Development of a Simplified Protocol for Respiratory Muscle Segmentation in Unenhanced Chest CT and Identification of New Radiomic Biomarkers of Sarcopenia in Lung Diseases: A Retrospective Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Analysis of Muscle Density and Radiomics and Correlation with Demographics
2.3. Definition of a Simplified Segmentation Protocol for Respiratory Muscles
3. Results
3.1. Patients’ Characteristic
3.2. Density Characterization
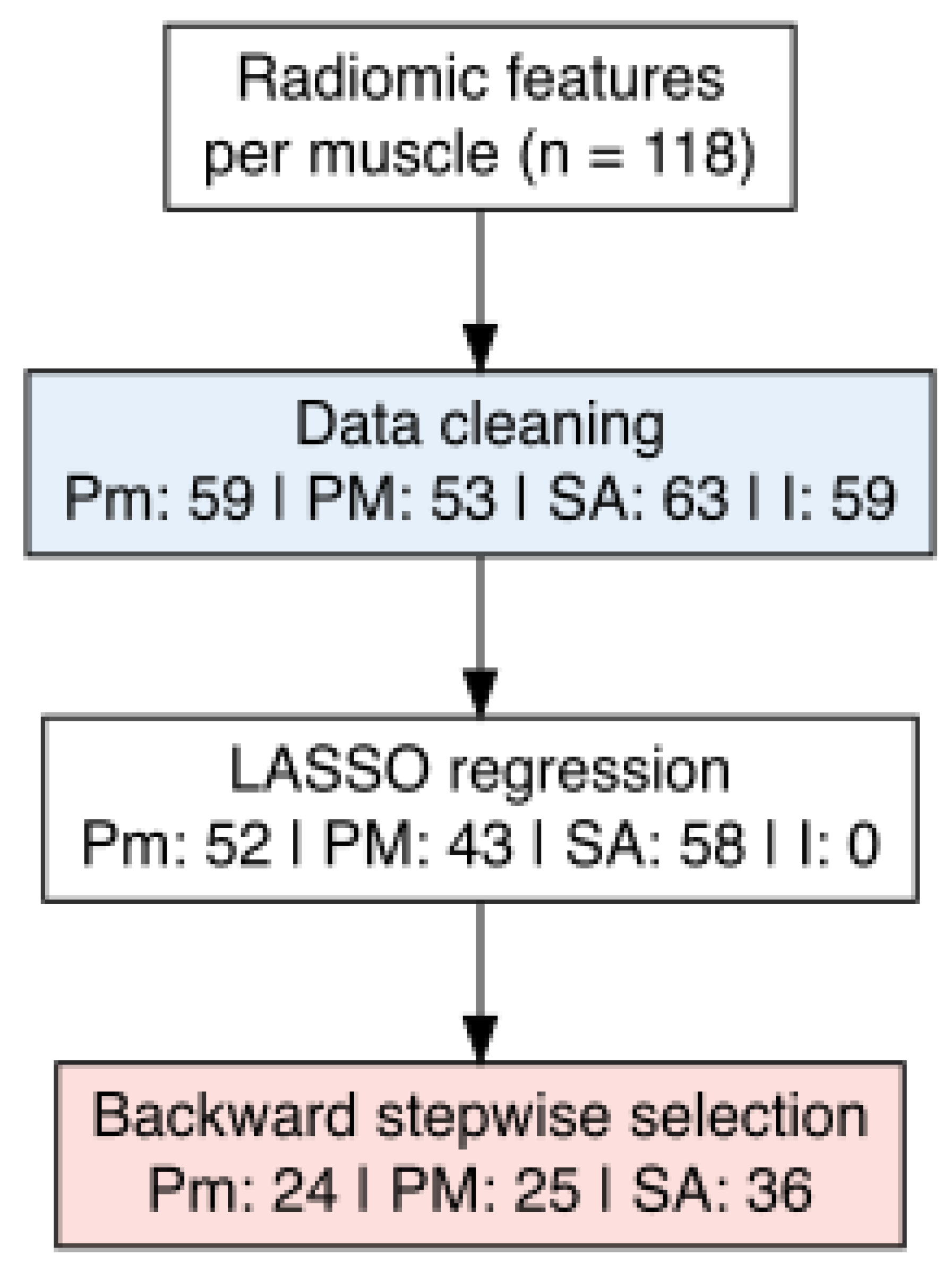
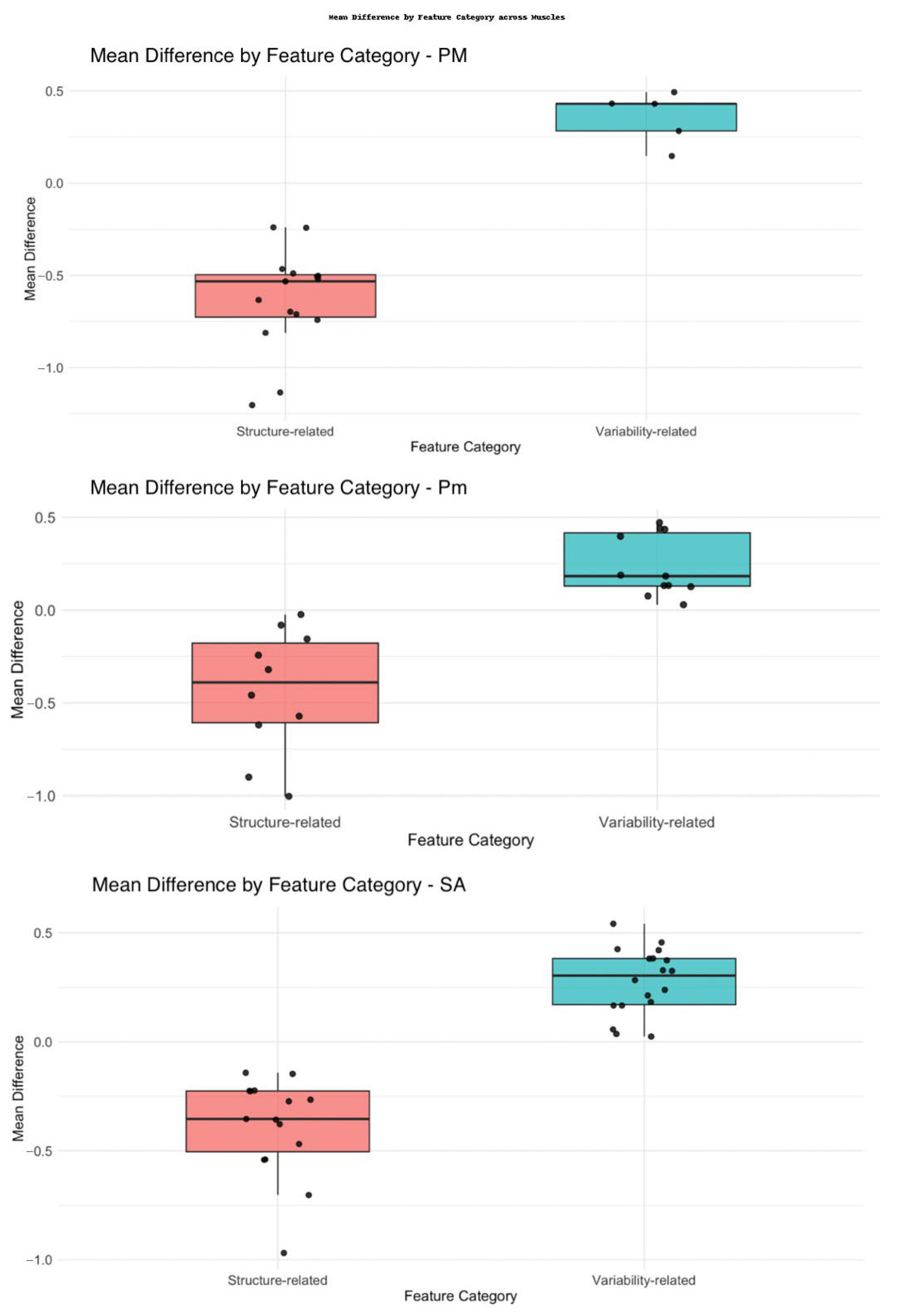
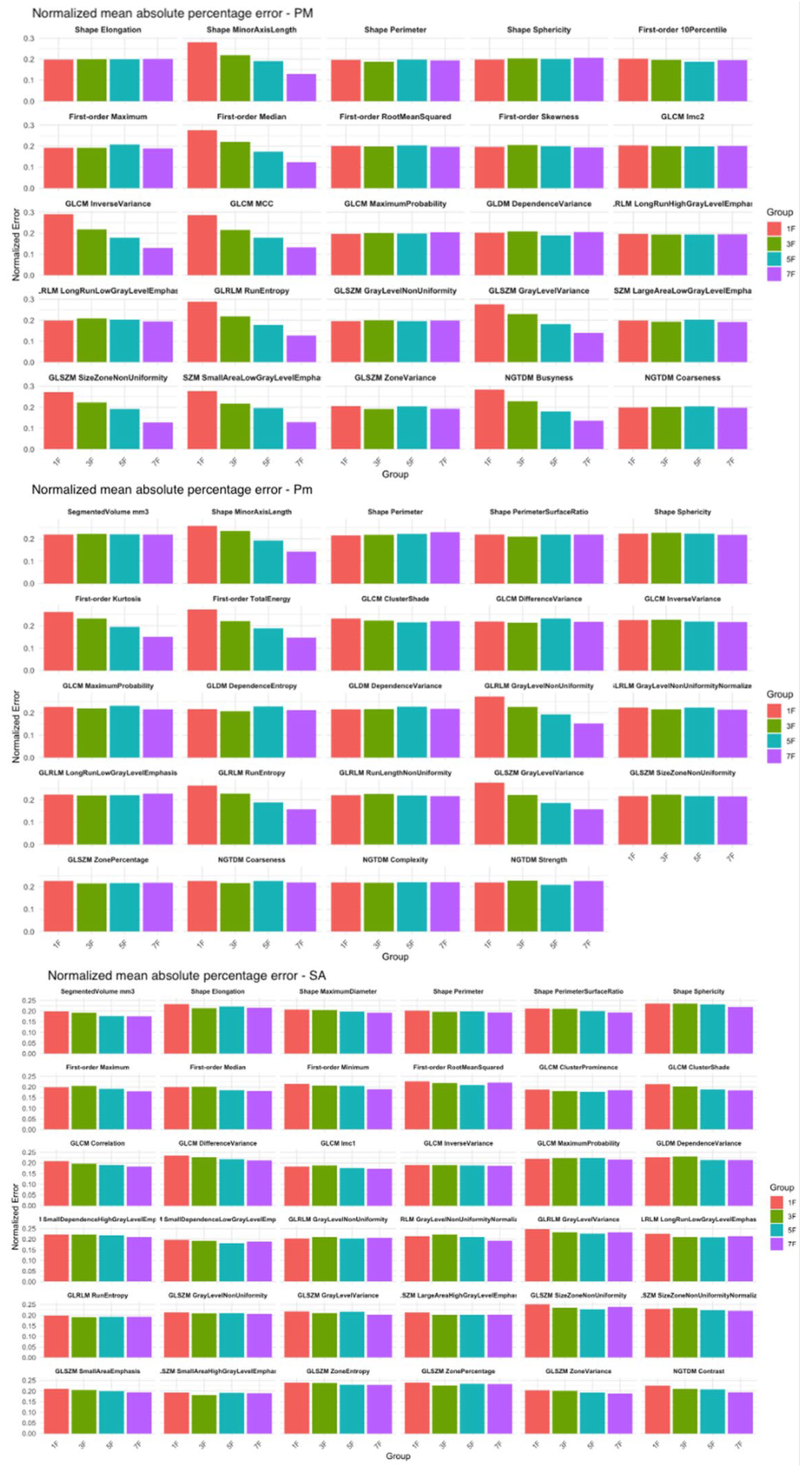
3.3. Radiomics Features Analysis
3.4. Identification of Anatomic Landmarks and Definition of Segmentation Protocol
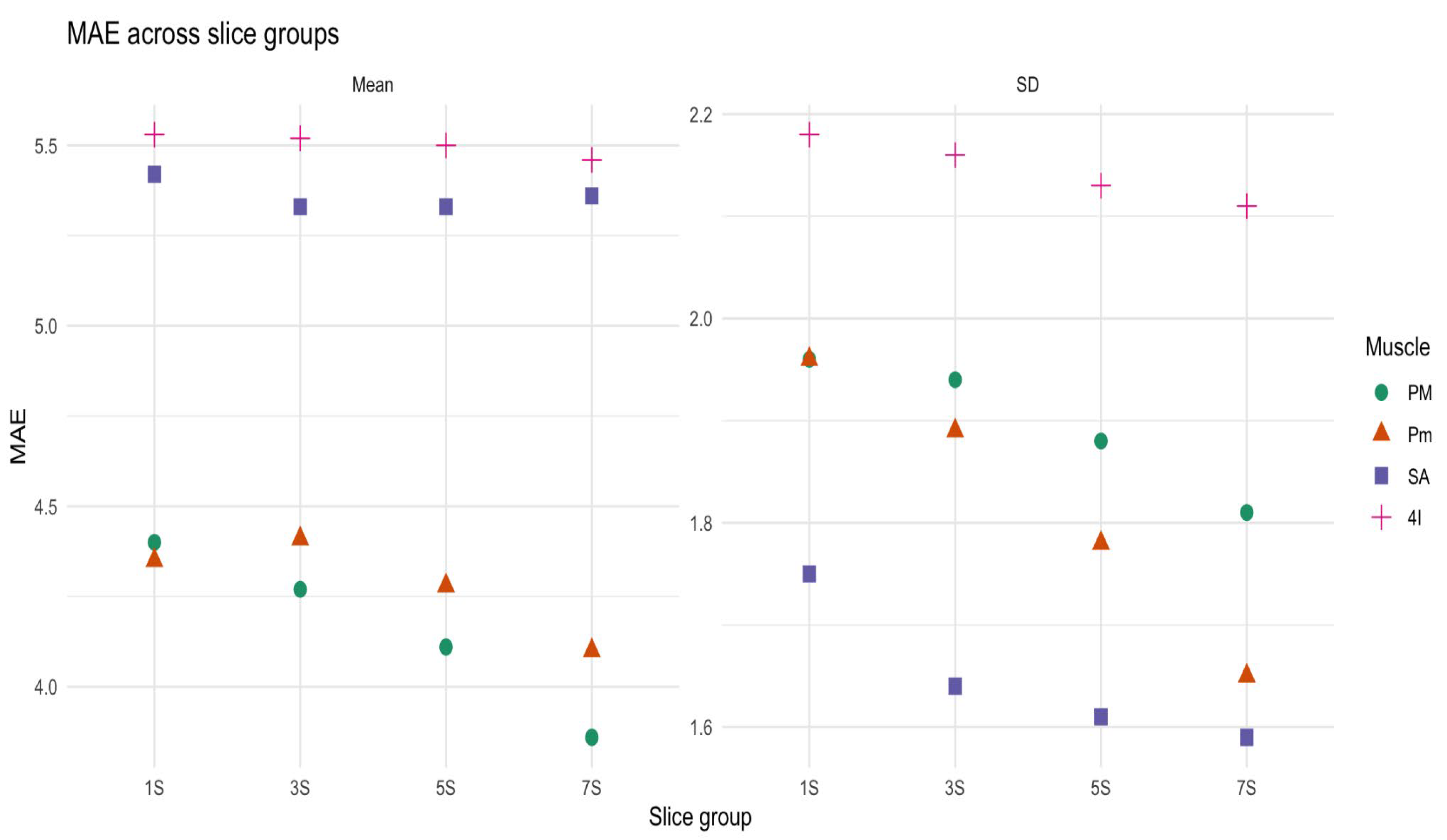
3.5. Comparison of Density and Radiomics in Small Slice Sets and in the Entire Muscle Volume
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | Body Mass Index |
| COPD | Chronic Obstructive Pulmonary Disease |
| CT | Computed Tomography |
| HU | Hounsfield Units |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| MAE | Mean Absolute Error |
| MAPE | Mean Absolute Percentage Error |
| OR | Odds Ratio |
| Pm | Pectoralis Minor |
| PM | Pectoralis Major |
| SA | Serratus Anterior |
| SD | Standard Deviation |
| 4I | Fourth intercostal muscle |
References
