A Distinct Intestinal Domination Fingerprint in Patients Undergoing Allo-HSCT: Dynamics, Predictors and Implications on Clinical Outcomes
Abstract
1. Introduction
2. Patients, Material and Methods
2.1. Study Design, Ethical Aspects and Sample Collections
2.2. Intestinal Microbiota 16S Sequencing and Bioinformatics Pipeline
2.3. Statistical Analysis
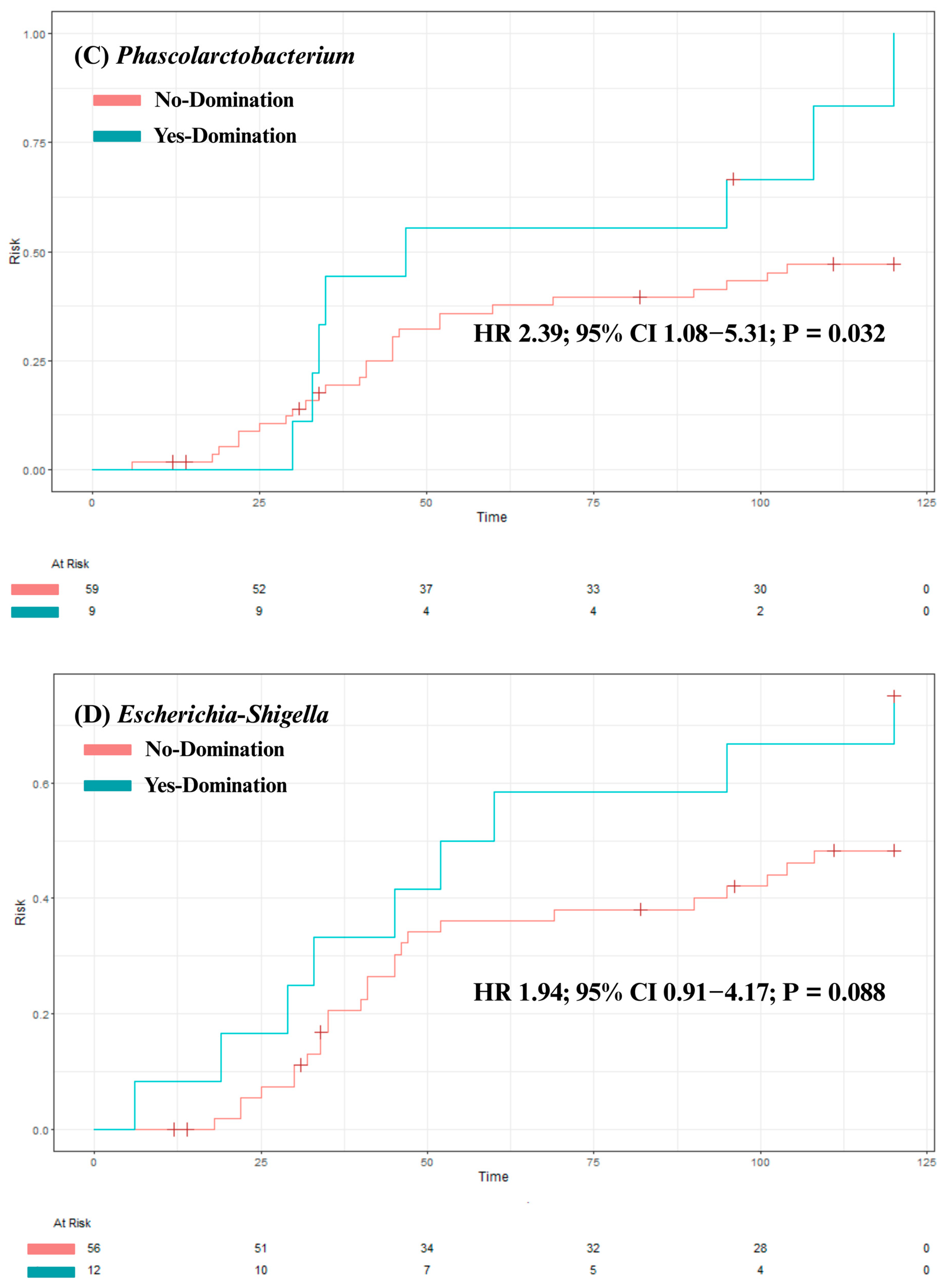
3. Results
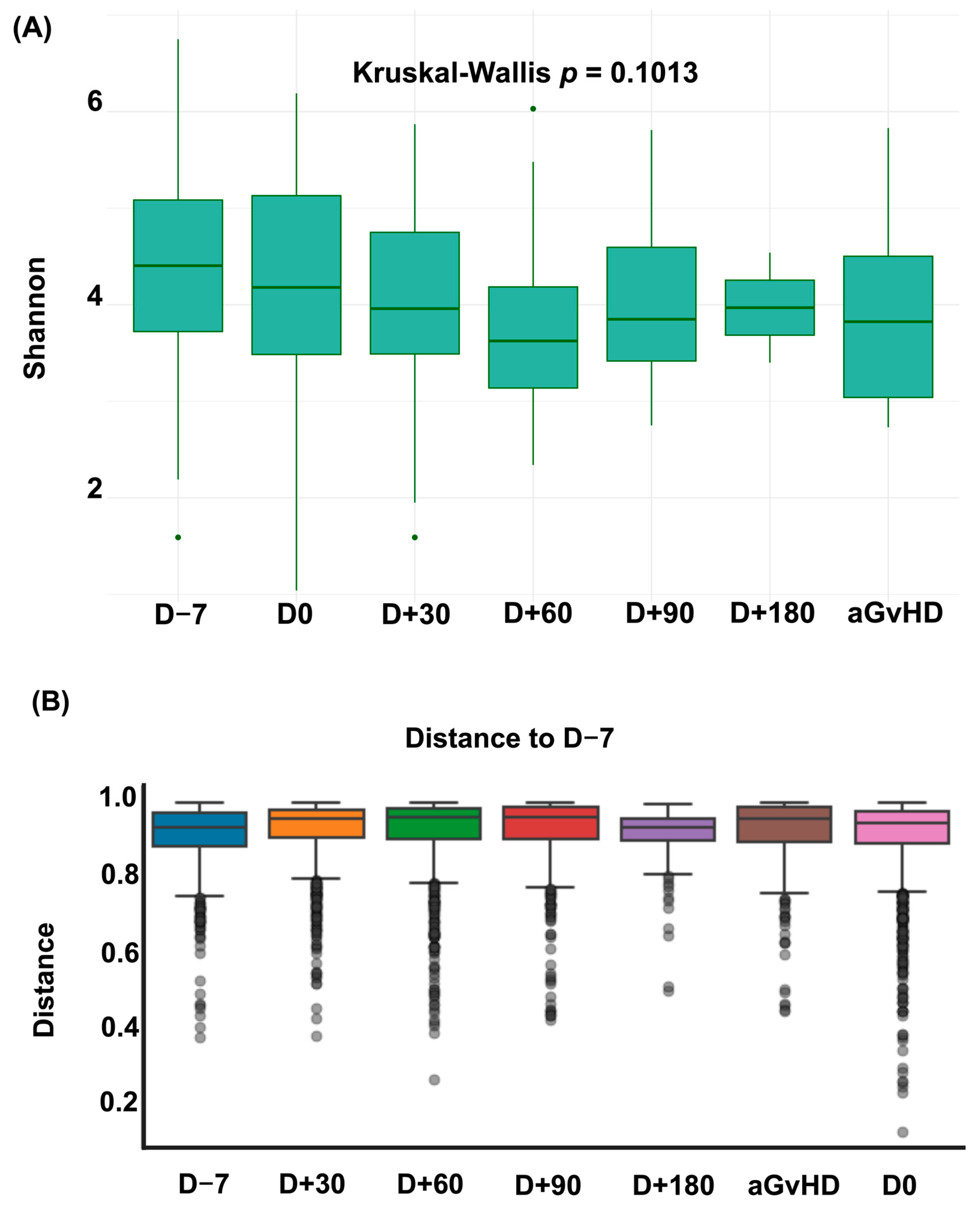
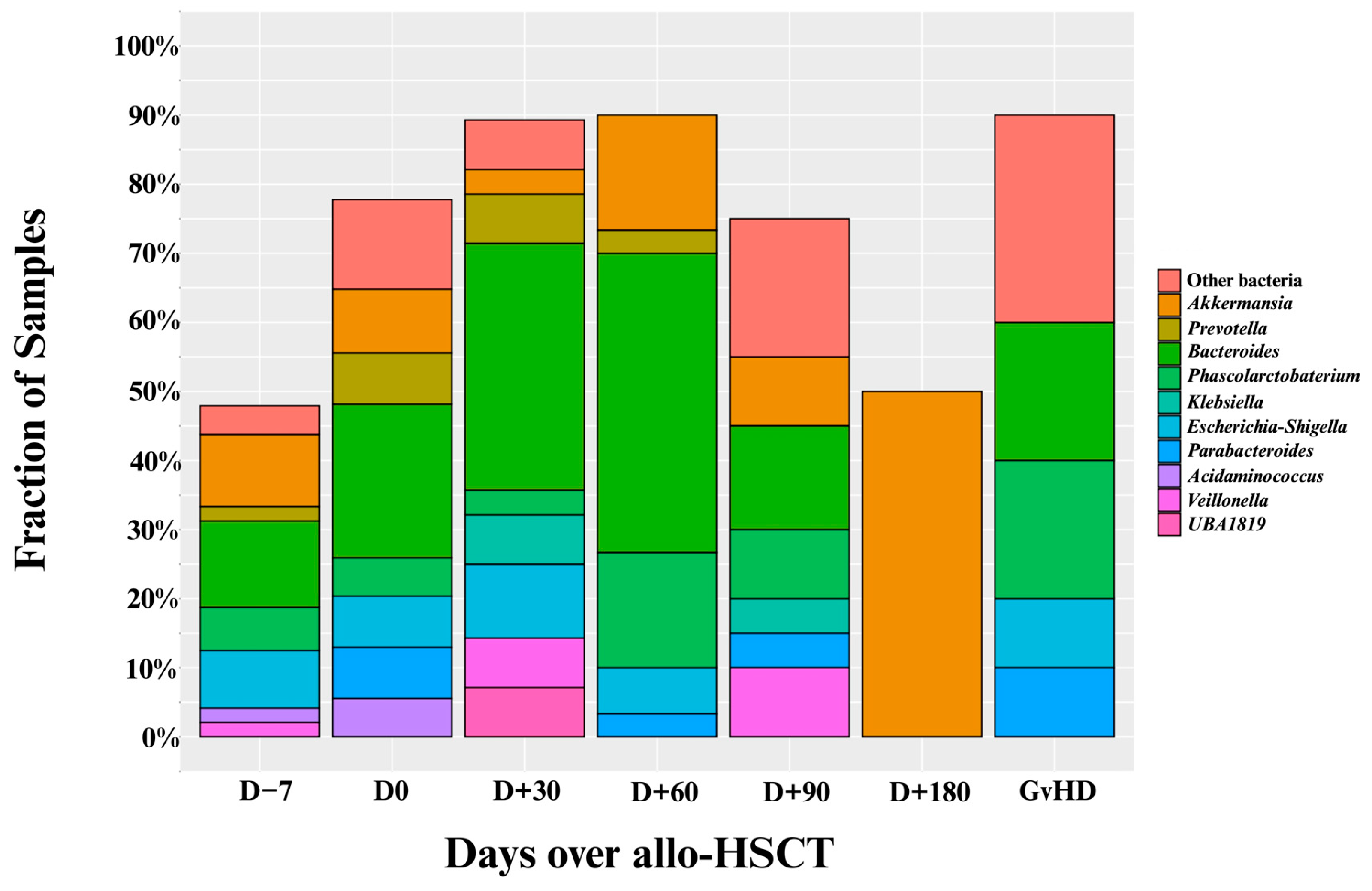
3.1. Diversity Metrics and Prevalence of Intestinal Domination
3.2. Patient Characteristics by Intestinal Domination Status
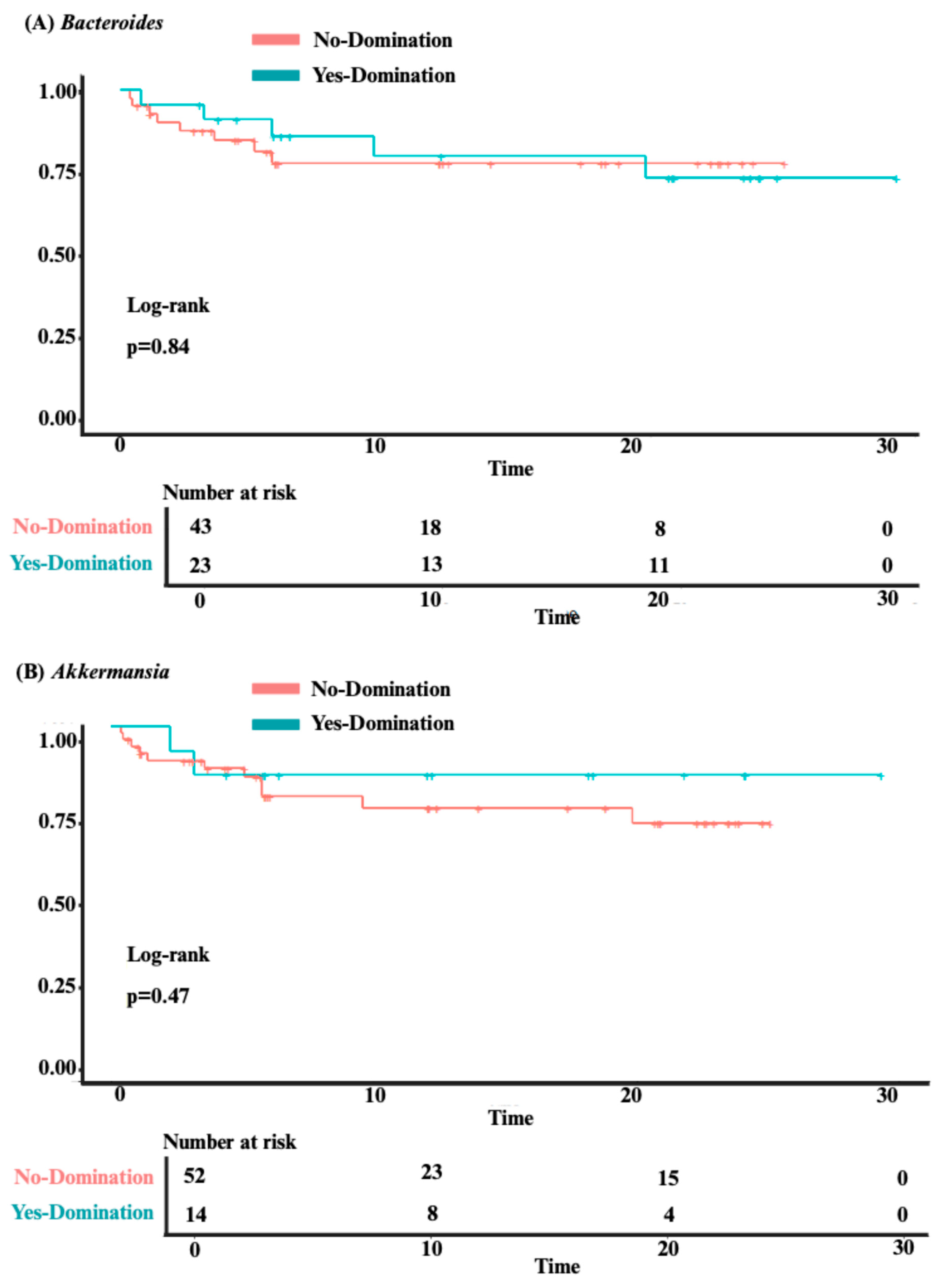
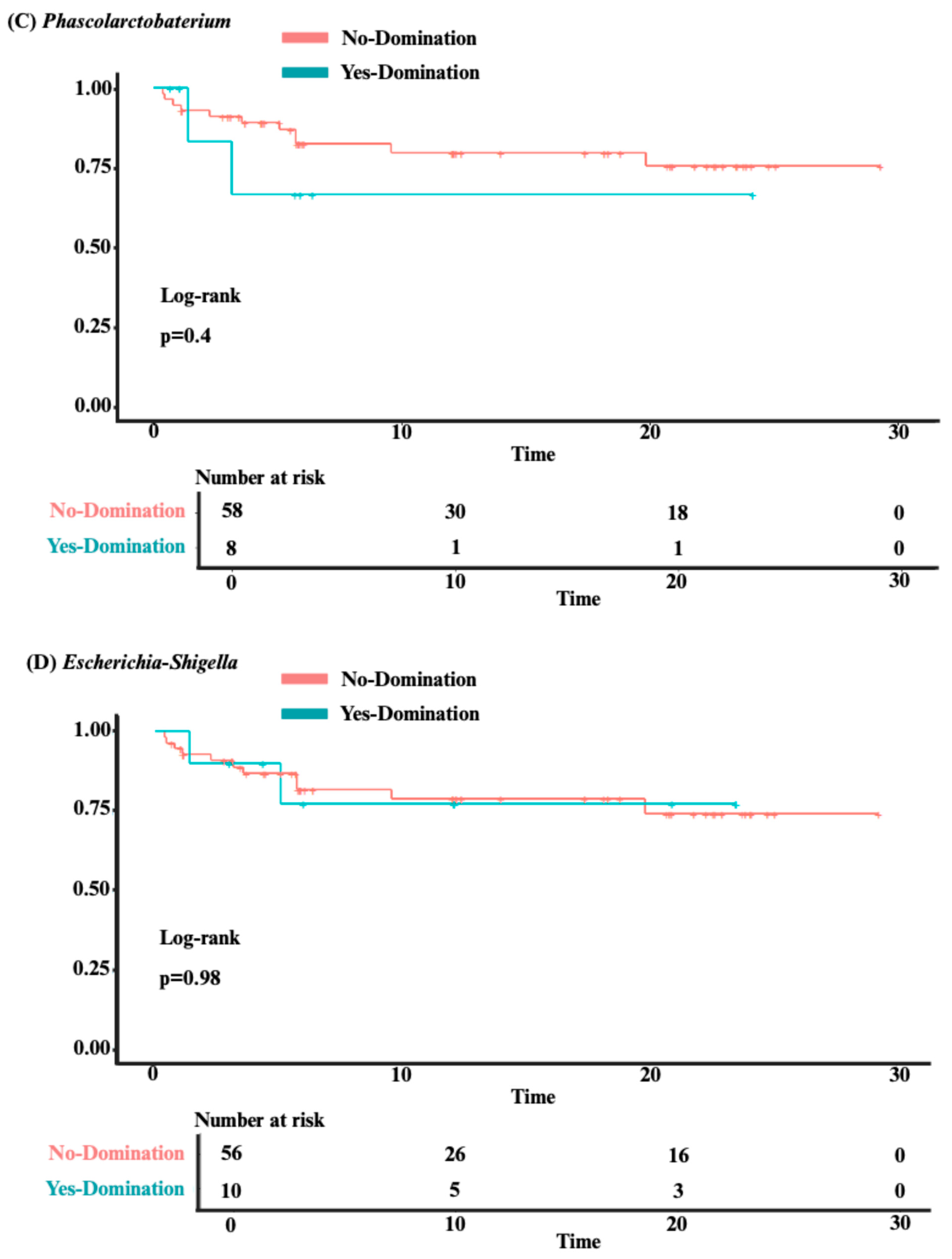
3.3. Analysis of Clinical Outcomes
3.4. Predictors of Intestinal Domination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, G.R.; Betts, B.C.; Tkachev, V.; Kean, L.S.; Blazar, B.R. Current Concepts and Advances in Graft-Versus-Host Disease Immunology. Annu. Rev. Immunol. 2021, 39, 19–49. [Google Scholar] [CrossRef]
- Ferrara, J.L.; Levine, J.E.; Reddy, P.; Holler, E. Graft-versus-Host Disease. Lancet 2009, 373, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- Jagasia, M.; Arora, M.; Flowers, M.E.D.; Chao, N.J.; McCarthy, P.L.; Cutler, C.S.; Urbano-Ispizua, A.; Pavletic, S.Z.; Haagenson, M.D.; Zhang, M.-J.; et al. Risk Factors for Acute GVHD and Survival after Hematopoietic Cell Transplantation. Blood 2012, 119, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Ilett, E.E.; Jørgensen, M.; Noguera-Julian, M.; Nørgaard, J.C.; Daugaard, G.; Helleberg, M.; Paredes, R.; Murray, D.D.; Lundgren, J.; MacPherson, C.; et al. Associations of the Gut Microbiome and Clinical Factors with Acute GVHD in Allogeneic HSCT Recipients. Blood Adv. 2020, 4, 5797–5809. [Google Scholar] [CrossRef]
- Nesher, L.; Rolston, K.V.I. Febrile Neutropenia in Transplant Recipients. In Principles and Practice of Transplant Infectious Diseases; Safdar, A., Ed.; Springer: New York, NY, USA, 2019; pp. 185–198. ISBN 978-1-4939-9032-0. [Google Scholar]
- Barrett, A.J.; Battiwalla, M. Relapse after Allogeneic Stem Cell Transplantation. Expert. Rev. Hematol. 2010, 3, 429–441. [Google Scholar] [CrossRef]
