Salivary MicroRNAs as Innovative Biomarkers for Diagnosis and Prediction of the Oral Potentially Malignant Disorders Transition Towards Oral Cancer: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
- (1)
- Studies that analyzed the salivary microRNAs expression in OPMD and oral cancer patients;
- (2)
- Text available in full format;
- (3)
- Articles written in English;
- (4)
- Respecting the structure of IMRAD (introduction, material and method, results, discussions);
- (1)
- Reviews, meta-analysis, case reports, case series, opinion articles and letters to the editor;
- (2)
- Articles that incorporate various forms of treatment administration for OPMD a priori to saliva sample acquisition;
- (3)
- Studies that include patients with other oral cavity tumors (non-OSCC) or in other regions of the organism, patients with history or undergoing chemo/radiotherapy and patients with immunosuppression;
- (4)
- Duplicated publications;
- (5)
- Animal experiments;
2.2. Publication Search Strategy
2.3. Qualitative Evaluation
3. Results
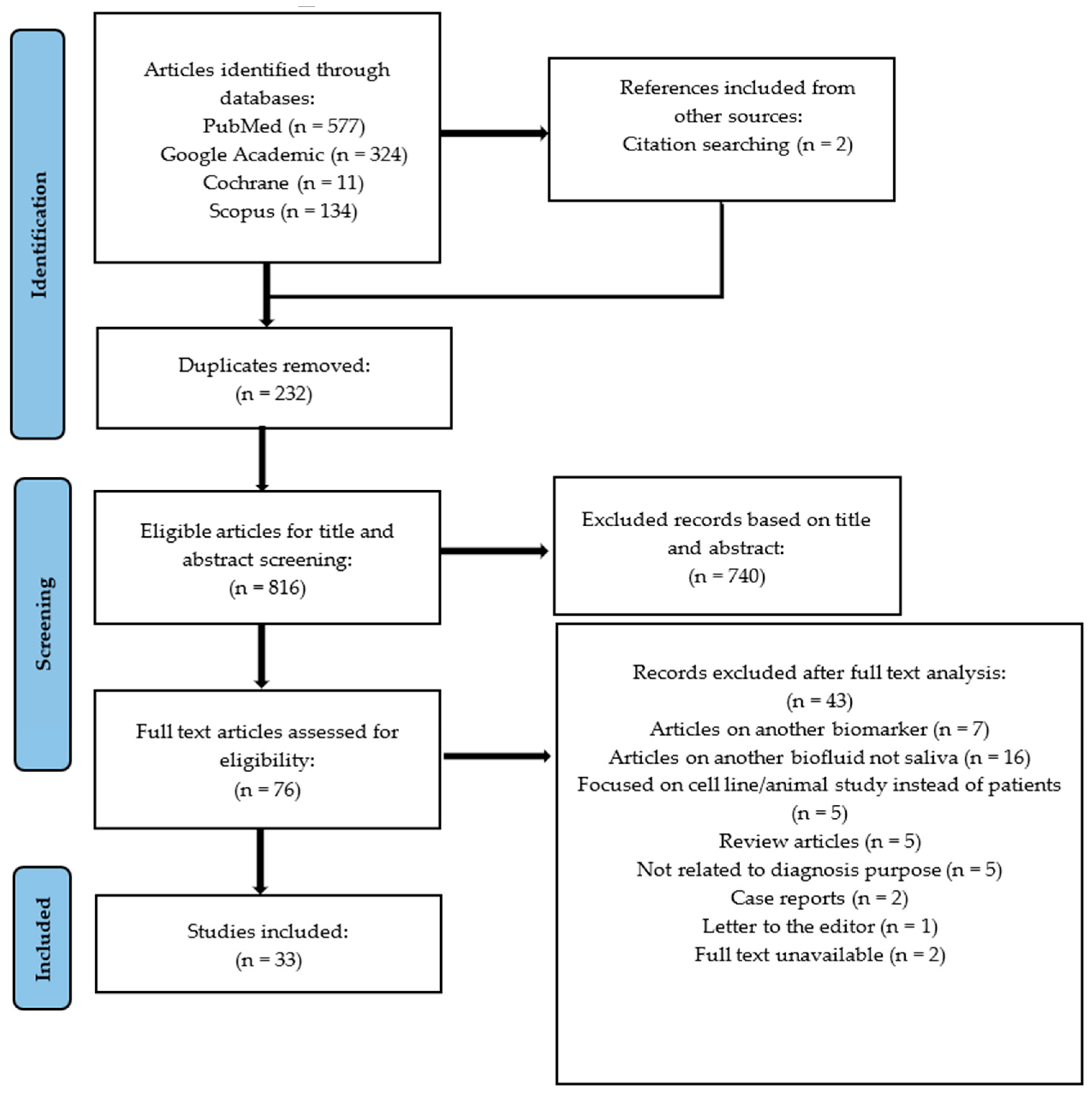
3.1. Literature Search and Study Selection
3.2. Characteristics of the Included Studies
3.3. Quality Assessment
3.4. Saliva Characteristics and Investigative Techniques
3.5. Salivary MicroRNAs Expression in OPMD vs. Healthy Controls
4. Discussions
4.1. Salivary MicroRNAs as Novel Biomarkers for Diagnosis of OPMD and OSCC
4.2. Salivary MicroRNAs Pathogenesis in Oral Premalignant Lesions Causing Malignant Transformation of OPMD
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OPMD | Oral potentially malignant diseases |
| QUADAS-2 | Diagnostic Accuracy Research Tool |
| OSCC | Oral squamous cell carcinoma |
| OSMF | Oral submucous fibrosis |
| OLP | Oral lichen planus |
| OLL | Oral lichenoid lesion |
| OED | Oral epithelial dysplasia |
| Interleukin-6 | Interleukin-6 |
| CRP | C-reactive protein |
| TME | Tumor microenvironment |
| ECM | Extracellular matrix |
| CAF | Cancer-associated fibroblasts |
| GRα | Glucocorticoid receptor α |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Haefner, M.F.; Lang, K.; Krug, D.; Koerber, S.A.; Uhlmann, L.; Kieser, M.; Debus, J.; Sterzing, F. Prognostic factors, patterns of recurrence and toxicity for patients with esophageal cancer undergoing definitive radiotherapy or chemo-radiotherapy. J. Radiat. Res. 2015, 56, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S.; Kujan, O.; Aguirre-Urizar, J.M.; Bagan, J.V.; Gonzalez-Moles, M.A.; Kerr, A.R.; Lodi, G.; Mello, F.W.; Monteiro, L.; Ogden, G.R.; et al. Oral potentially malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2021, 27, 1862–1880. [Google Scholar] [CrossRef]
- Rangel, R.; Pickering, C.R.; Sikora, A.G.; Spiotto, M.T. Genetic Changes Driving Immunosuppressive Microenvironments in Oral Premalignancy. Front. Immunol. 2022, 13, 840923. [Google Scholar] [CrossRef]
- Sathasivam, H.P.; Casement, J.; Bates, T.; Sloan, P.; Thomson, P.; Robinson, M.; Kist, R. Gene expression changes associated with malignant transformation of oral potentially malignant disorders. J. Oral Pathol. Med. 2021, 50, 60–67. [Google Scholar] [CrossRef]
