MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review
Abstract
1. Introduction
1.1. General Insight into MicroRNAs
1.2. MiRNAs Linked to Brain Function
1.3. MiRNAs and Skin Disorders
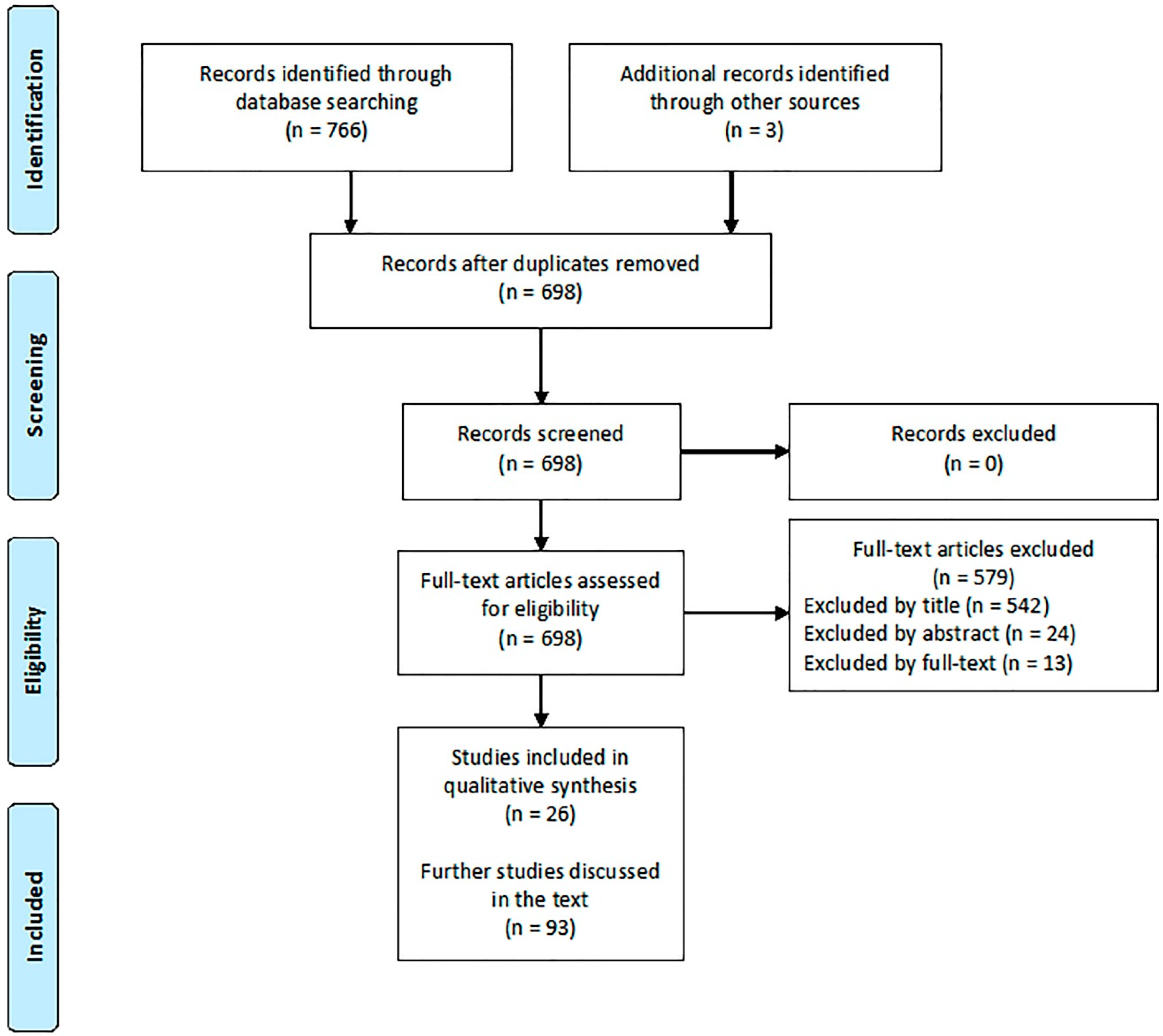
2. Materials and Methods
2.1. Studies about ASD
2.2. Studies about AD
3. Results
3.1 Studies about ASD
3.2 Studies about AD
4. Discussion
4.1. The Overlap between Atopy and Autism
4.2. Role for Overlapping MiRNAs in ASD and AD
5. Conclusions
Author Contributions
Funding
Acknowledgment
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Barger, B.D.; Campbell, J.M.; McDonough, J.D. Prevalence and onset of regression within autism spectrum disorders: A meta-analytic review. J. Autism Dev. Disord. 2015, 43, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Fombonne, E. Pervasive developmental disorders in preschool children: Confirmation of high prevalence. Am. J. Psychiatry 2005, 162, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Fombonne, E. Editorial: The rising prevalence of autism. J. Child Psychol. Psychiatry 2018, 59, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Gaugler, T.; Klei, L.; Sanders, S.J.; Bodea, C.A.; Goldberg, A.P.; Lee, A.B.; Mahajan, M.; Manaa, D.; Pawitan, Y.; Reichert, J.; et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014, 46, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.R.; State, M.W. Recent advances in the genetics of autism. Biol. Psychiatry 2007, 61, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Beaudet, A.L. Autism: Highly heritable but not inherited. Nat. Med. 2007, 13, 534–536. [Google Scholar] [CrossRef]
- Colvert, E.; Tick, B.; McEwen, F.; Stewart, C.; Curran, S.R.; Woodhouse, E.; Gillan, N.; Hallett, V.; Lietz, S.; Garnett, T.; et al. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry 2015, 72, 415–423. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Daly, M.J.; O’Donovan, M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012, 13, 537–551. [Google Scholar] [CrossRef]
- Ozonoff, S.; Young, G.S.; Carter, A.; Messinger, D.; Yirmiya, N.; Zwaigenbaum, L.; Bryson, S.; Carver, L.J.; Constantino, J.N.; Dobkins, D.; et al. Recurrence risk for autism spectrum disorders: A Baby Siblings Research Consortium study. Pediatrics 2011, 128, e488–e495. [Google Scholar] [CrossRef]
- Sanders, S.J.; He, X.; Willsey, A.J.; Ercan-Sencicek, A.G.; Samocha, K.E.; Cicek, A.E.; Murtha, M.T.; Bal, V.H.; Bishop, S.L.; Dong, S.; et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 2015, 87, 1215–1233. [Google Scholar] [CrossRef]
- Vorstman, J.A.S.; Parr, J.R.; Moreno-De-Luca, D.; Anney, R.J.L.; Nurnberger, J.I., Jr.; Hallmayer, J.F. Autism genetics: Opportunities and challenges for clinical translation. Nat. Rev. Genet. 2017, 18, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Al-Mubarak, B.; Abouelhoda, M.; Omar, A.; AlDhalaan, H.; Aldosari, M.; Nester, M.; Alshamrani, H.A.; El-Kalioby, M.; Goljan, E.; Albar, R.; et al. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: A trio study from Saudi families. Sci Rep. 2017, 7, 5679. [Google Scholar] [CrossRef] [PubMed]
- Bourgeron, T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015, 16, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Thiruvahindrapuram, B.; Merico, D.; Walker, S.; Tammimies, K.; Hoang, N.; Chrysler, C.; Nalpathamkalam, T.; Pellecchia, G.; Liu, Y.; et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015, 21, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Toma, C.; Torrico, B.; Hervás, A.; Valdés-Mas, R.; Tristán-Noguero, A.; Padillo, V.; Maristany, M.; Salgado, M.; Arenas, C.; Puente, X.S.; et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol. Psychiatry 2014, 19, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Hertz-Picciotto, I.; Schmidt, R.J.; Krakowiak, P. Understanding environmental contributions to autism: Causal concepts and the state of science. Autism Res. 2018, 11, 554–586. [Google Scholar] [CrossRef] [PubMed]
- Meek, S.E.; Lemery-Chalfant, K.; Jahromi, L.B.; Valiente, C. A review of gene-environment correlations and their implications for autism: A conceptual model. Psychol. Rev. 2013, 120, 497–521. [Google Scholar] [CrossRef]