- Wang, S.; Yue, X.; Zhou, H.; Chen, X.; Chen, H.; Hu, L.; Pan, W.; Zhao, X.; Xiao, H. The Association of Intestinal Microbiota Diversity and Outcomes of Allogeneic Hematopoietic Cell Transplantation: A Systematic Review and Meta-Analysis. Ann. Hematol. 2023, 102, 3555–3566. [Google Scholar] [CrossRef]
- Staffas, A.; Burgos da Silva, M.; van den Brink, M.R.M. The Intestinal Microbiota in Allogeneic Hematopoietic Cell Transplant and Graft-versus-Host Disease. Blood 2017, 129, 927–933. [Google Scholar] [CrossRef]
- Peled, J.U.; Gomes, A.L.C.; Devlin, S.M.; Littmann, E.R.; Taur, Y.; Sung, A.D.; Weber, D.; Hashimoto, D.; Slingerland, A.E.; Slingerland, J.B.; et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2020, 382, 822–834. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, W.; Liang, Z.; Wang, J.; Zeng, Z.; Kołat, D.; Li, X.; Zhou, D.; Xu, X.; Zhao, L. Critical Role of the Gut Microbiota in Immune Responses and Cancer Immunotherapy. J. Hematol. Oncol. 2024, 17, 33. [Google Scholar] [CrossRef]
- Masetti, R.; Leardini, D.; Muratore, E.; Fabbrini, M.; D’Amico, F.; Zama, D.; Baccelli, F.; Gottardi, F.; Belotti, T.; Ussowicz, M.; et al. Gut Microbiota Diversity before Allogeneic Hematopoietic Stem Cell Transplantation as a Predictor of Mortality in Children. Blood 2023, 142, 1387–1398. [Google Scholar] [CrossRef]
- Mancini, N.; Greco, R.; Pasciuta, R.; Barbanti, M.C.; Pini, G.; Morrow, O.B.; Morelli, M.; Vago, L.; Clementi, N.; Giglio, F.; et al. Enteric Microbiome Markers as Early Predictors of Clinical Outcome in Allogeneic Hematopoietic Stem Cell Transplant: Results of a Prospective Study in Adult Patients. Open Forum Infect. Dis. 2017, 4, ofx215. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Jenq, R.R.; Perales, M.-A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The Effects of Intestinal Tract Bacterial Diversity on Mortality Following Allogeneic Hematopoietic Stem Cell Transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Payen, M.; Nicolis, I.; Robin, M.; Michonneau, D.; Delannoye, J.; Mayeur, C.; Kapel, N.; Berçot, B.; Butel, M.-J.; Le Goff, J.; et al. Functional and Phylogenetic Alterations in Gut Microbiome Are Linked to Graft-versus-Host Disease Severity. Blood Adv. 2020, 4, 1824–1832. [Google Scholar] [CrossRef] [PubMed]