- Maltais, F.; Decramer, M.; Casaburi, R.; Barreiro, E.; Burelle, Y.; Debigaré, R.; Dekhuijzen, P.N.; Franssen, F.; Gayan-Ramirez, G.; Gea, J.; et al. An Official American Thoracic Society/European Respiratory Society Statement: Update on Limb Muscle Dysfunction in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2014, 189, e15–e62. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.; Yelin, E.H.; Katz, P.P.; Sanchez, G.; Iribarren, C.; Eisner, M.D.; Blanc, P.D. Respiratory and Skeletal Muscle Strength in Chronic Obstructive Pulmonary Disease: Impact on Exercise Capacity and Lower Extremity Function. J. Cardiopulm. Rehabil. Prev. 2011, 31, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.S.; Seo, J.H.; Ko, M.H.; Park, S.H.; Kang, S.W.; Won, Y.H. Respiratory Muscle Strength in Patients with Chronic Obstructive Pulmonary Disease. Ann. Rehabil. Med. 2017, 41, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, R.I.N.; Azambuja, A.M.; Cureau, F.V.; Sbruzzi, G. Inspiratory Muscle Training in COPD. Respir. Care 2020, 65, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.L.; Diaz, A.A.; Ross, J.C.; San Jose Estepar, R.; Zhou, L.; Regan, E.A.; Eckbo, E.; Muralidhar, N.; Come, C.E.; Cho, M.H.; et al. Quantitative Computed Tomography Measures of Pectoralis Muscle Area and Disease Severity in Chronic Obstructive Pulmonary Disease: A Cross-Sectional Study. Ann. Am. Thorac. Soc. 2014, 11, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Mai, D.V.C.; Drami, I.; Pring, E.T.; Gould, L.E.; Lung, P.; Popuri, K.; Chow, V.; Beg, M.F.; Athanasiou, T.; Jenkins, J.T.; et