- Deng, S.; Wang, S.; Shi, X.; Zhou, H. Microenvironment in Oral Potentially Malignant Disorders: Multi-Dimensional Characteristics and Mechanisms of Carcinogenesis. Int. J. Mol. Sci. 2022, 23, 8940. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Udaltsova, N.; Engels, E.A.; Katzel, J.A.; Yanik, E.L.; Katki, H.A.; Lingen, M.W.; Silverberg, M.J. Oral Leukoplakia and Risk of Progression to Oral Cancer: A Population-Based Cohort Study. J. Natl. Cancer Inst. 2020, 112, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, S.; Datta, S.; Ray, J.G.; Chaudhuri, K.; Chatterjee, R. Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 2019, 58, 137–145. [Google Scholar] [CrossRef]
- Sarode, G.; Maniyar, N.; Sarode, S.C.; Jafer, M.; Patil, S.; Awan, K.H. Epidemiologic aspects of oral cancer. Dis. Mon. 2020, 66, 100988. [Google Scholar] [CrossRef]
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral Cancer and Precancer: A Narrative Review on the Relevance of Early Diagnosis. Int. J. Environ. Res. Public Health 2020, 17, 9160. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef]
- Chamoli, A.; Gosavi, A.S.; Shirwadkar, U.P.; Wangdale, K.V.; Behera, S.K.; Kurrey, N.K.; Kalia, K.; Mandoli, A. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol. 2021, 121, 105451. [Google Scholar] [CrossRef]
- Osan, C.; Chira, S.; Nutu, A.M.; Braicu, C.; Baciut, M.; Korban, S.S.; Berindan-Neagoe, I. The Connection between MicroRNAs and Oral Cancer Pathogenesis: Emerging Biomarkers in Oral Cancer Management. Genes 2021, 12, 1989. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jiang, Y.; Singh, A.P.; Takeshita, F. MicroRNAs: Emerging novel targets of cancer therapies. BioMed Res. Int. 2015, 2015, 506323. [Google Scholar] [CrossRef]
- Elewaily, M.I.; Elsergany, A.R. Emerging role of exosomes and exosomal microRNA in cancer: Pathophysiology and clinical potential. J. Cancer Res. Clin. Oncol. 2021, 147, 637–648. [Google Scholar] [CrossRef]
- Tastan, B.; Tarakcioglu, E.; Birinci, Y.; Park, Y.; Genc, S. Role of Exosomal MicroRNAs in Cell-to-Cell Communication. Methods Mol. Biol. 2022, 2257, 269–292. [Google Scholar]
- Mariner, P.D.; Korst, A.; Karimpour-Fard, A.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Improved Detection of Circulating miRNAs in Serum and Plasma Following Rapid Heat/Freeze Cycling. Microrna 2018, 7, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, S.; Liu, X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol. Lett. 2019, 17, 5601–5607. [Google Scholar] [CrossRef]
- Hagiwara, K.; Katsuda, T.; Gailhouste, L.; Kosaka, N.; Ochiya, T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015, 589, 4071–4078. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Axmann, M.; Meier, S.M.; Karner, A.; Strobl, W.; Stangl, H.; Plochberger, B. Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes 2018, 9, 533. [Google Scholar] [CrossRef]