- Golob, J.L.; Pergam, S.A.; Srinivasan, S.; Fiedler, T.L.; Liu, C.; Garcia, K.; Mielcarek, M.; Ko, D.; Aker, S.; Marquis, S.; et al. Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs. Host Disease After Hematopoietic Cell Transplantation. Clin. Infect. Dis. 2017, 65, 1984–1991. [Google Scholar] [CrossRef]
- Haak, B.W.; Littmann, E.R.; Chaubard, J.-L.; Pickard, A.J.; Fontana, E.; Adhi, F.; Gyaltshen, Y.; Ling, L.; Morjaria, S.M.; Peled, J.U.; et al. Impact of Gut Colonization with Butyrate Producing Microbiota on Respiratory Viral Infection Following Allo-HCT. Blood 2018, 132, 2978–2986. [Google Scholar] [CrossRef]
- Jenq, R.R.; Taur, Y.; Devlin, S.M.; Ponce, D.M.; Goldberg, J.D.; Ahr, K.F.; Littmann, E.R.; Ling, L.; Gobourne, A.C.; Miller, L.C.; et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2015, 21, 1373–1383. [Google Scholar] [CrossRef]
- Meedt, E.; Hiergeist, A.; Gessner, A.; Dettmer, K.; Liebisch, G.; Ghimire, S.; Poeck, H.; Edinger, M.; Wolff, D.; Herr, W.; et al. Prolonged Suppression of Butyrate-Producing Bacteria Is Associated with Acute Gastrointestinal Graft-vs.-Host Disease and Transplantation-Related Mortality After Allogeneic Stem Cell Transplantation. Clin. Infect. Dis. 2022, 74, 614–621. [Google Scholar] [CrossRef]
- Gu, Z.; Xiong, Q.; Wang, L.; Wang, L.; Li, F.; Hou, C.; Dou, L.; Zhu, B.; Liu, D. The Impact of Intestinal Microbiota in Antithymocyte Globulin–Based Myeloablative Allogeneic Hematopoietic Cell Transplantation. Cancer 2022, 128, 1402–1410. [Google Scholar] [CrossRef]
- Chhabra, S.; Szabo, A.; Clurman, A.; McShane, K.; Waters, N.; Eastwood, D.; Samanas, L.; Fei, T.; Armijo, G.; Abedin, S.; et al. Mitigation of Gastrointestinal Graft-versus-Host Disease with Tocilizumab Prophylaxis Is Accompanied by Preservation of Microbial Diversity and Attenuation of Enterococcal Domination. Haematologica 2022, 108, 250–256. [Google Scholar] [CrossRef]
- Stein-Thoeringer, C.K.; Nichols, K.B.; Lazrak, A.; Docampo, M.D.; Slingerland, A.E.; Slingerland, J.B.; Clurman, A.G.; Armijo, G.; Gomes, A.L.C.; Shono, Y.; et al. Lactose Drives Enterococcus Expansion to Promote Graft-versus-Host Disease. Science 2019, 366, 1143–1149. [Google Scholar] [CrossRef]