al. A Systematic Review of Automated Segmentation of 3D Computed-Tomography Scans for Volumetric Body Composition Analysis. J. Cachexia Sarcopenia Muscle 2023, 14, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- Ciciliot, S.; Rossi, A.C.; Dyar, K.A.; Blaauw, B.; Schiaffino, S. Muscle Type and Fiber Type Specificity in Muscle Wasting. Int. J. Biochem. Cell Biol. 2013, 45, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-To” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- de Jong, E.E.C.; Sanders, K.J.C.; Deist, T.M.; van Elmpt, W.; Jochems, A.; van Timmeren, J.E.; Leijenaar, R.T.H.; Degens, J.H.R.J.; Schols, A.M.W.J.; Dingemans, A.C.; et al. Can Radiomics Help to Predict Skeletal Muscle Response to Chemotherapy in Stage IV Non-Small Cell Lung Cancer? Eur. J. Cancer 2019, 120, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, E.M.; Boone, J.M. Determination of Height, Weight, Body Mass Index, and Body Surface Area with a Single Abdominal CT Image. Radiology 2003, 228, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Derstine, B.A.; Holcombe, S.A.; Ross, B.E.; Wang, N.C.; Su, G.L.; Wang, S.C. Skeletal Muscle Cutoff Values for Sarcopenia Diagnosis Using T10 to L5 Measurements in a Healthy U.S. Population. Sci. Rep. 2018, 8, 11369. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European Consensus on Definition and Diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Shi, W.; Peng, X.; Zhan, Y.; Zhou, L.; Wang, Y.; Feng, M.; Zhao, J.; Shan, F.; Liu, L. Development and Validation of a Nomogram Incorporating CT Radiomics Signatures and Radiological Features for Differentiating Invasive Adenocarcinoma from Adenocarcinoma in Situ and Minimally Invasive Adenocarcinoma Presenting as Ground-Glass Nodules Measuring 5–10 mm in Diameter. Front. Oncol. 2021, 11, 618677. [Google Scholar] [CrossRef]
- Qiao, J.; Zhang, X.; Du, M.; Wang, P.; Xin, J. 18F-FDG PET/CT Radiomics Nomogram for Predicting Occult Lymph Node Metastasis of Non-Small Cell Lung Cancer. Front. Oncol. 2022, 12, 974934. [Google Scholar] [CrossRef] [PubMed]
- Fiz, F.; Costa, G.; Gennaro, N.; La Bella, L.; Boichuk, A.; Sollini, M.; Politi, L.S.; Balzarini, L.; Torzilli, G.; Chiti, A.; et al. Contrast Administration Impacts CT-Based Radiomics of Colorectal Liver Metastases and Non-Tumoral Liver Parenchyma Revealing the “Radiological” Tumour Microenvironment. Diagnostics 2021, 11, 1162. [Google Scholar] [CrossRef] [PubMed]
- Carpani, G.; Susi, M.E.; Picasso, R.; Marcenaro, G.; Maccio, M.; Mongelli, M.; Martinoli, C.; Paglialonga, A. Analysis of Respiratory Muscles Density in Different Age Groups Using CT Scans. Stud Health Technol Inform. 2025, 15, 153–157. [Google Scholar] [CrossRef]