- Gorini, F.; Muratori, F.; Morales, M.A. The Role of Heavy Metal Pollution in Neurobehavioral Disorders: A Focus on Autism. Rev. J. Autism Dev. Disord. 2014, 1, 354–372. [Google Scholar] [CrossRef]
- Kalkbrenner, A.E.; Schmidt, R.J.; Penlesky, A.C. Environmental chemical exposures and autism spectrum disorders: A review of the epidemiological evidence. Curr. Probl. Pediatr. Adolesc. Health Care 2014, 44, 277–318. [Google Scholar] [CrossRef]
- Tonacci, A.; Billeci, L.; Ruta, L.; Tartarisco, G.; Pioggia, G.; Gangemi, S. A systematic review of the association between allergic asthma and autism. Minerva Pediatr. 2017, 69, 538–550. [Google Scholar]
- Billeci, L.; Tonacci, A.; Tartarisco, G.; Ruta, L.; Pioggia, G.; Gangemi, S. Association Between Atopic Dermatitis and Autism Spectrum Disorders: A Systematic Review. Am. J. Clin. Dermatol. 2015, 16, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Billeci, L.; Tonacci, A.; Tartarisco, G.; Ruta, L.; Pioggia, G.; Gangemi, S. Reply to Fluegge: Association Between Atopic Dermatitis and Autism Spectrum Disorders: A Systematic Review. Am. J. Clin. Dermatol. 2016, 17, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRnas Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef]
- Bjorklund, G.; Saad, K.; Chirumbolo, S.; Kern, J.K.; Geier, D.A.; Geier, M.R.; Urbina, M.A. Immune dysfunction and neuroinflammation in autism spectrum disorder. Acta Neurobiol. Exp. 2016, 76, 257–268. [Google Scholar] [CrossRef]
- De Jong, M.; Punt, M.; De Groot, E.; Minderaa, R.B.; Hadders-Algra, M. Minor neurological dysfunction in children with autism spectrum disorder. Dev. Med. Child Neurol. 2011, 53, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Du, L.; Shan, L.; Jia, F.Y. Research advances in immunological dysfunction in children with autism spectrum disorders. Zhongguo Dang Dai Er Ke Za Zhi 2014, 16, 1289–1293. [Google Scholar]
- Jašarević, E.; Howerton, C.L.; Howard, C.D.; Bale, T.L. Alterations in the vaginal microbiome by maternal stress are associated with metabolic reprogramming of the offspring gut and brain. Endocrinology 2015, 156, 3265–3276. [Google Scholar] [CrossRef]
- Young, A.M.; Chakrabarti, B.; Roberts, D.; Lai, M.C.; Suckling, J.; Baron-Cohen, S. From molecules to neural morphology: Understanding neuroinflammation in autism spectrum condition. Mol. Autism 2016, 7, 9. [Google Scholar] [CrossRef]
- Sakamoto, A.; Moriuchi, H.; Matsuzaki, J.; Motoyama, K.; Moriuchi, M. Retrospective diagnosis of congenital cytomegalovirus infection in children with autism spectrum disorder but no other major neurologic deficit. Brain Dev. 2015, 37, 200–205. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, J.M. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience 2016, 324, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Kern, J.K.; Geier, D.A.; Sykes, L.K.; Homme, K.G.; Geier, M.R. Medical conditions in autism and events associated with initial onset of autism. OA Autism 2014, 2, 9. [Google Scholar]
- Ruggeri, B.; Sarkans, U.; Schumann, G.; Persico, A.M. Biomarkers in autism spectrum disorder: The old and the new. Psychopharmacology 2014, 231, 1201–1216. [Google Scholar] [CrossRef]
- Chan, J.K.; Roth, J.; Oppenheim, J.J.; Tracey, K.J.; Vogl, T.; Feldmann, M.; Horwood, N.; Nanchahal, J. Alarmins: Awaiting a clinical response. J. Clin. Investig. 2012, 122, 2711–2719. [Google Scholar] [CrossRef]
- Di Salvo, E.; Casciaro, M.; Quartuccio, S.; Genovese, L.; Gangemi, S. Do Alarmins Have a Potential Role in Autism Spectrum Disorders Pathogenesis and Progression? Biomolecules 2019, 9, 2. [Google Scholar] [CrossRef]
- Simpson, E.L. Atopic dermatitis: A review of topical treatment options. Curr. Med. Res. Opin. 2010, 26, 633–640. [Google Scholar] [CrossRef]
- Tonelli, D.D.P.; Pulvers, J.N.; Haffner, C.; Murchison, E.P.; Hannon, G.J.; Huttner, W.B. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 2008, 135, 3911–3921. [Google Scholar] [CrossRef]
- Dai, R.; Ahmed, S.A. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 2011, 157, 163–179. [Google Scholar] [CrossRef]
- Mannucci, C.; Casciaro, M.; Minciullo, P.L.; Calapai, G.; Navarra, M.; Gangemi, S. Involvement of microRNAs in skin disorders: A literature review. Allergy Asthma Proc. 2017, 38, 9–15. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Radojicic, J.; Zaravinos, A.; Vrekoussis, T.; Kafousi, M.; Spandidos, D.A.; Stathopoulos, E.N. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011, 10, 507–517. [Google Scholar] [CrossRef]