- Chong, Z.X.; Yeap, S.K.; Ho, W.Y. Roles of circulating microRNA(s) in human breast cancer. Arch. Biochem. Biophys. 2020, 695, 108583. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Group, P.-P. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; Group, Q. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Trachtenberg, A.J.; Kuo, W.P.; Cheng, Y.S. Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer. J. Dent. Res. 2014, 93 (Suppl 7), 86S–93S. [Google Scholar] [CrossRef]
- Lundegard, M.; Nylander, K.; Danielsson, K. Difficulties detecting miRNA-203 in human whole saliva by the use of PCR. Med. Oral Patol. Oral Cir. Bucal 2015, 20, e130–e134. [Google Scholar] [CrossRef] [PubMed]
- Zahran, F.; Ghalwash, D.; Shaker, O.; Al-Johani, K.; Scully, C. Salivary microRNAs in oral cancer. Oral Dis. 2015, 21, 739–747. [Google Scholar] [CrossRef]
- Byun, J.S.; Hong, S.H.; Choi, J.K.; Jung, J.K.; Lee, H.J. Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients. Oral Dis. 2015, 21, 987–993. [Google Scholar] [CrossRef]
- Hung, K.F.; Liu, C.J.; Chiu, P.C.; Lin, J.S.; Chang, K.W.; Shih, W.Y.; Kao, S.Y.; Tu, H.F. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016, 53, 42–47. [Google Scholar] [CrossRef]
- Duz, M.B.; Karatas, O.F.; Guzel, E.; Turgut, N.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: A pilot study. Cell. Oncol. 2016, 39, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, M.; Jafari, S.; Barati, M.; Mahdipour, M.; Gholami, M.S. Predictive value of salivary microRNA-320a, vascular endothelial growth factor receptor 2, CRP and IL-6 in Oral lichen planus progression. Inflammopharmacology 2017, 25, 577–583. [Google Scholar] [CrossRef]
- Maheswari, T.N.U.; Nivedhitha, M.S. Study to explore the significance of saliva as a diagnostic tool to detect microRNA in oral potentially malignant disorders. J. Adv. Pharm. Educ. Res. 2017, 7, 278–282. [Google Scholar]
- Aghbari, S.M.H.; Gaafar, S.M.; Shaker, O.G.; Ashiry, S.E.; Zayed, S.O. Evaluating the accuracy of microRNA27b and microRNA137 as biomarkers of activity and potential malignant transformation in oral lichen planus patients. Arch. Dermatol. Res. 2018, 310, 209–220. [Google Scholar] [CrossRef]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Patel, S.; Modi, B.; Shah, F.; Rawal, R. Uncovering the potential of CD44v/SYNE1/miR34a axis in salivary fluids of oral cancer patients. J. Oral Pathol. Med. 2018, 47, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Mehdipour, M.; Shahidi, M.; Manifar, S.; Jafari, S.; Mashhadi Abbas, F.; Barati, M.; Mortazavi, H.; Shirkhoda, M.; Farzanegan, A.; Elmi Rankohi, Z. Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus—A short report. Cell. Oncol. 2018, 41, 329–334. [Google Scholar] [CrossRef]
- Yap, T.; Koo, K.; Cheng, L.; Vella, L.J.; Hill, A.F.; Reynolds, E.; Nastri, A.; Cirillo, N.; Seers, C.; McCullough, M. Predicting the Presence of Oral Squamous Cell Carcinoma Using Commonly Dysregulated MicroRNA in Oral Swirls. Cancer Prev. Res. 2018, 11, 491–502. [Google Scholar] [CrossRef]
- Yap, T.; Seers, C.; Koo, K.; Cheng, L.; Vella, L.J.; Hill, A.F.; Reynolds, E.; Nastri, A.; Cirillo, N.; McCullough, M. Non-invasive screening of a microRNA-based dysregulation signature in oral cancer and oral potentially malignant disorders. Oral Oncol. 2019, 96, 113–120. [Google Scholar] [CrossRef]