- Yao, L.; Petrosyan, A.; Chaudhari, A.J.; Lenchik, L.; Boutin, R.D. Clinical, Functional, and Opportunistic CT Metrics of Sarcopenia at the Point of Imaging Care: Analysis of All-Cause Mortality. Skeletal Radiol. 2024, 53, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wu, G. Chest CT Radiomics Is Feasible in Evaluating Muscle Change in Diabetes Patients. Curr. Med. Imaging 2024, 20, e15734056268543. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; An, Y.; Liu, P.; Mu, S.; Zhou, W.; Jia, H.; Huang, W.; Li, J.; Wang, R. Pectoralis Muscle Predicts Distant Metastases in Breast Cancer by Deep Learning Radiomics. Acta Radiol. 2023, 64, 2561–2569. [Google Scholar] [CrossRef] [PubMed]
- Ackermans, L.G.C.; Volmer, L.; Timmermans, Q.M.M.A.; Brecheisen, R.; Olde Damink, S.M.W.; Dekker, A.; Loeffen, D.; Poeze, M.; Blokhuis, T.J.; Wee, L.; et al. Clinical Evaluation of Automated Segmentation for Body Composition Analysis on Abdominal L3 CT Slices in Polytrauma Patients. Injury 2022, 53 (Suppl. S3), S30–S41. [Google Scholar] [CrossRef] [PubMed]
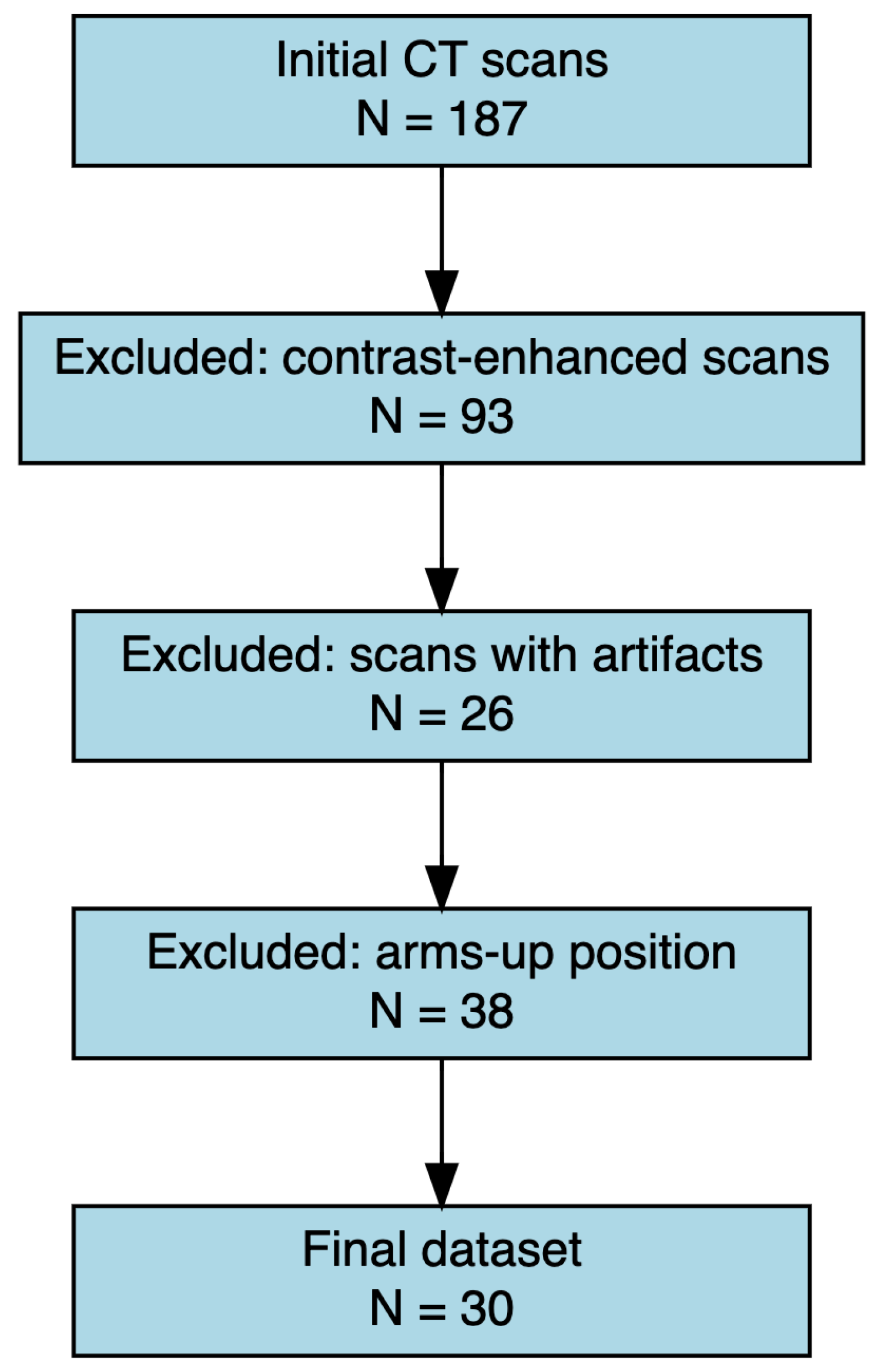

| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Unenhanced CT scans including thoracic region from T1 to L1 | Presence of metallic artifacts (e.g., pacemakers, prostheses, sternal wires) |
| Patient positioned with both arms raised above the head | Age < 18 years |
| CT acquired with the same scanner model | Poor image quality |
| Kilovoltage set between 120 and 130 kV | |
| Slice thickness of 2 mm |
| Variable | Total (n = 30) | Sarcopenic (n = 17) | Non-Sarcopenic (n = 13) |
