Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Definition of Bacteraemia
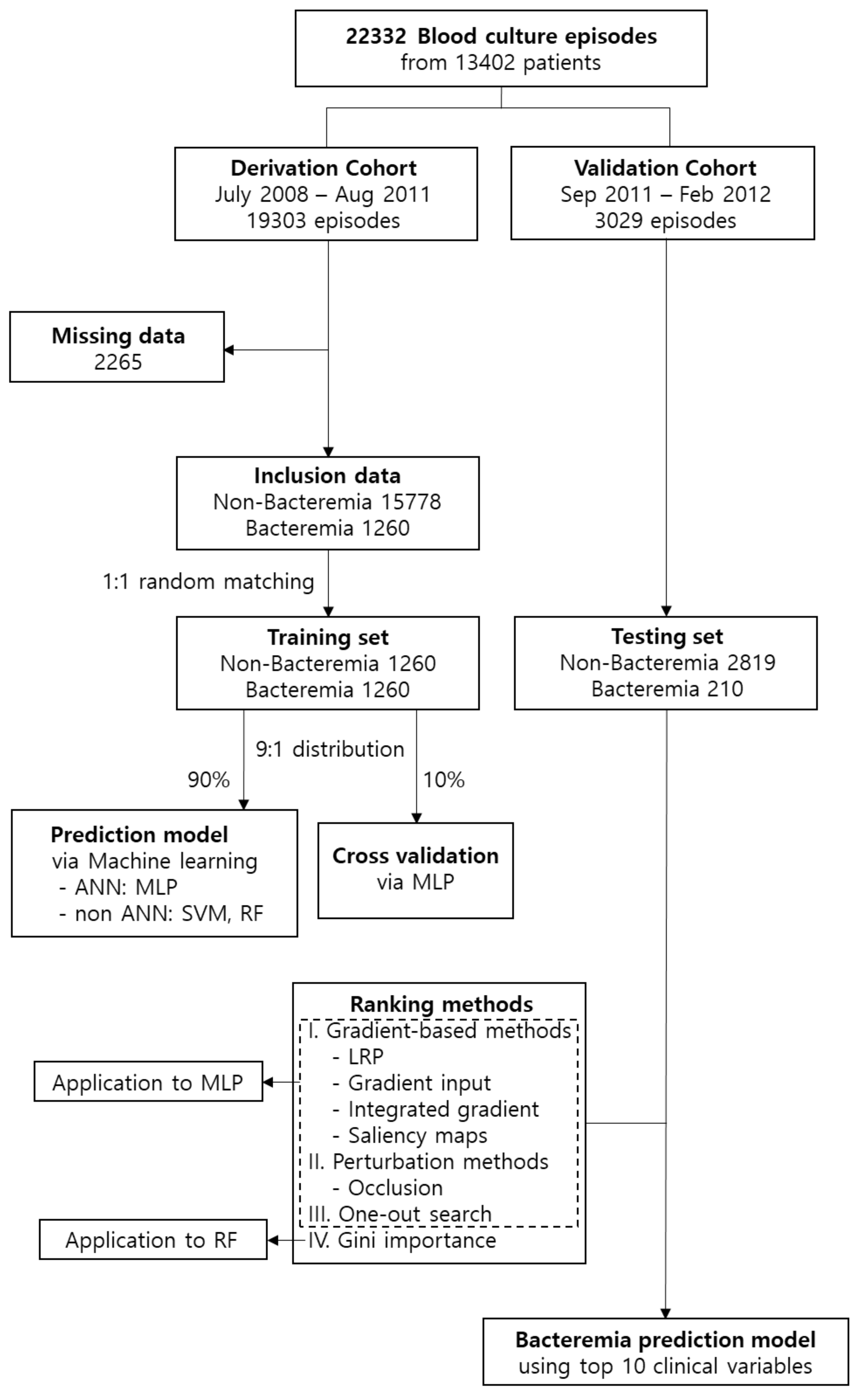
2.2. Study Population
2.3. Clinical Variables for Training
2.4. Statistical Analysis
2.5. Ethics Approval and Consent to Participate
3. Results
3.1. Bacteraemia and Non-Bacteraemia Groups
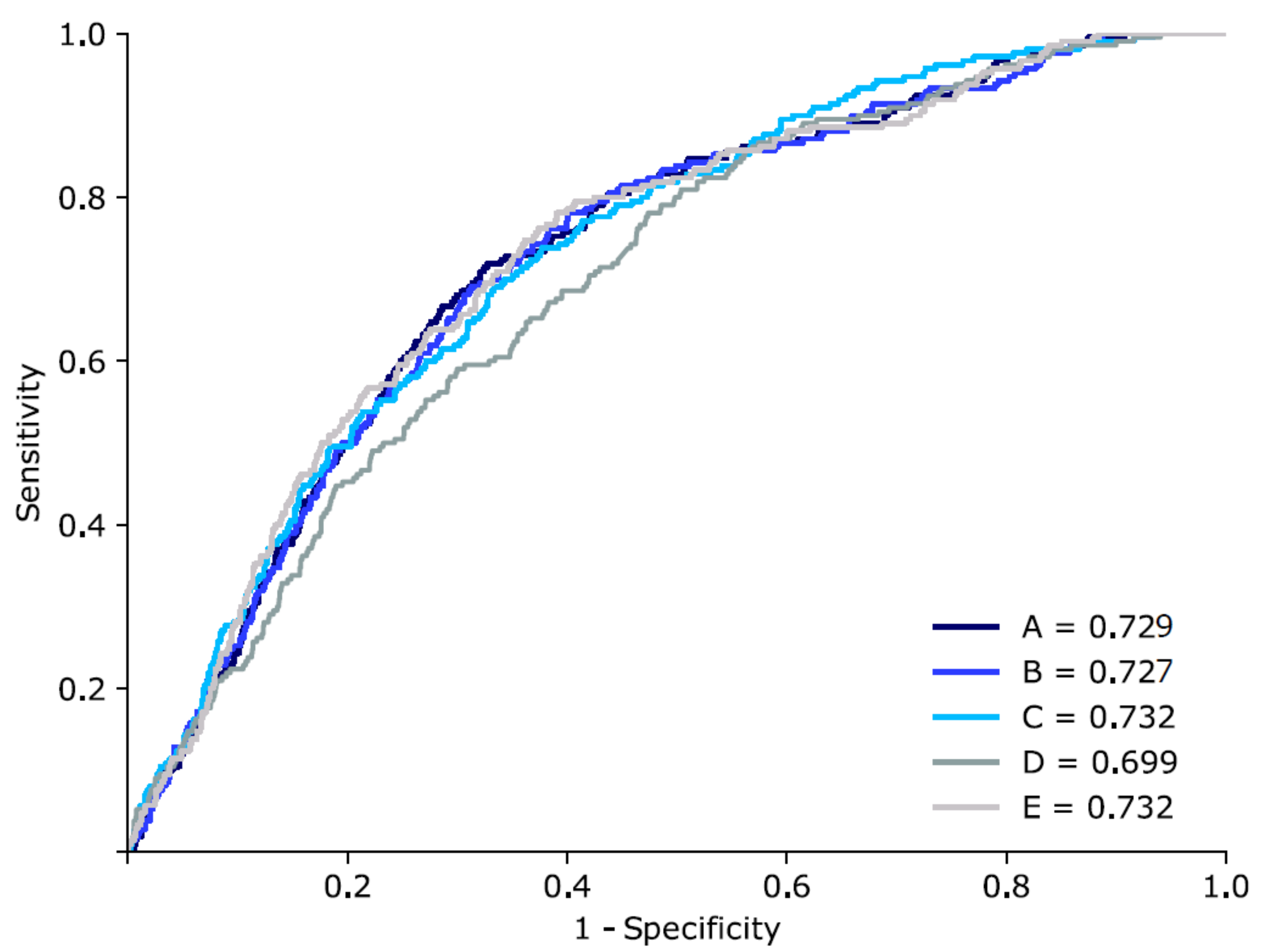
3.2. Bacteraemia Prediction with Various Machine Learning Techniques
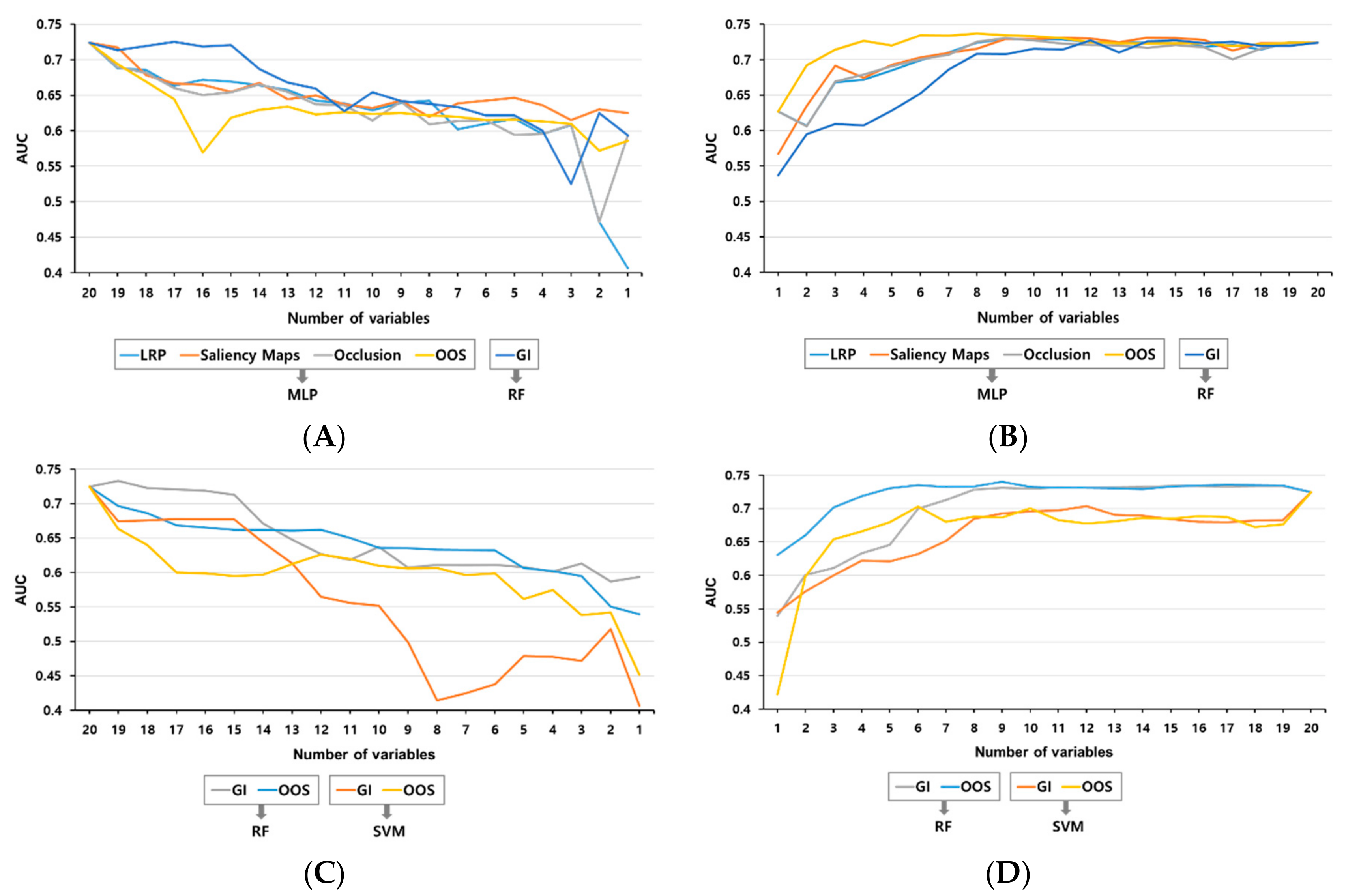
3.3. Methods for the Ranking of the Variables
3.4. Validation by Addition or Subtraction of the Ranking Order
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sackett, D.L.; Rosenberg, W.M.; Gray, J.A.; Haynes, R.B.; Richardson, W.S. Evidence based medicine: What it is and what it isn’t. BMJ Clin. Res. Ed. 1996, 312, 71–72. [Google Scholar] [CrossRef] [PubMed]
- McCue, M.E.; McCoy, A.M. The scope of big data in one medicine: Unprecedented opportunities and challenges. Front. Vet. Sci. 2017, 4, 194. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pinto, L.N.; Luo, Y.; Churpek, M.M. Big data and data science in critical care. Chest 2018, 154, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.X.; Vu, G.T.; Ha, G.H.; Vuong, Q.H.; Ho, M.T.; Vuong, T.T.; La, V.P.; Ho, M.T.; Nghiem, K.P.; Nguyen, H.L.T.; et al. Global evolution of research in artificial intelligence in health and medicine: A bibliometric study. J. Clin. Med. 2019, 8, 360. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future-big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Khazaei, H.; Mench-Bressan, N.; McGregor, C.; Pugh, J.E. Health informatics for neonatal intensive care units: An analytical modeling perspective. IEEE J. Transl. Eng. Health Med. 2015, 3, 1–9. [Google Scholar] [CrossRef]
- Taylor, R.A.; Moore, C.L.; Cheung, K.H.; Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PLoS ONE 2018, 13, e0194085. [Google Scholar] [CrossRef]
- Beeler, C.; Dbeibo, L.; Kelley, K.; Thatcher, L.; Webb, D.; Bah, A.; Monahan, P.; Fowler, N.R.; Nicol, S.; Judy-Malcolm, A.; et al. Assessing patient risk of central line-associated bacteremia via machine learning. Am. J. Infect. Control 2018, 46, 986–991. [Google Scholar] [CrossRef]