- Fujimoto, K.; Hayashi, T.; Yamamoto, M.; Sato, N.; Shimohigoshi, M.; Miyaoka, D.; Yokota, C.; Watanabe, M.; Hisaki, Y.; Kamei, Y.; et al. An Enterococcal Phage-Derived Enzyme Suppresses Graft-versus-Host Disease. Nature 2024, 632, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Xavier, J.B.; Lipuma, L.; Ubeda, C.; Goldberg, J.; Gobourne, A.; Lee, Y.J.; Dubin, K.A.; Socci, N.D.; Viale, A.; et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin. Infect. Dis. 2012, 55, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.A.; Tan, C.Y.; Ren, Y.; Hill, L.; Bush, A.; Lew, M.; Andermann, T.; Peled, J.U.; Gomes, A.; Van Den Brink, M.R.M.; et al. Enterococcus Intestinal Domination Is Associated with Increased Mortality in the Acute Leukemia Chemotherapy Population. Clin. Infect. Dis. 2024, 78, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Kusakabe, S.; Fukushima, K.; Yokota, T.; Hino, A.; Fujita, J.; Motooka, D.; Nakamura, S.; Shibayama, H.; Kanakura, Y. Enterococcus: A Predictor of Ravaged Microbiota and Poor Prognosis after Allogeneic Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2020, 26, 1028–1033. [Google Scholar] [CrossRef]
- Harris, A.C.; Young, R.; Devine, S.; Hogan, W.J.; Ayuk, F.; Bunworasate, U.; Chanswangphuwana, C.; Efebera, Y.A.; Holler, E.; Litzow, M.; et al. International, Multicenter Standardization of Acute Graft-versus-Host Disease Clinical Data Collection: A Report from the Mount Sinai Acute GVHD International Consortium. Biol. Blood Marrow Transplant. 2016, 22, 4–10. [Google Scholar] [CrossRef]
- Hsieh, F.Y.; Lavori, P.W. Sample-Size Calculations for the Cox Proportional Hazards Regression Model with Nonbinary Covariates. Control Clin. Trials 2000, 21, 552–560. [Google Scholar] [CrossRef]
- Álvares-da-Silva, M.R.; Oliveira, C.P.; Fagan, A.; Longo, L.; Thoen, R.U.; Yoshimura Zitelli, P.M.; Tanaka Ferreira, R.M.; Mcgeorge, S.; Shamsaddini, A.; Farias, A.Q.; et al. Interaction of Microbiome, Diet, and Hospitalizations Between Brazilian and American Patients with Cirrhosis. Clin. Gastroenterol. Hepatol. 2022, 20, 930–940. [Google Scholar] [CrossRef]