|---|---|---|---|
| Age (years) | 64.7 ± 17.2 | 70.4 ± 12.5 | 56.1 ± 20.0 |
| BMI (kg/m2) | 23.1 ± 3.3 | 22.0 ± 2.9 | 24.9 ± 3.2 |
| Females (n) | 16 (53%) | 11 (65%) | 5 (38%) |
| Males (n) | 14 (47%) | 6 (35%) | 8 (62%) |
| Caucasian | 28 (93.3%) | 17 (100%) | 11 (85%) |
| Muscle | Total Density (HU) | Non-Sarcopenic Density (HU) | Sarcopenic Density (HU) |
|---|---|---|---|
| PM | 25.5 ± 19.9 | 36.9 ± 14.1 | 15.2 ± 18.8 |
| Pm | 27.6 ± 15.4 | 32.8 ± 15.9 | 23.3 ± 13.6 |
| SA | 15.0 ± 21.5 | 23.9 ± 16.6 | 3.1 ± 21.6 |
| 4I | −27.8 ± 26.3 | −18.4 ± 25.3 | −38.8 ± 23.0 |
| Muscle | Density 18–45 Years (HU) | Density 46–69 Years (HU) | Density ≥ 70 Years (HU) |
|---|---|---|---|
| PM | 44.3 ± 9.2 | 17.1 ± 23.9 | 14.2 ± 10.9 |
| Pm | 41.3 ± 11.8 | 22.9 ± 12.3 | 20.7 ± 9.1 |
| SA | 34.2 ± 7.7 | 9.1 ± 13.5 | 8.7 ± 18.7 |
| 4I | 7.1 ± 11.9 | −37.9 ± 12.6 | −35.4 ± 20 |
| Muscle | Variability-Related Features | Density-Related Features |
|---|---|---|
| Pectoralis Major (PM) | GLCM Inverse Variance, GLCM MCC, GLCM Maximum Probability, GLDM Dependence Variance, GLRLM Run Entropy, GLSZM Gray Level Non-Uniformity, GLSZM Gray Level Variance, GLSZM Zone Variance, GLSZM Size Zone Non-Uniformity, NGTDM Busyness, NGTDM Coarseness, RLM Long Run Low Gray Level Emphasis, GLCM Imc2 | First-order Maximum, First-order Median, First-order Root Mean Squared, First-order Skewness, First-order 10Percentile |