- Zaravinos, A.; Radojicic, J.; Lambrou, G.I.; Volanis, D.; Delakas, D.; Stathopoulos, E.N.; Spandidos, D.A. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J. Urol. 2012, 188, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Sabina, S.; Vecoli, C.; Borghini, A.; Guarino, R.; Andreassi, M.G. Analysis of miRNAs Targeting 3’UTR of H2AFX Gene: A General in Silico Approach. Microrna 2015, 4, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Furdui, C. Ionizing radiation: Mechanisms and therapeutics. Antioxid. Redox Signal. 2014, 21, 218–220. [Google Scholar] [CrossRef]
- Li, X.Y.; Luo, Q.F.; Wei, C.K.; Li, D.F.; Li, J.; Fang, L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int. J. Clin. Exp. Med. 2014, 7, 32–40. [Google Scholar]
- Wojtas, B.; Ferraz, C.; Stokowy, T.; Hauptmann, S.; Lange, D.; Dralle, H.; Musholt, T.; Jarzab, B.; Paschke, R.; Eszlinger, M. Differential miRNA expression defines migration and reduced apoptosis in follicular thyroid carcinomas. Mol. Cell. Endocrinol. 2014, 388, 1–9. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Adlakha, Y.K.; Saini, N. Brain miRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol. Cancer 2014, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.K.; Xu, X.M. MicroRNA in central nervous system trauma and degenerative disorders. Physiol. Genom. 2011, 43, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Ziats, M.N.; Rennert, O.M. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 2014, 19, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Hutsler, J.J.; Zhang, H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2000, 1309, 83–94. [Google Scholar] [CrossRef]
- Glantz, L.A.; Lewis, D.A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry 2000, 57, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kolomeets, N.S.; Orlovskaya, D.D.; Rachmanova, V.I.; Uranova, N.A. Ultrastructural alterations in hippocampal mossy fiber synapses in Schizophrenia: A postmortem morphometric study. Synapse 2005, 57, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Sweet, R.A.; Henteleff, R.A.; Zhang, W.; Sampson, A.R.; Lewis, D.A. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology 2009, 34, 374–389. [Google Scholar] [CrossRef]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.-F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by fmrp-associated micrornas mir-125b and mir-132. Neuron 2010, 68, 161. [Google Scholar] [CrossRef]
- Impey, S.; Davare, M.; Lasiek, A.; Fortin, D.; Ando, H.; Varlamova, O.; Obrietan, K.; Soderling, T.R.; Goodman, R.H.; Wayman, G.A. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 2010, 43, 146–156. [Google Scholar] [CrossRef]
- Smrt, R.D.; Szulwach, K.E.; Pfeiffer, R.L.; Li, X.; Guo, W.; Pathania, M.; Teng, Z.-Q.; Luo, Y.; Peng, J.; Bordey, A.; et al. Microrna mir-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010, 28, 1060–1070. [Google Scholar] [CrossRef]
- Siegel, G.; Obernosterer, G.; Fiore, R.; Oehmen, M.; Bicker, S.; Christensen, M.; Khudayberdiev, S.; Leuschner, P.F.; Busch, C.J.L.; Kane, C.; et al. A functional screen implicates microrna-138-dependent regulation of the depalmitoylation enzyme apt1 in dendritic spine morphogenesis. Nat. Cell Biol. 2009, 11, 705–716. [Google Scholar] [CrossRef]
- Davis, T.H.; Cuellar, T.L.; Koch, S.M.; Barker, A.J.; Harfe, B.D.; McManus, M.T.; Ullian, E.M. Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008, 28, 4322–4330. [Google Scholar] [CrossRef]
- Fiorucci, G.; Chiantore, M.V.; Mangino, G.; Percario, Z.A.; Affabris, E.; Romeo, G. Cancer regulator microRNA: Potential relevance in diagnosis, prognosis and treatment of cancer. Curr. Med. Chem. 2012, 19, 461–474. [Google Scholar] [CrossRef]
- Sonkoly, E.; Janson, P.; Majuri, M.-L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. Mir-155 is overexpressed in patients with atopic dermatitis and modulates t-cell proliferative responses by targeting cytotoxic t lymphocyte–associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589.e20. [Google Scholar] [CrossRef]