- Mani, S.; Ozdas, A.; Aliferis, C.; Varol, H.A.; Chen, Q.; Carnevale, R.; Chen, Y.; Romano-Keeler, J.; Nian, H.; Weitkamp, J.H. Medical decision support using machine learning for early detection of late-onset neonatal sepsis. J. Am. Med Inform. Assoc. JAMIA 2014, 21, 326–336. [Google Scholar] [CrossRef]
- Hsieh, M.H.; Hsieh, M.J.; Chen, C.M.; Hsieh, C.C.; Chao, C.M.; Lai, C.C. An artificial neural network model for predicting successful extubation in intensive care units. J. Clin. Med. 2018, 7, 240. [Google Scholar] [CrossRef] [PubMed]
- Blount, M.; Ebling, M.R.; Eklund, J.M.; James, A.G.; McGregor, C.; Percival, N.; Smith, K.P.; Sow, D. Real-time analysis for intensive care: Development and deployment of the artemis analytic system. IEEE Eng. Med. Biol. Mag. Q. Mag. Eng. Med. Biol. Soc. 2010, 29, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, T.B.; Detsky, A.S. The inevitable application of big data to health care. JAMA 2013, 309, 1351–1352. [Google Scholar] [CrossRef] [PubMed]
- Shankar-Hari, M.; Phillips, G.S.; Levy, M.L.; Seymour, C.W.; Liu, V.X.; Deutschman, C.S.; Angus, D.C.; Rubenfeld, G.D.; Singer, M. Developing a new definition and assessing new clinical criteria for septic shock: For the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016, 315, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Wacker, C.; Prkno, A.; Brunkhorst, F.M.; Schlattmann, P. Procalcitonin as a diagnostic marker for sepsis: A systematic review and meta-analysis. Lancet Infect. Dis. 2013, 13, 426–435. [Google Scholar] [CrossRef]
- Wu, J.; Hu, L.; Zhang, G.; Wu, F.; He, T. Accuracy of presepsin in sepsis diagnosis: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0133057. [Google Scholar] [CrossRef] [PubMed]
- Larsen, F.F.; Petersen, J.A. Novel biomarkers for sepsis: A narrative review. Eur. J. Intern. Med. 2017, 45, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.J.; Kim, M.; Yoon, J.H.; Song, Y.G. A new statistical approach to predict bacteremia using electronic medical records. Scand. J. Infect. Dis. 2013, 45, 672–680. [Google Scholar] [CrossRef]