- Dogra, S.K.; Doré, J.; Damak, S. Gut Microbiota Resilience: Definition, Link to Health and Strategies for Intervention. Front. Microbiol. 2020, 11, 572921. [Google Scholar] [CrossRef]
- Safarchi, A.; Al-Qadami, G.; Tran, C.D.; Conlon, M. Understanding Dysbiosis and Resilience in the Human Gut Microbiome: Biomarkers, Interventions, and Challenges. Front. Microbiol. 2025, 16, 1559521. [Google Scholar] [CrossRef]
- Tresoldi, A.T.; Cardoso, L.G.O.; Castilho, G.V.; Dantas, S.R.P.E.; Von Nowakonski, A.; Pereira, R.M.; Trabasso, P. Low Prevalence of Vancomycin Resistant Enterococci Colonization in Intensive Care Patients in a Brazilian Teaching Hospital. Braz. J. Infect. Dis. 2006, 10, 239–241. [Google Scholar] [CrossRef]
- MetaHIT Consortium (additional members); Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Huttenhower, G.; Gevers, D.; Knight, R.; Abubucker, S.; Hallsworth-Pepin, P.; Madupu, R.; Magrini, V.; Mitrev, M.; Versalovic, J.; Zeng, O.; et al. The Human Microbiome Project Consortium Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Li, P.; Lei, Q.; Yu, X.; Shen, Y.; Chen, Y.; Hou, C.; Hu, B.; Cui, Y.; Liu, Z.; Qin, Y.; et al. Commensal Bacteroides T6SS Alleviate GI-aGVHD via Mediating Gut Microbiota Composition and Bile Acids Metabolism. Gut 2025, 74, 334565. [Google Scholar] [CrossRef] [PubMed]
- Hayase, E.; Hayase, T.; Mukherjee, A.; Stinson, S.C.; Jamal, M.A.; Ortega, M.R.; Sanchez, C.A.; Ahmed, S.S.; Karmouch, J.L.; Chang, C.-C.; et al. Bacteroides Ovatus Alleviates Dysbiotic Microbiota-Induced Graft-versus-Host Disease. Cell Host Microbe 2024, 32, 1621–1636.e6. [Google Scholar] [CrossRef] [PubMed]
- Sofi, M.H.; Wu, Y.; Ticer, T.; Schutt, S.; Bastian, D.; Choi, H.-J.; Tian, L.; Mealer, C.; Liu, C.; Westwater, C.; et al. A Single Strain of Bacteroides Fragilis Protects Gut Integrity and Reduces GVHD. JCI Insight 2021, 6, e136841. [Google Scholar] [CrossRef]
- Facchin, S.; Calgaro, M.; Savarino, E.V. Rethinking Short-Chain Fatty Acids: A Closer Look at Propionate in Inflammation, Metabolism, and Mucosal Homeostasis. Cells 2025, 14, 1130. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nagai, F.; Morotomi, M. Characterization of Phascolarctobacterium succinatutens Sp. Nov., an Asaccharolytic, Succinate-Utilizing Bacterium Isolated from Human Feces. Appl. Env. Microbiol. 2012, 78, 511–518. [Google Scholar] [CrossRef]