- Wei, T.; Orfanidis, K.; Xu, N.; Janson, P.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. The expression of microrna-203 during human skin morphogenesis. Exp. Dermatol. 2010, 19, 854–856. [Google Scholar] [CrossRef]
- Hawkes, J.E.; Nguyen, G.H.; Fujita, M.; Florell, S.R.; Callis Duffin, K.; Krueger, G.G.; O’Connell, R.M. microRNAs in psoriasis. J. Investig. Dermatol. 2016, 136, 365–371. [Google Scholar] [CrossRef]
- Nutten, S. Atopic dermatitis: Global epidemiology and risk factors. Ann. Nutr. Metab. 2015, 66, 8–16. [Google Scholar] [CrossRef]
- Abu-Elneel, K.; Liu, T.; Gazzaniga, F.S.; Nishimura, Y.; Wall, D.P.; Geschwind, D.H.; Lao, K.; Kosik, K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008, 9, 153–161. [Google Scholar] [CrossRef]
- Sarachana, T.; Zhou, R.; Chen, G.; Manji, H.K.; Hu, V.W. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010, 2, 23. [Google Scholar] [CrossRef]
- Talebizadeh, Z.; Butler, M.G.; Theodoro, M.F. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res. 2008, 1, 240–250. [Google Scholar] [CrossRef]
- Mundalil Vasu, M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef]
- Popov, N.T.; Madjirova, N.P.; Minkov, I.N.; Vachev, T.I. Micro RNA HSA-486-3P Gene Expression Profiling in the Whole Blood of Patients with Autism. Biotechnol. Biotechnol. Equip. 2012, 26, 3385–3388. [Google Scholar] [CrossRef]
- Weber, F.; Teresi, R.E.; Broelsch, C.E.; Frilling, A.; Eng, C. A limited set of human microRNA is deregulated in follicular thyroid carcinoma. J. Clin. Endocrinol. Metab. 2006, 91, 3584–3591. [Google Scholar] [CrossRef]
- Mor, M.; Nardone, S.; Sams, D.S.; Elliott, E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 2015, 6, 46. [Google Scholar] [CrossRef]
- Ander, B.P.; Barger, N.; Stamova, B.; Sharp, F.R.; Schumann, C.M. Atypical miRNA expression in temporal cortex associated with dysregulation of immune, cell cycle, and other pathways in autism spectrum disorders. Mol. Autism 2015, 6, 37. [Google Scholar] [CrossRef]
- Wu, Y.E.; Parikshak, N.N.; Belgard, T.G.; Geschwind, D.H. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat. Neurosci. 2016, 19, 1463–1476. [Google Scholar] [CrossRef]
- Huang, F.; Long, Z.; Chen, Z.; Li, J.; Hu, Z.; Qiu, R.; Zhuang, W.; Tang, B.; Xia, K.; Jiang, H. Investigation of gene regulatory networks associated with autism spectrum disorder based on mirna expression in china. PLoS ONE 2015, 10, e0129052. [Google Scholar] [CrossRef]
- Toma, C.; Torrico, B.; Hervás, A.; Salgado, M.; Rueda, I.; Valdés-Mas, R.; Buitelaar, J.K.; Rommelse, N.; Franke, B.; Freitag, C.; et al. Common and rare variants of microRNA genes in autism spectrum disorders. World J. Biol. Psychiatry 2015, 16, 376–386. [Google Scholar] [CrossRef]
- Hicks, S.D.; Ignacio, C.; Gentile, K.; Middleton, F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016, 16, 52. [Google Scholar] [CrossRef]
- Nguyen, L.S.; Lepleux, M.; Makhlouf, M.; Martin, C.; Fregeac, J.; Siquier-Pernet, K.; Philippe, A.; Feron, F.; Gepner, B.; Rougeulle, C.; et al. Profiling olfactory stem cells from living patients identifies mirnas relevant for autism pathophysiology. Mol. Autism 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kichukova, T.M.; Popov, N.T.; Ivanov, I.S.; Vachev, T.I. Profiling of Circulating Serum MicroRNAs in Children with Autism Spectrum Disorder using Stem-loop qRT-PCR Assay. Folia Med. 2017, 59, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Jyonouchi, H.; Geng, L.; Streck, D.L.; Dermody, J.J.; Toruner, G.A. MicroRNA expression changes in association with changes in interleukin-1ß/interleukin10 ratios produced by monocytes in autism spectrum disorders: Their association with neuropsychiatric symptoms and comorbid conditions (observational study). J. Neuroinflamm. 2017, 14, 229. [Google Scholar] [CrossRef] [PubMed]
- Pagan, C.; Goubran-Botros, H.; Delorme, R.; Benabou, M.; Lemière, N.; Murray, K.; Amsellem, F.; Callebert, J.; Chaste, P.; Jamain, S.; et al. Disruption of melatonin synthesis is associated with impaired 14-3-3 and miR-451 levels in patients with autism spectrum disorders. Sci. Rep. 2017, 7, 2096. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.S.; Fregeac, J.; Bole-Feysot, C.; Cagnard, N.; Iyer, A.; Anink, J.; Aronica, E.; Alibeu, O.; Nitschke, P.; Colleaux, L.; et al. Role of miR-146a in neural stem cell differentiation and neural lineage determination: Relevance for neurodevelopmental disorders. Mol. Autism 2018, 9, 38. [Google Scholar] [CrossRef]
