Hussein, H.; El Mawella, S.A.; Ahmed, T. Folate, vitamin B12, and negative symptoms in schizophrenia. Egypt. J. Psychiatry 2018, 39, 89. [Google Scholar] [CrossRef]
- Hill, M.; Shannahan, K.; Jasinski, S.; Macklin, E.A.; Raeke, L.; Roffman, J.L.; Goff, D.C. Folate supplementation in schizophrenia: A possible role for MTHFR genotype. Schizophr. Res. 2011, 127, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Maguire, Á.; Hargreaves, A.; Gill, M. Coenzyme Q10 and neuropsychiatric and neurological disorders: Relevance for schizophrenia. Nutr. Neurosci. 2018, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schmelzer, C.; Döring, F. Identification of LPS-inducible genes downregulated by ubiquinone in human THP-1 monocytes. BioFactors 2010, 36, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Bubber, P.; Hartounian, V.; Gibson, G.E.; Blass, J.P. Abnormalities in the tricarboxylic acid (TCA) cycle in the brains of schizophrenia patients. Eur. Neuropsychopharmacol. 2011, 21, 254–260. [Google Scholar] [CrossRef]
- Castillo, R.I.; Rojo, L.E.; Henríquez-Henríquez, M.; Silva, H.; Maturana, A.; Villar, M.J.; Fuentes, M.; Gaspar, P.A. From Molecules to the Clinic: Linking Schizophrenia and Metabolic Syndrome through Sphingolipids Metabolism. Front. Mol. Neurosci. 2016, 10, 488. [Google Scholar] [CrossRef]
- Weston-Green, K.; Babic, I.; De Santis, M.; Pan, B.; Montgomery, M.K.; Mitchell, T.; Huang, X.-F.; Nealon, J. Disrupted sphingolipid metabolism following acute clozapine and olanzapine administration. J. Biomed. Sci. 2018, 25, 40. [Google Scholar] [CrossRef]
- Liu, M.-L.; Zhang, X.-T.; Du, X.-Y.; Fang, Z.; Liu, Z.; Xu, Y.; Zheng, P.; Xu, X.-J.; Cheng, P.-F.; Huang, T.; et al. Severe disturbance of glucose metabolism in peripheral blood mononuclear cells of schizophrenia patients: A targeted metabolomic study. J. Transl. Med. 2015, 13, 226. [Google Scholar] [CrossRef]
- Yang, X.; Sun, L.; Zhao, A.; Hu, X.; Qing, Y.; Jiang, J.; Yang, C.; Xu, T.; Wang, P.; Liu, J.; et al. Serum fatty acid patterns in patients with schizophrenia: A targeted metabonomics study. Transl. Psychiatry 2017, 7, e1176. [Google Scholar] [CrossRef]
- Peters, B.D.; Machielsen, M.W.J.; Hoen, W.P.; Caan, M.W.A.; Malhotra, A.K.; Szeszko, P.R.; Duran, M.; Olabarriaga, S.D.; de Haan, L. Polyunsaturated fatty acid concentration predicts myelin integrity in early-phase psychosis. Schizophr. Bull. 2013, 39, 830–838. [Google Scholar] [CrossRef]
- Fernstrom, J.D. Branched-chain amino acids and brain function. J. Nutr. 2005, 135, 1539S–1546S. [Google Scholar] [CrossRef]
- Bevans, M.L.; Read, L.L.; Chao, H.M.; Clelland, J.D.; Suckow, R.F.; Maher, T.J.; Citrome, L.; Richardson, M.A. Efficacy of the Branched-Chain Amino Acids in the Treatment of Tardive Dyskinesia in Men. Am. J. Psychiatry 2003, 160, 1117–1124. [Google Scholar]