- Li, W.; Li, L.; Yang, F.; Hu, Q.; Xiong, D. Correlation between Gut Bacteria Phascolarctobacterium and Exogenous Metabolite α-Linolenic Acid in T2DM: A Case-Control Study. Ann. Transl. Med. 2022, 10, 1056. [Google Scholar] [CrossRef]
- Bacigalupo, A.; Ballen, K.; Rizzo, D.; Giralt, S.; Lazarus, H.; Ho, V.; Apperley, J.F.; Slavin, S.; Pasquini, M.C.; Sandmaier, B.M.; et al. Defining the intensity of conditioning regimens: Working definitions. Biol. Blood Marrow Transplant. 2009, 15, 1628–1633. [Google Scholar] [CrossRef]


| Variable | No-Intestinal Domination (n = 15) | Yes-Intestinal Domination (n = 54) | Total (N = 69) | p-Value |
|---|---|---|---|---|
| Age (years) | 0.6 | |||
| Mean (SD) | 44 (19) | 41 (15) | 40 (16) | |
| Median (IQR) | 40 (27–68) | 42 (31–51) | 40 (28–49) | |
| Range | 18–74 | 12–71 | 12–73 | |
| Weight (kg) | 0.12 | |||
| Mean (SD) | 79 (16) | 72 (17) | 74 (17) | |
| Median (IQR) | 79 (68–87) | 68 (62–82) | 74 (64–82) | |
| Range | 48–110 | 43–130 | 43–130 | |
| Height (cm) | 0.091 | |||
| Mean (SD) | 170 (10) | 166 (10) | 167 (10) | |
| Median (IQR) | 173 (160–178) | 164 (158–174) | 165 (160–175) | |
| Range | 150–185 | 146–189 | 146–189 | |
| Center | 0.2 | |||
| HB-FUNFARME | 4 (27%) | 5 (9.3%) | 9 (13%) | |
| HAC | 4 (27%) | 18 (33%) | 22 (32%) | |
| HCB | 0 (0%) | 8 (15%) | 8 (12%) | |
| BP | 7 (47%) | 23 (43%) | 30 (43%) | |
| Sex | 0.2 | |||
| Male | 9 (60%) | 22 (41%) | 31 (45%) | |
| Female | 6 (40%) | 32 (59%) | 38 (55%) | |
| Prior Allo-HSCT | 0 (0%) | 3 (6.4%) | 3 (5.1%) | >0.9 |
| Primary Disease | 0.4 | |||
| Acute Myeloid Leukemia | 6 (40%) | 23 (43%) | 29 (42%) | |
| Acute Lymphoid Leukemia | 1 (6.7%) | 15 (28%) | 16 (23%) | |
| Chronic Myeloid Leukemia | 1 (6.7%) | 2 (3.7%) | 3 (4.3%) | |
| Hodgkin’s Lymphoma | 1 (6.7%) | 1 (1.9%) | 2 (2.9%) | |
| Non-Hodgkin’s Lymphoma | 0 (0%) | 1 (1.9%) | 1 (1.4%) | |
| Aplastic Anemia | 2 (13%) | 5 (9.3%) | 7 (10%) | |
| Sickle Cell Disease | 1 (6.7%) | 2 (3.7%) | 3 (4.3%) | |
| Other | 3 (20%) | 5 (9.3%) | 8 (12%) | |
| Stem Cell Source | 0.4 | |||