- Yu, D.; Jiao, X.; Cao, T.; Huang, F. Serum miRNA expression profiling reveals miR-486-3p may play a significant role in the development of autism by targeting ARID1B. Neuroreport 2018, 29, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.M.; An, J.Y.; Edson, J.; Watts, M.; Murigneux, V.; Whitehouse, A.J.O.; Jackson, C.J.; Bellgrove, M.A.; Cristino, A.S.; Claudianos, C.; et al. An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol. Psychiatry 2018. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Qi, R.; Xu, J.; Di, Z.; Zheng, H.; Huo, W.; Zhang, L.; Chen, H.; Gao, X. Profiling of serum and urinary micrornas in children with atopic dermatitis. PLoS ONE 2014, 9, e115448. [Google Scholar] [CrossRef]
- Ralfkiaer, U.; Lindahl, L.M.; Litman, T.; Gjerdrum, L.M.; Ahler, C.B.; Gniadecki, R.; Marstrand, T.; Fredholm, S.; Iversen, L.; Wasik, M.A.; et al. MicroRNA expression in early mycosis fungoides is distinctly different from atopic dermatitis and advanced cutaneous T-cell lymphoma. Anticancer Res. 2014, 34, 7207–7217. [Google Scholar]
- Rebane, A.; Runnel, T.; Aab, A.; Maslovskaja, J.; Rückert, B.; Zimmermann, M.; Plaas, M.; Kärner, J.; Treis, A.; Pihlap, M.; et al. Microrna-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J. Allergy Clin. Immunol. 2014, 134, 836–847.e11. [Google Scholar] [CrossRef]
- Ma, L.; Xue, H.B.; Wang, F.; Shu, C.M.; Zhang, J.H. MicroRNA-155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin. Exp. Immunol. 2015, 181, 142–149. [Google Scholar] [CrossRef]
- Ding, Y.; Shao, X.; Li, X.; Zhai, Y.; Zhang, Y.; Wang, S.; Fang, H. Identification of candidate genes in atopic dermatitis based on bioinformatic methods. Int. J. Dermatol. 2016, 55, 791–800. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, B.; Wang, C.; Wang, H.; Huang, P.; Pan, Y. MicroRNA-124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell. Immunol. 2017, 319, 53–60. [Google Scholar] [CrossRef]
- Jyonouchi, H. Autism spectrum disorders and allergy: Observation from a pediatric allergy/immunology clinic. Exp. Rev. Clin. Immunol. 2010, 6, 397–411. [Google Scholar] [CrossRef]
- Bakkaloglu, B.; Anlar, B.; Anlar, F.Y.; Oktem, F.; Pehlivantürk, B.; Unal, F.; Ozbesler, C.; Gökler, B. Atopic features in early childhood autism. Eur. J. Paediatr. Neurol. 2008, 12, 476–479. [Google Scholar] [CrossRef]
- Chen, M.-H.; Su, T.-P.; Chen, Y.-S.; Hsu, J.-W.; Huang, K.-L.; Chang, W.-H.; Chen, T.-J.; Pan, T.-L.; Bai, Y.-M. Is atopy in early childhood a risk factor for adhd and asd? a longitudinal study. J. Psychosom. Res. 2014, 77, 316–321. [Google Scholar] [CrossRef]
- Zerbo, O.; Leong, A.; Barcellos, L.; Bernal, P.; Fireman, B.; Croen, L.A. Immune mediated conditions in autism spectrum disorders. Brain Behav. Immun. 2015, 46, 232–236. [Google Scholar] [CrossRef]
- Theoharides, T.C. Is a subtype of autism an allergy of the brain? Clin. Ther. 2013, 35, 584–591. [Google Scholar] [CrossRef]
- Shaw, C.A.; Seneff, S.; Kette, S.D.; Tomljenovic, L.; Oller, J.W., Jr.; Davidson, R.M. Aluminum-induced entropy in biological systems: Implications for neurological disease. J. Toxicol. 2014, 2014, 491316. [Google Scholar] [CrossRef]
- Verlaet, A.A.; Noriega, D.B.; Hermans, N.; Savelkoul, H.F. Nutrition, immunological mechanisms and dietary immunomodulation in ADHD. Eur. Child Adolesc. Psychiatry 2014, 23, 519–529. [Google Scholar] [CrossRef]
- Adams, J.B.; Baral, M.; Geis, E.; Mitchell, J.; Ingram, J.; Hensley, A.; Zappia, I.; Newmark, S.; Gehn, E.; Rubin, R.A.; et al. The severity of autism is associated with toxic metal body burden and red blood cell glutathione levels. J. Toxicol. 2009, 2009, 1–7. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Zhao, Y.; Cui, J.G. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008, 283, 31315–31322. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, J.S.; Cho, D.H.; Park, H.J. Molecular Mechanisms of Cutaneous Inflammatory Disorder: Atopic Dermatitis. Int. J. Mol. Sci. 2016, 17, 1234. [Google Scholar] [CrossRef]