- Uma Maheswari, T.N.; Nivedhitha, M.S.; Ramani, P. Expression profile of salivary micro RNA-21 and 31 in oral potentially malignant disorders. Braz. Oral Res. 2020, 34, e002. [Google Scholar] [CrossRef]
- Prasad, S.R.; Pai, A.; Shyamala, K.; Yaji, A. Expression of Salivary miRNA 21 in Oral Submucous Fibrosis (OSMF): An Observational Study. Microrna 2020, 9, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.J.; You, G.R.; Lee, C.J.; Lu, Y.C.; Tang, S.J.; Huang, Y.F.; Huang, Y.C.; Lee, L.Y.; Fan, K.H.; Chen, Y.C.; et al. Systemic Investigation Identifying Salivary miR-196b as a Promising Biomarker for Early Detection of Head-Neck Cancer and Oral Precancer Lesions. Diagnostics 2021, 11, 1411. [Google Scholar] [CrossRef] [PubMed]
- Romani, C.; Salviato, E.; Paderno, A.; Zanotti, L.; Ravaggi, A.; Deganello, A.; Berretti, G.; Gualtieri, T.; Marchini, S.; D’Incalci, M.; et al. Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma. Theranostics 2021, 11, 2987–2999. [Google Scholar] [CrossRef]
- Mehterov, N.; Vladimirov, B.; Sacconi, A.; Pulito, C.; Rucinski, M.; Blandino, G.; Sarafian, V. Salivary miR-30c-5p as Potential Biomarker for Detection of Oral Squamous Cell Carcinoma. Biomedicines 2021, 9, 1079. [Google Scholar] [CrossRef]
- Kumari, P.; Syed, S.A.; Wahid, M.; Qureshi, M.A.; Kumar, R. Expression of miR-31 in saliva-liquid biopsy in patients with oral squamous cell carcinoma. J. Taibah Univ. Med. Sci. 2021, 16, 733–739. [Google Scholar] [CrossRef]
- Khan, S.; Butt, S.A.; Hassan, S.; Khan, R. Expression of Salivary miRNA-31 in Oral Submucous fibrosis. J. Adv. Med. Med. Res. 2021, 33, 137–144. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, H.; Li, L.; Wang, K. Clinical significance of miR-142-3p in oral lichen planus and its regulatory role in keratinocyte proliferation. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2021, 132, 441–447. [Google Scholar] [CrossRef]
- Tu, H.F.; Lin, L.H.; Chang, K.W.; Cheng, H.W.; Liu, C.J. Exploiting salivary miR-375 as a clinical biomarker of oral potentially malignant disorder. J. Dent. Sci. 2022, 17, 659–665. [Google Scholar] [CrossRef]
- Di Stasio, D.; Romano, A.; Boschetti, C.E.; Montella, M.; Mosca, L.; Lucchese, A. Salivary miRNAs Expression in Potentially Malignant Disorders of the Oral Mucosa and Oral Squamous Cell Carcinoma: A Pilot Study on miR-21, miR-27b, and miR-181b. Cancers 2022, 15, 291. [Google Scholar] [CrossRef]
- Scholtz, B.; Horvath, J.; Tar, I.; Kiss, C.; Marton, I.J. Salivary miR-31-5p, miR-345-3p, and miR-424-3p Are Reliable Biomarkers in Patients with Oral Squamous Cell Carcinoma. Pathogens 2022, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Patel, S.; Patel, P.; Mandlik, D.; Patel, K.; Tanavde, V. Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients. Int. J. Mol. Sci. 2022, 23, 10639. [Google Scholar] [CrossRef]
- Faur, C.I.; Roman, R.C.; Jurj, A.; Raduly, L.; Almasan, O.; Rotaru, H.; Chirila, M.; Moldovan, M.A.; Hedesiu, M.; Dinu, C. Salivary Exosomal MicroRNA-486-5p and MicroRNA-10b-5p in Oral and Oropharyngeal Squamous Cell Carcinoma. Medicina 2022, 58, 1478. [Google Scholar] [CrossRef]