- Mozes, B.; Milatiner, D.; Block, C.; Blumstein, Z.; Halkin, H. Inconsistency of a model aimed at predicting bacteremia in hospitalized patients. J. Clin. Epidemiol. 1993, 46, 1035–1040. [Google Scholar] [CrossRef]
- Bates, D.W.; Sands, K.; Miller, E.; Lanken, P.N.; Hibberd, P.L.; Graman, P.S.; Schwartz, J.S.; Kahn, K.; Snydman, D.R.; Parsonnet, J.; et al. Predicting bacteremia in patients with sepsis syndrome. Academic medical center consortium sepsis project working group. J. Infect. Dis. 1997, 176, 1538–1551. [Google Scholar] [CrossRef]
- Horan, T.C.; Andrus, M.; Dudeck, M.A. Cdc/nhsn surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 2008, 36, 309–332. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2008, 21, 1263–1284. [Google Scholar]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. IJCAI 1995, 14, 1137–1145. [Google Scholar]
- Arlot, S.; Celisse, A. A survey of cross-validation procedures for model selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Jaimes, F.; Arango, C.; Ruiz, G.; Cuervo, J.; Botero, J.; Velez, G.; Upegui, N.; Machado, F. Predicting bacteremia at the bedside. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2004, 38, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Falguera, M.; Trujillano, J.; Caro, S.; Menendez, R.; Carratala, J.; Ruiz-Gonzalez, A.; Vila, M.; Garcia, M.; Porcel, J.M.; Torres, A. A prediction rule for estimating the risk of bacteremia in patients with community-acquired pneumonia. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2009, 49, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Krogh, A.; Hertz, J.A. A simple weight decay can improve generalization. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 1992; pp. 950–957. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. Coursera Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Smale, S.; Zhou, D.-X. Learning theory estimates via integral operators and their approximations. Constr. Approx. 2007, 26, 153–172. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In European Conference on Computer Vision; Springer: Berlin, Germany, 2014; pp. 818–833. [Google Scholar]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.