- Włodarczyk, A.; Wiglusz, M.S.; Cubała, W.J. Ketogenic diet for schizophrenia: Nutritional approach to antipsychotic treatment. Med. Hypotheses 2018, 118, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Penttilä, M.; Jaaskelainen, E.; Hirvonen, N.; Isohanni, M.; Miettunen, J. Duration of untreated psychosis as predictor of long-term outcome in schizophrenia: Systematic review and meta-analysis. Br. J. Psychiatry 2014, 205, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Lin, S.-C.; Huang, Y.-H.; Wang, F.-C.; Huang, C.-J. Early prediction of olanzapine-induced weight gain for schizophrenia patients. Psychiatry Res. 2018, 263, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Castro-Nallar, E.; Bendall, M.L.; Pérez-Losada, M.; Sabuncyan, S.; Severance, E.G.; Dickerson, F.B.; Schroeder, J.R.; Yolken, R.H.; Crandall, K.A. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ 2015, 3, e1140. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Bahr, S.M.; Tyler, B.C.; Wooldridge, N.; Butcher, B.D.; Burns, T.L.; Teesch, L.M.; Oltman, C.L.; Azcarate-Peril, M.A.; Kirby, J.R.; Calarge, C.A. Use of the second-generation antipsychotic, risperidone, and secondary weight gain are associated with an altered gut microbiota in children. Transl. Psychiatry 2015, 5, e652. [Google Scholar] [CrossRef]
- Marlicz, W.; Yung, D.E.; Skonieczna-Żydecka, K.; Loniewski, I.; Van Hemert, S.; Loniewska, B.; Koulaouzidis, A. From clinical uncertainties to precision medicine: The emerging role of the gut barrier and microbiome in small bowel functional diseases. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 961–978. [Google Scholar] [CrossRef]
- Skonieczna-Żydecka, K.; Łoniewski, I.; Misera, A.; Stachowska, E.; Maciejewska, D.; Marlicz, W.; Galling, B. Second-generation antipsychotics and metabolism alterations: A systematic review of the role of the gut microbiome. Psychopharmacology 2018, 236, 1491–1512. [Google Scholar] [CrossRef]
- Rao, T.S.S.; Asha, M.R.; Ramesh, B.N.; Rao, K.S.J. Understanding nutrition, depression and mental illnesses. Indian J. Psychiatry 2008, 50, 77–82. [Google Scholar]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
| Variables | Median (1st Q–3rd Q)/ n (%) |
|---|---|
| Sex (F/M) | 9 (45%)/11 (55%) |
| Age (years) | 33.5 (31–39) |
| BMI (kg/m2) | 28.91 (24.82–31.27) |
| Olanzapine maximum dose (mg) | 20.00 (20.0–20.0) |
| Olanzapine average dose per day (mg) | 15.54 (13.50–16.34) |
| Disease duration (months) | 90 (32–114) |
| Duration of untreated psychosis (months) | 4.5 (1.75–12.0) |
| Smoking (number of cigarettes per day) a | 1.5 (1.0–3.0) |
| Coffee (number of cups) | 2.0 (0.0–3.0) |
| Tea (number of cups) | 2.5 (1.0–3.0) |
| Variables (Females + Males) | Cluster Type 2 (n = 5) | Cluster Type 3 (n = 12) | p/qa |
| BMI (kg/m2) W0 | 28.7 (27–29.9) | 29.6 (24.4–32) | 0.874/0.874 |
| PANNS W0 | 95 (94–98) | 68 (62.8–74.2) | 0.007/0.047 |
| PANNS N subscale W0 | 28 (23–28) | 20.5 (17–22.2) | 0.020/0.070 |
| PANNS P subscale W0 | 24 (23–26) | 20 (15.8–22) | 0.026/0.073 |
| PANNS G subscale W0 | 46 (43–47) | 32 (27.8–37) | 0.010/0.047 |
| SF36 W0 | 90 (83–97) | 76.5 (72.5–83.8) | 0.102/0.238 |
| CGI-S W0 | 7 (6–7) | 5 (5–6) | 0.009/0.047 |