- Mehdipour, M.; Shahidi, M.; Anbari, F.; Mirzaei, H.; Jafari, S.; Kholghi, A.; Lotfi, E.; Manifar, S.; Mashhadiabbas, F. Salivary level of microRNA-146a and microRNA-155 biomarkers in patients with oral lichen planus versus oral squamous cell carcinoma. BMC Oral Health 2023, 23, 433. [Google Scholar] [CrossRef]
- Garg, A.; Urs, A.B.; Koner, B.C.; Augustine, J.; Guru, S.A. Evaluation of Diagnostic Significance of Salivary miRNA-184 and miRNA-21 in Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders. Head Neck Pathol. 2023, 17, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Tarrad, N.A.F.; Hassan, S.; Shaker, O.G.; AbdelKawy, M. Salivary LINC00657 and miRNA-106a as diagnostic biomarkers for oral squamous cell carcinoma, an observational diagnostic study. BMC Oral Health 2023, 23, 994. [Google Scholar] [CrossRef] [PubMed]
- Farshbaf, A.; Mohajertehran, F.; Aghaee-Bakhtiari, S.H.; Ayatollahi, H.; Douzandeh, K.; Pakfetrat, A.; Mohtasham, N. Downregulation of salivary miR-3928 as a potential biomarker in patients with oral squamous cell carcinoma and oral lichen planus. Clin. Exp. Dent. Res. 2024, 10, e877. [Google Scholar] [CrossRef] [PubMed]
- Rocchetti, F.; Tenore, G.; Macali, F.; Vicidomini, T.; Podda, G.M.; Fantozzi, P.J.; Silvestri, V.; Porzio, V.; Valentini, V.; Ottini, L.; et al. Expression Analysis of Circulating microRNAs in Saliva and Plasma for the Identification of Clinically Relevant Biomarkers for Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders. Cancers 2024, 16, 2990. [Google Scholar] [CrossRef]
- MM, E.L.; Korien, I.A.; Rashwan, W.A.; Shaker, O.G. The oncogenic potential of salivary microRNA-93 and microRNA-412-3p in oral lichen planus: A case-control study. BDJ Open 2024, 10, 98. [Google Scholar]
- Carpenter, G.H. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Chiamulera, M.M.A.; Zancan, C.B.; Remor, A.P.; Cordeiro, M.F.; Gleber-Netto, F.O.; Baptistella, A.R. Salivary cytokines as biomarkers of oral cancer: A systematic review and meta-analysis. BMC Cancer 2021, 21, 205. [Google Scholar] [CrossRef]
- Aziz, S.; Ahmed, S.S.; Ali, A.; Khan, F.A.; Zulfiqar, G.; Iqbal, J.; Khan, A.A.; Shoaib, M. Salivary Immunosuppressive Cytokines IL-10 and IL-13 Are Significantly Elevated in Oral Squamous Cell Carcinoma Patients. Cancer Invest 2015, 33, 318–328. [Google Scholar] [CrossRef]
- Coon, J.; Kingsley, K.; Howard, K.M. miR-365 (microRNA): Potential Biomarker in Oral Squamous Cell Carcinoma Exosomes and Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 5317. [Google Scholar] [CrossRef]
- Manzano-Moreno, F.J.; Costela-Ruiz, V.J.; Garcia-Recio, E.; Olmedo-Gaya, M.V.; Ruiz, C.; Reyes-Botella, C. Role of Salivary MicroRNA and Cytokines in the Diagnosis and Prognosis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 12215. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Gomar-Vercher, S.; Simon-Soro, A.; Montiel-Company, J.M.; Almerich-Silla, J.M.; Mira, A. Stimulated and unstimulated saliva samples have significantly different bacterial profiles. PLoS ONE 2018, 13, e0198021. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.; Vella, L.J.; Seers, C.; Nastri, A.; Reynolds, E.; Cirillo, N.; McCullough, M. Oral swirl samples—A robust source of microRNA protected by extracellular vesicles. Oral Dis. 2017, 23, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef]