-R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef] [PubMed]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. arXiv 2017, arXiv:1704.02685. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. arXiv 2017, arXiv:1703.01365. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Menze, B.H.; Kelm, B.M.; Masuch, R.; Himmelreich, U.; Bachert, P.; Petrich, W.; Hamprecht, F.A. A comparison of random forest and its gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009, 10, 213. [Google Scholar] [CrossRef]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. Apache ii: A severity of disease classification system. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonca, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The sofa (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the european society of intensive care medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Esbenshade, A.J.; Zhao, Z.; Aftandilian, C.; Saab, R.; Wattier, R.L.; Beauchemin, M.; Miller, T.P.; Wilkes, J.J.; Kelly, M.J.; Fernbach, A.; et al. Multisite external validation of a risk prediction model for the diagnosis of blood stream infections in febrile pediatric oncology patients without severe neutropenia. Cancer 2017, 123, 3781–3790. [Google Scholar] [CrossRef]
- Webb, B.J.; Healy, R.; Majers, J.; Burr, Z.; Gazdik, M.; Lopansri, B.; Hoda, D.; Petersen, F.B.; Ford, C. Prediction of bloodstream infection due to vancomycin-resistant enterococcus in patients undergoing leukemia induction or hematopoietic stem-cell transplantation. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017, 64, 1753–1759. [Google Scholar] [CrossRef]
- Sullivan, T.; Ichikawa, O.; Dudley, J.; Li, L.; Aberg, J. The rapid prediction of carbapenem resistance in patients with klebsiella pneumoniae bacteremia using electronic medical record data. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2018; Volume 5. [Google Scholar]
- Verghese, A.; Shah, N.H.; Harrington, R.A. What this computer needs is a physician: Humanism and artificial intelligence. JAMA 2018, 319, 19–20. [Google Scholar] [CrossRef]