- Srivastava, A.; Nikamo, P.; Lohcharoenkal, W.; Li, D.; Meisgen, F.; Xu Landén, N.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 550–561. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Etzrodt, M.; Cortez-Retamozo, V.; Newton, A.; Zhao, J.; Ng, A.; Wildgruber, M.; Romero, P.; Wurdinger, T.; Xavier, R.; Geissmann, F.; et al. Regulation of monocyte functional heterogeneity by mir-146a and relb. Cell Rep. 2012, 1, 317–324. [Google Scholar] [CrossRef]
- Crone, S.; Jacobsen, A.; Federspiel, B.; Bardram, L.; Krogh, A.; Lund, A.H.; Friis-Hansen, L. Microrna-146a inhibits g protein-coupled receptor-mediated activation of nf-κb by targeting card10 and cops8 in gastric cancer. Mol. Cancer 2012, 11, 71. [Google Scholar] [CrossRef]
- Zhao, J.L.; Rao, D.S.; O’Connell, R.M.; Garcia-Flores, Y.; Baltimore, D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. Elife 2013, 2, e00537. [Google Scholar] [CrossRef]
- Kanda, N.; Hau, C.S.; Tada, Y.; Sato, S.; Watanabe, S. Decreased serum LL-37 and vitamin D3 levels in atopic dermatitis: Relationship between IL-31 and oncostatin M. Allergy 2012, 67, 804–812. [Google Scholar] [CrossRef]
- Pioggia, G.; Tonacci, A.; Tartarisco, G.; Billeci, L.; Muratori, F.; Ruta, L.; Gangemi, S. Autism and lack of d3 vitamin: A systematic review. Res. Autism Spectr. Disord. 2014, 8, 1685–1698. [Google Scholar] [CrossRef]
- Banerjee, S.; Meng, J.; Das, S.; Krishnan, A.; Haworth, J.; Charboneau, R.; Zeng, Y.; Ramakrishnan, S.; Roy, S. Morphine induced exacerbation of sepsis is mediated by tempering endotoxin tolerance through modulation of miR-146a. Sci. Rep. 2013, 3, 1977. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.-F.; Thai, T.-H.; Calado, D.P.; Chaudhry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewsky, K.; et al. Foxp3-dependent microrna155 confers competitive fitness to regulatory t cells by targeting socs1 protein. Immunity 2009, 30, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lin, C.; Wu, Z.; Gong, H.; Zeng, Y.; Wu, J.; Li, M.; Li, J. Mir-18a impairs dna damage response through downregulation of ataxia telangiectasia mutated (atm) kinase. PLoS ONE 2011, 6, e25454. [Google Scholar] [CrossRef] [PubMed]
- Sand, M.; Skrygan, M.; Georgas, D.; Sand, D.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. The miRNA machinery in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases and benign melanocytic nevi. Cell Tissue Res. 2012, 350, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Ståhle, M.; Pivarcsi, A. MicroRNAs: Novel regulators in skin inflammation. Clin. Exp. Dermatol. 2008, 33, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Rajewsky, K. MicroRNA control in the immune system: Basic principles. Cell 2009, 136, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J. Allergy Clin. Immunol. 2013, 132, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Rebane, A.; Akdis, C.A. MicroRNAs: Essential players in the regulation of inflammation. J. Allergy Clin. Immunol. 2013, 132, 15–26. [Google Scholar] [CrossRef] [PubMed]
| Study | N (Case/Control) | Design | Findings | |
|---|---|---|---|---|
| Up-Regulated miRNA | Down-Regulated miRNA | |||
| Abu-Elneel et al. (2008) [70] | 26 (13/13) | Measure of the expression level of 466 human miRNAs from postmortem cerebellar tissue by multiplex real-time PCR, with 377 miRNAs detected and used for further analysis | miR-106a, miR-106b, miR-140, miR-146b, miR-181d, miR-193b, miR-320a, miR-381, miR-432, miR-539, miR-550, miR-652 | miR-7, miR-15a, miR-15b, miR-21, miR-23a, miR-27a, miR-93, miR-95, miR-128, miR-129, miR-132, miR-148b, miR-212, miR-431, miR-484, miR-598 |
| Sarachana et al. (2010) [71] | 14 (5/9) | Lymphoblasts derived from peripheral lymphocytes were obtained; miRNA expression profiling performed by high-throughput miRNA microarray analysis. Differentially expressed miRNAs confirmed by qRT-PCR analysis, putative target genes of two of the confirmed miRNA validated by knockdown and overexpression of the respective miRNAs | miR-16-2, miR-106b, miR-132, miR-133b, miR-136, miR-139, miR-148b, miR-153, miR-182, miR-189, miR-190, miR-199b, miR-211, miR-219, miR-326, miR-367, miR-455, miR-495, miR-518a, miR-520b | miR-23a, miR-23b, miR-25, miR-29b, miR-30e, miR-93, miR-103, miR-107, miR-185, miR-186, miR-191, miR-194, miR-195, miR-205, miR-342, miR-346, miR-376a-AS, miR-451, miR-519c, miR-524 |