- Chen, Y.F.; Wei, Y.Y.; Yang, C.C.; Liu, C.J.; Yeh, L.Y.; Chou, C.H.; Chang, K.W.; Lin, S.C. miR-125b suppresses oral oncogenicity by targeting the anti-oxidative gene PRXL2A. Redox Biol. 2019, 22, 101140. [Google Scholar] [CrossRef]
- Speight, P.M.; Khurram, S.A.; Kujan, O. Oral potentially malignant disorders: Risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 612–627. [Google Scholar] [CrossRef]
- Iocca, O.; Sollecito, T.P.; Alawi, F.; Weinstein, G.S.; Newman, J.G.; De Virgilio, A.; Di Maio, P.; Spriano, G.; Pardinas Lopez, S.; Shanti, R.M. Potentially malignant disorders of the oral cavity and oral dysplasia: A systematic review and meta-analysis of malignant transformation rate by subtype. Head Neck 2020, 42, 539–555. [Google Scholar] [CrossRef]
- Bugshan, A.; Farooq, I. Oral squamous cell carcinoma: Metastasis, potentially associated malignant disorders, etiology and recent advancements in diagnosis. F1000Research 2020, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kim, J.W.; Paeng, J.Y. Clinical analysis of neck node metastasis in oral cavity cancer. J. Korean Assoc. Oral Maxillofac. Surg. 2018, 44, 282–288. [Google Scholar] [CrossRef]
- Mallinger, W.D.; Quick, C.M. Benign and Premalignant Lesions of the Endometrium. Surg. Pathol. Clin. 2019, 12, 315–328. [Google Scholar] [CrossRef]
- Denisov, E.V.; Schegoleva, A.A.; Gervas, P.A.; Ponomaryova, A.A.; Tashireva, L.A.; Boyarko, V.V.; Bukreeva, E.B.; Pankova, O.V.; Perelmuter, V.M. Premalignant lesions of squamous cell carcinoma of the lung: The molecular make-up and factors affecting their progression. Lung Cancer 2019, 135, 21–28. [Google Scholar] [CrossRef]
- Ali, M.; Gupta, G.; Silu, M.; Chand, D.; Samor, V. Narrow band imaging in early diagnosis of laryngopharyngeal malignant and premalignant lesions. Auris Nasus Larynx 2022, 49, 676–679. [Google Scholar] [CrossRef]
- Woo, S.B. Oral Epithelial Dysplasia and Premalignancy. Head Neck Pathol. 2019, 13, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Dhanuthai, K.; Rojanawatsirivej, S.; Thosaporn, W.; Kintarak, S.; Subarnbhesaj, A.; Darling, M.; Kryshtalskyj, E.; Chiang, C.P.; Shin, H.I.; Choi, S.Y.; et al. Oral cancer: A multicenter study. Med. Oral Patol. Oral Cir. Bucal 2018, 23, e23–e29. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, D.; Huber, M.A.; Kerr, A.R. Oral Potentially Malignant Disorders and Oral Cavity Cancer. Dermatol. Clin. 2020, 38, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Matse, J.H.; Yoshizawa, J.; Wang, X.; Elashoff, D.; Bolscher, J.G.; Veerman, E.C.; Leemans, C.R.; Pegtel, D.M.; Wong, D.T.; Bloemena, E. Human Salivary Micro-RNA in Patients with Parotid Salivary Gland Neoplasms. PLoS ONE 2015, 10, e0142264, Correction in PLoS ONE 2016, 11, e0146701. [Google Scholar] [CrossRef]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef]