| Pectoralis Minor (Pm) | GLCM Cluster Shade, GLCM Difference Variance, GLCM Inverse Variance, GLCM Maximum Probability, GLDM Dependence Entropy, GLDM Dependence Variance, GLRLM Gray Level Non-Uniformity, GLRLM Run Entropy, GLRLM Run Length Non-Uniformity, GLSZM Gray Level Variance, GLSZM Size Zone Non Uniformity, GLSZM Zone Percentage, NGTDM Coarseness, NGTDM Complexity, NGTDM Strength, LRLM Gray Level Non Uniformity Normalized | First-order Kurtosis, First-order Total Energy, Segmented Volume mm3 |
| Serratus Anterior (SA) | GLCM Cluster Prominence, GLCM Cluster Shade, GLCM Correlation, GLCM Difference Variance, GLCM Inverse Variance, GLCM Maximum Probability, GLDM Dependence Variance, GLRLM Gray Level Non-Uniformity, GLSZM Gray Level Variance, GLSZM Zone Entropy, GLSZM Zone Variance, GLSZM Size Zone Non-Uniformity, GLSZM Zone Percentage, NGTDM Contrast, NGTDM Coarseness | First-order Maximum, First-order Median, First-order Minimum, First-order Root Mean Squared, Segmented Volume mm3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picasso, R.; Susi, M.E.; Marcenaro, G.; Macciò, M.; Zaottini, F.; Pistoia, F.; La Grutta, L.; Sollami, G.; Maggio, A.; Bagnasco, D.; et al. Development of a Simplified Protocol for Respiratory Muscle Segmentation in Unenhanced Chest CT and Identification of New Radiomic Biomarkers of Sarcopenia in Lung Diseases: A Retrospective Study. J. Clin. Med. 2025, 14, 8712. https://doi.org/10.3390/jcm14248712