- Tung, C.B.; Tung, C.F.; Yang, D.Y.; Hu, W.H.; Hung, D.Z.; Peng, Y.C.; Chang, C.S. Extremely high levels of alkaline phosphatase in adult patients as a manifestation of bacteremia. Hepato-Gastroenterology 2005, 52, 1347–1350. [Google Scholar] [PubMed]
- Sasaki, S.; Hasegawa, T.; Kawarazaki, H.; Nomura, A.; Uchida, D.; Imaizumi, T.; Furusho, M.; Nishiwaki, H.; Fukuma, S.; Shibagaki, Y.; et al. Development and validation of a clinical prediction rule for bacteremia among maintenance hemodialysis patients in outpatient settings. PLoS ONE 2017, 12, e0169975. [Google Scholar]
- Kanai, S.; Honda, T.; Uehara, T.; Matsumoto, T. Liver function tests in patients with bacteremia. J. Clin. Lab. Anal. 2008, 22, 66–69. [Google Scholar] [CrossRef] [PubMed]
| Bacteremia, Yes (n = 1260) | Bacteremia, No (n = 1260) | p-Value | |
|---|---|---|---|
| Age, years | 64.4 ± 15.3 | 60.1 ± 17.3 | <0.001 |
| Sex, male | 558 (44.3%) | 533 (42.3%) | 0.306 |
| Vital sign | |||
| SBP, mmHg | 91.6 ± 23.2 | 99.4 ± 22.8 | <0.001 |
| Body temperature (min), °C | 36.1 ± 0.4 | 36.1 ± 0.3 | 0.578 |
| Body temperature (max), °C | 38.1 ± 0.9 | 37.6 ± 0.9 | <0.001 |
| Heart rate, beats/min | 115.7 ± 29.2 | 105.7 ± 28.5 | <0.001 |
| Respiratory rate, /min | 25.5 ± 8.6 | 23.6 ± 8.9 | <0.001 |
| Laboratory data | |||
| Creatinine, mg/dL | 1.6 ± 1.5 | 1.2 ± 1.3 | 0.317 |
| Albumin, g/dL | 2.8 ± 0.6 | 3.2 ± 0.7 | <0.001 |
| CRP, mg/L | 1.6 ± 1.5 | 1.2 ± 1.3 | <0.001 |
| Alkaline phosphatase, IU/L | 191.3 ± 213.7 | 136.0 ± 141.0 | <0.001 |
| WBC count (min), /μL | 10.3 ± 6.2 | 9.3 ± 5.5 | <0.001 |
| WBC count (max), /μL | 14.8 ± 8.9 | 11.9 ± 7.3 | <0.001 |
| Platelet count, /μL | 185.9 ± 134.8 | 218.7 ± 129.8 | <0.001 |
| Prothrombin time, s | 18.5 ± 8.9 | 15.9 ± 6.6 | <0.001 |
| Clinical information | |||
| Hospital day to blood culture, days | 24.6 ± 75.3 | 10.3 ± 17.6 | <0.001 |
| ICU care, yes | 410 (32.6%) | 235 (18.7%) | <0.001 |
| Central venous catheter, yes | 114 (9.1%) | 0 (0%) | <0.001 |
| Steroid therapy, yes | 477 (37.9) | 429 (34.0) | 0.045 |
| Antibiotic therapy, yes | 591 (46.9%) | 526 (41.7%) | 0.009 |
| Model | Algorithms | AUC (95% CI) | Sensitivity | Specificity |
|---|---|---|---|---|