| BMI (kg/m2) | −0.53 (−1.33–0.72) | 0.35 (−0.23–0.90) | 0.562/0.656 |
| PANNS | −44 (−65–−31) | −37 (−39.5–−21.8) | 0.342/0.749 |
| PANNS N subscale | −10 (−17–−9) | −6 (−8.75–−4.5) | 0.205/0.410 |
| PANNS P subscale | −12 (−18–−6) | −11 (−15–−7.75) | 0.874/0.874 |
| PANNS G subscale | −22 (−26–−16) | −16 (−17.8–−8.75) | 0.315/0.479 |
| SF36 | −5 (−18–−4) | −3 (−12.2–6) | 0.245/0.429 |
| CGI-I | 4 (3–4) | 3.5 (3–4) | 0.452/0.575 |
| Variable (Males) | Cluster Type 2 (n = 5) | Cluster Type 3 (n = 4) | p/qa |
| BMI (kg/m2) W0 | 28.7 (27–29.9) | 30.5 (27.5–32.2) | 0.713/0.768 |
| PANNS W0 | 95 (94–98) | 67 (59.2–76.5) | 0.037/0.198 |
| PANNS N subscale W0 | 28 (23–28) | 20 (18.8–22.2) | 0.084/0.198 |
| PANNS P subscale W0 | 24 (23–26) | 17 (14–19) | 0.027/0.198 |
| PANNS G subscale W0 | 46 (43–47) | 30.5 (24.8–37.5) | 0.065/0.198 |
| SF36 W0 | 90 (83–97) | 83.5 (76.2–90) | 0.391/0.547 |
| CGI-S W0 | 7 (6–7) | 5.5 (5–6) | 0.050/0.198 |
| BMI (kg/m2) | −0.53 (−1.33–0.72) | −0.92 (−1.97–−0.30) | 0.713/0.768 |
| PANNS | −44 (−65–−31) | −20.5 (−26.2–−17.8) | 0.140/0.280 |
| PANNS N subscale | −10 (−17–−9) | −6 (−7.5–−5.75) | 0.389/0.547 |
| PANNS P subscale | −12 (−18–−6) | −7.5 (−8.25–−6) | 0.389/0.547 |
| PANNS G subscale | −22 (−26–−16) | −8.5 (−11–−7) | 0.085/0.198 |
| SF36 | −5 (−18–−4) | −11 (−13.5–−7) | 1.0/1.0 |
| CGI-I | 4 (3–4) | 4 (4–4) | 0.661/0.768 |
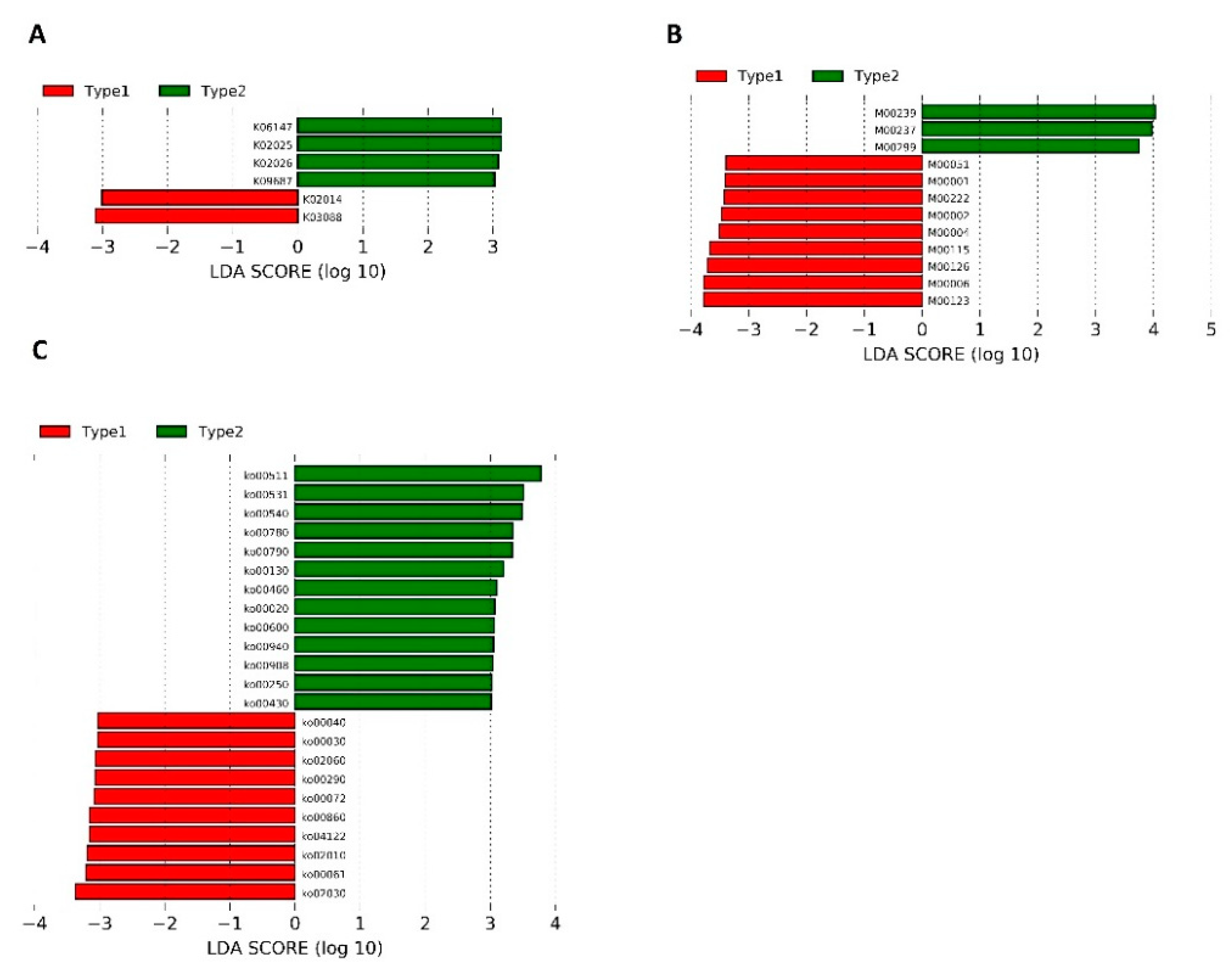
| Pathways | Physiological Function | Potential Roles in SZ | References |
|---|---|---|---|
| Pathways found to be more active in patients with significantly less severe symptoms (according to PANNS and CGI-S) | |||
| ko00430: Taurine and hypotaurine metabolism | Taurine: N-methyl-D-aspartate (NDMA) receptor inhibition and stem cell activation; a neurotransmitter and an inhibitory neuromodulator in the central nervous system (CNS); a potential immunomodulating compound, and an attenuator of apoptosis | Taurine supplementation was found to alleviate SZ symptoms significantly | [46,47,48] |
| Ko00250: Alanine (ALA), aspartate (ASP), and glutamate metabolism | ALA: An agonist that binds to the glycine site of NMDA receptors and improves the positive and cognitive symptoms of patients with SZ; ASP: Binding to the agonist site of NMDARs | NMDAR hypofunction in schizophrenia pathogenesis | [49,50] |
| Ko00790: Folate biosynthesis | Folate: Production of adenosylmethionine (SAM) | Schizophrenia patients may have lower folate levels (negative correlation with negative symptoms of SZ) | [51,52,53] |
| Ko00130: Ubiquinone and other terpenoids–quinone biosynthesis | Ubiquinone: ATP production, mitochondrial function, and reduction of proinflammatory mediators | Mitochondrial dysfunction as a part of SZ etiology | [54,55] |
| Ko00020: Citrate cycle (TCA cycle) | TCA: Normal energy metabolism of the brain | Abnormalities in energy metabolism were found to play a role in SZ pathophysiology | [56] |