Picasso R, Susi ME, Marcenaro G, Macciò M, Zaottini F, Pistoia F, La Grutta L, Sollami G, Maggio A, Bagnasco D, et al. Development of a Simplified Protocol for Respiratory Muscle Segmentation in Unenhanced Chest CT and Identification of New Radiomic Biomarkers of Sarcopenia in Lung Diseases: A Retrospective Study. Journal of Clinical Medicine. 2025; 14(24):8712. https://doi.org/10.3390/jcm14248712
Chicago/Turabian StylePicasso, Riccardo, Maria Elena Susi, Giovanni Marcenaro, Marta Macciò, Federico Zaottini, Federico Pistoia, Ludovico La Grutta, Giulia Sollami, Arianna Maggio, Diego Bagnasco, and et al. 2025. "Development of a Simplified Protocol for Respiratory Muscle Segmentation in Unenhanced Chest CT and Identification of New Radiomic Biomarkers of Sarcopenia in Lung Diseases: A Retrospective Study" Journal of Clinical Medicine 14, no. 24: 8712. https://doi.org/10.3390/jcm14248712
APA StylePicasso, R., Susi, M. E., Marcenaro, G., Macciò, M., Zaottini, F., Pistoia, F., La Grutta, L., Sollami, G., Maggio, A., Bagnasco, D., Braido, F., Ferraris, M., Napoli, L., Bondi, B., Carpani, G., Vettori, G., Mongelli, M., Paglialonga, A., & Martinoli, C. (2025). Development of a Simplified Protocol for Respiratory Muscle Segmentation in Unenhanced Chest CT and Identification of New Radiomic Biomarkers of Sarcopenia in Lung Diseases: A Retrospective Study. Journal of Clinical Medicine, 14(24), 8712. https://doi.org/10.3390/jcm14248712