| Talebizadeh et al. (2008) [72] | 12 (6/6) | Six subject with Autism Spectrum Disorders (ASD) (3 males, aged 5, 12, and 14 years, and 3 females, aged 6, 11, and 13 years), 6 age- and gender-matched TD controls. Lymphoblastoid cell lines, quantitative PCR, | miR-23a, miR-23b, miR-132, miR-146a, miR-146b, miR-663 | miR-92, miR-320, miR-363 |
| Mundalil Vasu et al. (2014) [73] | 110 (55/55) | 55 ASD (48 males, 6 females, aged 11.29 ± 5.45 years), 55 TD controls (41 males, 14 females, aged 11.3 ± 2.37 years). RNA extracted from serum, mature miRNAs selectively converted into cDNA. The expression of 125 mature miRNAs was compared between pooled control and ASD samples. The differential expression of 14 miRNAs further validated by SYBR Green quantitative PCR of individual samples. Target genes and pathways of miRNAs predicted by DIANA mirPath software | miR-19b-3p, miR-27a-3p, miR-101-3p, miR-106-5p, miR-130a-3p, miR-195b-5p | miR-151a-3p, miR-181b-5p, miR-320a, miR-328, miR-433, miR-489, miR-572, miR-663a |
| Popov et al. (2012) [74] | 55 (30/25) | Thirty ASD (24 males, 6 females, aged 3–20), 25 TD controls (20 males, 5 females, aged 3–20 years). Whole blood collection, analysis of gene expression changes applying LC expression profiling service, using pooled whole blood-derived total RNA samples | miR-486-3p | |
| Seno et al. (2011) [75] | 42 (20/22) | 20 severe ASD (13 males and 7 females), 22 unaffected siblings (19 males and 3 females). Lymphoblastoid cell lines, RNA was extracted and assayed using Illumina gene and miRNA expression arrays. Control quality in BeadStudio (Illumina) | miR-10a, miR-30a, miR-181a, miR-181b, miR-181c, miR-199b-5p, miR-338-3p, miR-486-3p, miR-486-5p, miR-500, miR-502-3p, miR-548 | miR-199a-5p, miR-455-3p, miR-577, miR-656 |
| Mor et al. (2015) [76] | 24 (12/12) | Brain tissue samples taken from postmortem Brodmann’s area 10 | miR-7-5p, miR-19a-3p, miR-19b-3p, miR-21-3p, miR-21-5p, miR-142-3p, miR-142-5p, miR-144-3p, miR-146a-5p, miR-155-5p, miR-219-5p, miR-338-5p, miR-379-5p, miR-451a, miR-494, miR-3168 | miR-34a-5p, miR-92b-3p, miR-211-5p, miR-3960 |
| Ander et al. (2015) [77] | 18 (10/8) | Brain tissue samples taken from postmortem Brodmann’s areas 22, 41, 42 | miR-664-3p, miR-4709-3p, miR-4753-5p | miR-1, miR-297, miR-4742-3p |
| Wu et al. (2016) [78] | 56 (28/28) | Tissue samples taken from postmortem cerebellar cortex, Brodmann area 9 | miR-10a-5p, miR-18b-5p, miR-20b-5p, miR-21-3p, miR-23a-3p, miR-107, miR-129-2-3p, miR-130b-5p, miR-148a-3p, miR-155-5p, miR-218-2-3p, miR-221-3p, miR-223-3p, miR-335-3p, miR-363-3p, miR-424-3p, miR-424-5p, miR-425-3p, miR-449b-5p, miR-450b-5p, miR-484, miR-629-5p, miR-651-5p, miR-708-5p, miR-766-3p, miR-874-3p, miR-887-3p, miR-940, miR-1277-3p, miR-3938, miR-2277-5p, let-7g-3p | miR-204-3p, miR-491-5p, miR-619-5p, miR-3687, miR-5096 |
| Huang et al. (2015) [79] | 40 (20/20) | Peripheral blood sample taken, microarray (5 ASD/5 controls), and quantitative Real-Time PCR (15 ASD/15 controls) | miR-34b-3p, miR-34c-3p, miR-483-5p, miR-494, miR-564, miR-642a-3p, miR-574-5p, miR-575, miR-921, miR-1246, miR-1249, miR-1273c, miR-4270, miR-4299, miR-4436a, miR-4443, miR-4516, miR-4669, miR-4721, miR-4728-5p, miR-4788, miR-5739, miR-6086, miR-6125 | miR-15a-5p, miR-15b-5p, miR-16-5p, miR-19b-3p, miR-20a-5p, miR-92a-3p, miR-103a-3p, miR-195-5p, miR-451a, miR-574-3p, miR-940, miR-1228-3p, miR-3613-3p, miR-3935, miR-4436b-5p, miR-4665-5p, miR-4700-3p, let-7a-5p, let-7d-5p, let-7f-5p |
| Toma et al. (2015) [80] | 1309 (636/673) | Genomic DNA isolated from blood lymphocytes, or from saliva | miR-133b/miR-206 cluster; pooled analysis: miR-133b/miR-206 and miR-17/miR-18a/miR-19a/miR-20a/miR- 19b-1/miR92a-1. | N/A |
| Hicks et al. (2016) [81] | 45 (24/21) | Salivary samples | miR-7-5p, miR-28-5p, miR-127-3p, miR-140-3p, miR-191-5p, miR-218-5p, miR-335-3p, miR-628-5p, miR-2467-5p, miR-3529-3p | miR-23a-3p, miR-27a-3p, miR-30e-5p, miR-32-5p |
| Nguyen et al. (2016) [82] | 14 (8/6) | Samples taken from olfactory mucosal stem cells and skin fibroblasts or Peripheral Blood Mononuclear Cells. Measured through microarray and quantitative Real-Time PCR validation | miR-146a | miR-221, miR-654-5p, miR-656 |