| Previous | Naive Bayesian | 0.7 | - | - |
| A | MLP (128) | 0.729 (0.712–0.728) | 0.810 | 0.589 |
| B | MLP (256) | 0.727 (0.713–0.727) | 0.810 | 0.532 |
| C | RF | 0.732 (0.722–0.733) | 0.682 | 0.655 |
| D | SVM | 0.699 (0.687–0.700) | 0.692 | 0.553 |
| E | MLP (128) + MLP (256) + RF | 0.732 (0.728–0.735) | 0.793 | 0.571 |
| Rank | One-Out Search | Gini Importance |
|---|---|---|
| 1 | ALP | Age |
| 2 | Platelet | Prothrombin time |
| 3 | Body temperature (max) | Heart rate |
| 4 | Systolic BP | Hospital day to blood culture |
| 5 | WBC count (min) | Albumin |
| 6 | ICU stay | ALP |
| 7 | CRP | Platelet |
| 8 | Central venous catheter | Body temperature (max) |
| 9 | Prothrombin time | WBC count (max) |
| 10 | Albumin | Creatinine |
| 11 | Steroid use | systolic BP |
| 12 | Sex | WBC count (min) |
| 13 | Antibiotic | Respiratory rate |
| 14 | Body temperature (min) | ICU stay |
| 15 | Hospital day to blood culture | Steroid |
| 16 | WBC count (max) | Sex |
| 17 | Heart rate | Body temperature (min) |
| 18 | Respiratory rate | Antibiotics |
| 19 | Age | CRP |
| 20 | Creatinine | Central venous catheter |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.H.; Dong, J.J.; Jeong, S.J.; Chae, M.-H.; Lee, B.S.; Kim, H.J.; Ko, S.H.; Song, Y.G. Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach. J. Clin. Med. 2019, 8, 1592. https://doi.org/10.3390/jcm8101592
Lee KH, Dong JJ, Jeong SJ, Chae M-H, Lee BS, Kim HJ, Ko SH, Song YG. Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach. Journal of Clinical Medicine. 2019; 8(10):1592. https://doi.org/10.3390/jcm8101592
Chicago/Turabian StyleLee, Kyoung Hwa, Jae June Dong, Su Jin Jeong, Myeong-Hun Chae, Byeong Soo Lee, Hong Jae Kim, Sung Hun Ko, and Young Goo Song. 2019. "Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach" Journal of Clinical Medicine 8, no. 10: 1592. https://doi.org/10.3390/jcm8101592
APA StyleLee, K. H., Dong, J. J., Jeong, S. J., Chae, M.-H., Lee, B. S., Kim, H. J., Ko, S. H., & Song, Y. G. (2019). Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach. Journal of Clinical Medicine, 8(10), 1592. https://doi.org/10.3390/jcm8101592