| Peripheral Blood | 9 (60%) | 39 (72%) | 48 (70%) | |
| Bone Marrow | 6 (40%) | 15 (28%) | 21 (30%) | |
| Donor Type | 0.11 | |||
| Matched Related | 3 (20%) | 17 (31%) | 20 (29%) | |
| Matched Unrelated | 4 (27%) | 3 (5.6%) | 7 (10%) | |
| Mismatched Related | 0 (0%) | 3 (5.6%) | 3 (4.3%) | |
| Haploidentical | 7 (47%) | 30 (56%) | 37 (54%) | |
| Mismatched Unrelated | 1 (6.7%) | 1 (1.9%) | 2 (2.9%) | |
| Donor Sex | 0.4 | |||
| Male | 7 (47%) | 33 (61%) | 40 (58%) | |
| Female | 8 (53%) | 21 (39%) | 29 (42%) | |
| Intensity of Conditioning Regimen | 0.8 | |||
| Ablative | 4 (27%) | 21 (39%) | 25 (36%) | |
| Reduced Intensity | 7 (47%) | 20 (37%) | 27 (39%) | |
| Nonmyeloablative | 4 (27%) | 12 (22%) | 16 (23%) | |
| TBI-Conditioning Regimen | 6 (40%) | 29 (54%) | 35 (51%) | 0.3 |
| Any Genus | Bacteroides | Akkermansia | Phascolarctobacterium | Escherichia-Shigella | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | |
| Age | 0.99 (0.95–1.02) | 0.46 | 0.99 (0.96–1.02) | 0.67 | 0.99 (0.96–1.03) | 0.73 | 1.03 (0.99–1.08) | 0.18 | 1.00 (0.96–1.04) | 0.88 |
| BMI | 0.98 (0.88–1.09) | 0.69 | 1.01 (0.92–1.11) | 0.81 | 0.99 (0.89–1.11) | 0.90 | 1.05 (0.93–1.20) | 0.41 | 0.94 (0.82–1.06) | 0.32 |
| Sex | ||||||||||
| Female | - | - | - | - | - | - | - | - | - | - |
| Male | 0.46 (0.14–1.45) | 0.19 | 0.65 (0.24–1.75) | 0.40 | 0.41 (0.10–1.41) | 0.18 | 0.31 (0.04–1.39) | 0.16 | 0.85 (0.23–2.99) | 0.80 |
| Conditioning regimen | ||||||||||
| Reduced intensity | - | - | - | - | - | - | - | - | - | - |
| Myeloablative | 1.75 (0.46–7.53) | 0.42 | 1.20 (0.39–3.69) | 0.75 | 2.33 (0.61–10.1) | 0.23 | 1.14 (0.19–6.70) | 0.88 | 1.14 (0.24–5.39) | 0.86 |
| Non-myeloablative | 1.00 (0.25–4.47) | >0.99 | 1.08 (0.29–3.85) | 0.91 | 1.38 (0.24–7.24) | 0.70 | 1.92 (0.32–11.7) | 0.46 | 2.00 (0.41–9.87) | 0.38 |
| TBI-Conditioning Regimen | 1.74 (0.55–5.84) | 0.35 | 1.57 (0.59–4.26) | 0.37 | 1.38 (0.43–4.70) | 0.59 | 1.25 (0.30–5.48) | 0.76 | 0.97 (0.27–3.43) | 0.96 |
| Stem cell source | ||||||||||
| Peripheral | - | - | - | - | - | - | - | - | - | - |
| Bone marrow | 0.58 (0.18–1.98) | 0.37 | 1.03 (0.35–2.93) | 0.96 | 1.35 (0.37–4.58) | 0.63 | 0.25 (0.01–1.50) | 0.21 | 1.83 (0.48–6.60) | 0.36 |