| Kichukova et al. (2017) [83] | 60 (30/30) | Blood samples. Quantitative Real-Time PCR validation | miR-18b-3p, miR-106b-5p, miR-142-3p, miR-210-5p, miR-365a-3p, miR-374b-5p, miR-619-5p, miR-664a-3p, miR-3620-3p, miR-4489, miR-8052 | hsa-let-7i-3p, miR -15a-5p, miR -20b-3p, miR -29c-5p, miR -96-5p, miR -145-5p, miR -183-5p, miR -193b-3p, miR -197-5p, miR-199a-5p, miR -301a-3p, miR -328-3p, miR -424-5p, miR -486-3p, miR -487b-3p, miR -500a-5p, miR -504-5p, miR -576-5p, miR -587-3p, miR-589-3p, miR -664b-3p, miR -671-3p, miR -3064-5p, miR -3135a, miR -3674, miR -3687, miR-3909, miR -6799-3p, miR -6849-3p |
| Jyonouchi et al. (2017) [84] | 96 (69/27) | Peripheral blood monocytes samples, miRNA expression determined by high-throughput sequencing | hsa-let-7a-1, hsa-let-7a-2, hsa-let-7a-3, hsa-let-7f-1, hsa-let-7f-2, hsa-let-7g, hsa-let-7i, miR-17, miR-26a-2, miR-30b, miR-30c-1, miR-30c-2, miR-98, miR-106b, miR-130a, miR-148a, miR-148b, miR-150, miR-186, miR-301a, miR-374b, miR-494, miR-1248, miR-3607, miR-3609 | hsa-let-7b, miR-15a, miR-15b, miR-16-1, miR-16-2, miR-18a, miR-19a, miR-19b-1, miR-19b-2, miR-20a, miR-21, miR-27a, miR-27b, miR-29a, miR-29b-1, miR-29b-2, miR-29c, miR-30e, miR-93, miR-101-1, miR-101-2, miR-103a-1, miR-103a-2, miR-107, miR-126, miR-142, miR-145, miR-146a, miR-151a, miR-181a-1, miR-181a-2, miR-199b, miR-221, miR-222, miR-320a, miR-376c, miR-409, miR-423, miR-484, miR-625, miR-4433b, miR-5701-1, miR-5701-2 |
| Pagan et al. (2017) [85] | 517 (239/278) * | Post-mortem pineal glands (melatonin) in 9 patients and 22 controls; gut samples (serotonin) in 11 patients and 13 controls; blood platelets from 239 individuals with ASD, their first-degree relatives and 278 controls | Plasmatic and pineal miR-451 | N/A |
| Nguyen et al. (2018) [86] | 11 (5/6) | Post-mortem analysis of temporal lobe in ASD children and controls, miRNA expression performed using Taqman assay | miR-146a | N/A |
| Yu et al. (2018) [87] | 43 (20/23) | Serum samples, quantitative reverse transcription-PCR to examine miRNAs | miR-486-3p, miR-557 | N/A |
| Williams et al. (2018) [88] | 128 (48/80) * | Blood samples from 48 ASD and 80 parents | miR-873-5p | N/A |
| Study | N (Case/Control) | Design | Findings | |
|---|---|---|---|---|
| Up-Regulated miRNA | Down-Regulated miRNA | |||
| Sonkoly et al. (2010) [66] | 47 (18/29) | Skin samples | miR-155 | |
| Lv et al. (2014) [89] | 58 (30/28) | Serum and urine samples | miR-203, miR-483-5p (serum) | miR-203 (urine) |
| Ralfkiaer et al. (2014) [90] | 75 (20/55) * | Skin samples | miR-149, miR-Plus-C1070, miR-205, miR-141, miR-23b, miR-221, miR-27b, miR-203, miR-7b, miR-19b, miR-27a, miR-455-3p, miR-200a, miR-211, miR-23a, miR-214 | miR-181a, miR-342-5p, miR-766, miR-7i, miR-186, miR-342-3p, miR-664, miR-425, miR-9, miR-331-3p, miR-146b-5p, miR-10a, miR-663, miR-937, miR-361-3p, miR-605, miR-146a, miR-940, miR-150, miR-1913, miR-155, miR-302c |
| Rebane et al. (2014) [91] | 18 (9/9) | Skin samples | miR-146a | |
| Ma et al. (2015) [92] | 64 (33/31) | Skin samples | miR-155 | |
| Ding et al. (2016) [93] | 22 (14/8) | Skin samples | miR-148b, miR-152, miR-324 | |
| Yang et al. (2017) [94] | 37 (37/0) | Skin samples | miR-124 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonacci, A.; Bagnato, G.; Pandolfo, G.; Billeci, L.; Sansone, F.; Conte, R.; Gangemi, S. MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review. J. Clin. Med. 2019, 8, 88. https://doi.org/10.3390/jcm8010088
Tonacci A, Bagnato G, Pandolfo G, Billeci L, Sansone F, Conte R, Gangemi S. MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review. Journal of Clinical Medicine. 2019; 8(1):88. https://doi.org/10.3390/jcm8010088
Chicago/Turabian StyleTonacci, Alessandro, Gianluca Bagnato, Gianluca Pandolfo, Lucia Billeci, Francesco Sansone, Raffaele Conte, and Sebastiano Gangemi. 2019. "MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review" Journal of Clinical Medicine 8, no. 1: 88. https://doi.org/10.3390/jcm8010088
APA StyleTonacci, A., Bagnato, G., Pandolfo, G., Billeci, L., Sansone, F., Conte, R., & Gangemi, S. (2019). MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review. Journal of Clinical Medicine, 8(1), 88. https://doi.org/10.3390/jcm8010088