| Ko00600: Sphingolipid (SL) metabolism | Formation of membrane “lipid rafts” of myelin sheaths, especially in neurons and oligodendrocytes (crucial for normal synaptic neurotransmission, axon-myelin stability, and communication/connectivity) | Inflammatory, synaptic, and white matter changes that result in disconnectivity in SZ may be related to SL | [57,58] |
| Pathways found to be more active in patients with significantly more severe symptoms (according to PANNS and CGI-S) | |||
| Ko00030: Pentose phosphate pathway | Formation of NADPH for biosynthetic processes, cellular redox balance, and synthesis of ribose | Pentose phosphate pathway-related molecules in schizophrenia were found to be increased | [59] |
| Ko00061: Fatty acid biosynthesis | Component of membranes and myelination process mediator | Lipolysis and β-oxidation were found to be upregulated in SZ, as a result of insufficient brain energy supply | [60,61] |
| Ko00290: Valine, leucine, and isoleucine biosynthesis | Protein synthesis, production of energy, compartmentalization of glutamate synthesis of amine neurotransmitters, including serotonin, dopamine, and norepinephrine | Branched- chain amino acids when administered to patients with tardive dyskinesia—aberration of voluntary motor control in SZ patients treated with psychotropic drugs | [62,63] |
| Ko00072: Synthesis and degradation of ketone bodies | An alternative source of energy under fasting and starving; restrictive diets prolonged intense exercise | Ketones may change the ratio of GABA (glutamate in favor of GABA) to compensate GABA levels in the CNS in SZ patients | [64] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pełka-Wysiecka, J.; Kaczmarczyk, M.; Bąba-Kubiś, A.; Liśkiewicz, P.; Wroński, M.; Skonieczna-Żydecka, K.; Marlicz, W.; Misiak, B.; Starzyńska, T.; Kucharska-Mazur, J.; et al. Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study. J. Clin. Med. 2019, 8, 1605. https://doi.org/10.3390/jcm8101605
Pełka-Wysiecka J, Kaczmarczyk M, Bąba-Kubiś A, Liśkiewicz P, Wroński M, Skonieczna-Żydecka K, Marlicz W, Misiak B, Starzyńska T, Kucharska-Mazur J, et al. Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study. Journal of Clinical Medicine. 2019; 8(10):1605. https://doi.org/10.3390/jcm8101605
Chicago/Turabian StylePełka-Wysiecka, Justyna, Mariusz Kaczmarczyk, Agata Bąba-Kubiś, Paweł Liśkiewicz, Michał Wroński, Karolina Skonieczna-Żydecka, Wojciech Marlicz, Błażej Misiak, Teresa Starzyńska, Jolanta Kucharska-Mazur, and et al. 2019. "Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study" Journal of Clinical Medicine 8, no. 10: 1605. https://doi.org/10.3390/jcm8101605
APA StylePełka-Wysiecka, J., Kaczmarczyk, M., Bąba-Kubiś, A., Liśkiewicz, P., Wroński, M., Skonieczna-Żydecka, K., Marlicz, W., Misiak, B., Starzyńska, T., Kucharska-Mazur, J., Łoniewski, I., & Samochowiec, J. (2019). Analysis of Gut Microbiota and Their Metabolic Potential in Patients with Schizophrenia Treated with Olanzapine: Results from a Six-Week Observational Prospective Cohort Study. Journal of Clinical Medicine, 8(10), 1605. https://doi.org/10.3390/jcm8101605