| Underlying diagnosis | ||||||||||
| Acute leukemia | - | - | - | - | - | - | - | - | - | - |
| Others | 0.40 (0.12–1.33) | 0.13 | 0.76 (0.25–2.20) | 0.62 | 0.89 (0.22–3.10) | 0.87 | 0.25 (0.01–1.50) | 0.21 | 1.18 (0.28–4.29) | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares Ferreira Junior, A.; Amanda Niz Alvarez, D.; da Silva Souza, L.; Linares Silva, N.; Dias Machado, L.; Yoshio Hirai, W.; Mesquita Ciconelli, R.; Piccolo Feliciano, J.V.; Colturato, I.; Maurício Navarro Barros, G.; et al. A Distinct Intestinal Domination Fingerprint in Patients Undergoing Allo-HSCT: Dynamics, Predictors and Implications on Clinical Outcomes. J. Clin. Med. 2025, 14, 8351. https://doi.org/10.3390/jcm14238351
Soares Ferreira Junior A, Amanda Niz Alvarez D, da Silva Souza L, Linares Silva N, Dias Machado L, Yoshio Hirai W, Mesquita Ciconelli R, Piccolo Feliciano JV, Colturato I, Maurício Navarro Barros G, et al. A Distinct Intestinal Domination Fingerprint in Patients Undergoing Allo-HSCT: Dynamics, Predictors and Implications on Clinical Outcomes. Journal of Clinical Medicine. 2025; 14(23):8351. https://doi.org/10.3390/jcm14238351
Chicago/Turabian StyleSoares Ferreira Junior, Alexandre, Danielle Amanda Niz Alvarez, Larissa da Silva Souza, Nathalia Linares Silva, Luiza Dias Machado, Welinton Yoshio Hirai, Rozana Mesquita Ciconelli, Joao Victor Piccolo Feliciano, Iago Colturato, George Maurício Navarro Barros, and et al. 2025. "A Distinct Intestinal Domination Fingerprint in Patients Undergoing Allo-HSCT: Dynamics, Predictors and Implications on Clinical Outcomes" Journal of Clinical Medicine 14, no. 23: 8351. https://doi.org/10.3390/jcm14238351
APA StyleSoares Ferreira Junior, A., Amanda Niz Alvarez, D., da Silva Souza, L., Linares Silva, N., Dias Machado, L., Yoshio Hirai, W., Mesquita Ciconelli, R., Piccolo Feliciano, J. V., Colturato, I., Maurício Navarro Barros, G., Scheinberg, P., Chao, N. J. A., & Lelis Vilela de Oliveira, G. (2025). A Distinct Intestinal Domination Fingerprint in Patients Undergoing Allo-HSCT: Dynamics, Predictors and Implications on Clinical Outcomes. Journal of Clinical Medicine, 14(23), 8351. https://doi.org/10.3390/jcm14238351