- Machida, T.; Tomofuji, T.; Maruyama, T.; Yoneda, T.; Ekuni, D.; Azuma, T.; Miyai, H.; Mizuno, H.; Kato, H.; Tsutsumi, K.; et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 2016, 36, 2375–2381. [Google Scholar] [CrossRef]
- Liu, C.J.; Lin, S.C.; Yang, C.C.; Cheng, H.W.; Chang, K.W. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 2012, 34, 219–224. [Google Scholar] [CrossRef]
- Brito, J.A.; Gomes, C.C.; Guimaraes, A.L.; Campos, K.; Gomez, R.S. Relationship between microRNA expression levels and histopathological features of dysplasia in oral leukoplakia. J. Oral Pathol. Med. 2014, 43, 211–216. [Google Scholar] [CrossRef]
- Yap, T.; Celentano, A.; Seers, C.; McCullough, M.J.; Farah, C.S. Molecular diagnostics in oral cancer and oral potentially malignant disorders—A clinician’s guide. J. Oral Pathol. Med. 2020, 49, 1–8. [Google Scholar] [CrossRef]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Peacock, O.; Lee, A.C.; Cameron, F.; Tarbox, R.; Vafadar-Isfahani, N.; Tufarelli, C.; Lund, J.N. Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS ONE 2014, 9, e110267. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Okayama, H.; Schetter, A.J.; Harris, C.C. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig. Dis. 2012, 30 (Suppl. 2), 9–15. [Google Scholar] [CrossRef]
- Spannuth, W.A.; Nick, A.M.; Jennings, N.B.; Armaiz-Pena, G.N.; Mangala, L.S.; Danes, C.G.; Lin, Y.G.; Merritt, W.M.; Thaker, P.H.; Kamat, A.A.; et al. Functional significance of VEGFR-2 on ovarian cancer cells. Int. J. Cancer 2009, 124, 1045–1053. [Google Scholar] [CrossRef]
- Silva, S.R.; Bowen, K.A.; Rychahou, P.G.; Jackson, L.N.; Weiss, H.L.; Lee, E.Y.; Townsend, C.M., Jr.; Evers, B.M. VEGFR-2 expression in carcinoid cancer cells and its role in tumor growth and metastasis. Int. J. Cancer 2011, 128, 1045–1056. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Rezaei, F.; Mohammadi, H.; Heydari, M.; Sadeghi, M.; Mozaffari, H.R.; Khavid, A.; Godiny, M.; Brand, S.; M. Dürsteler, K.; Beatrix Brühl, A.; et al. Association between IL-8 (-251T/A) and IL-6 (-174G/C) Polymorphisms and Oral Cancer Susceptibility: A Systematic Review and Meta-Analysis. Medicina 2021, 57, 405. [Google Scholar] [CrossRef]
- Karakasheva, T.A.; Lin, E.W.; Tang, Q.; Qiao, E.; Waldron, T.J.; Soni, M.; Klein-Szanto, A.J.; Sahu, V.; Basu, D.; Ohashi, S.; et al. IL-6 Mediates Cross-Talk. between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment. Cancer Res. 2018, 78, 4957–4970. [Google Scholar] [CrossRef]
- Hart, P.C.; Rajab, I.M.; Alebraheem, M.; Potempa, L.A. C-Reactive Protein and Cancer-Diagnostic and Therapeutic Insights. Front. Immunol. 2020, 11, 595835. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.M.; Xing, R.D.; Zhang, F.M.; Duan, Y.Q. Serum soluble CD44v6 levels in patients with oral and maxillofacial malignancy. Oral Dis. 2009, 15, 570–572. [Google Scholar] [CrossRef]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Jian, X.; Guo, F.; Li, N.; Jiang, C.; Yin, P.; Min, A.J.; Huang, L. miR-203 inhibits arecoline-induced epithelial-mesenchymal transition by regulating secreted frizzled-related protein 4 and transmembrane-4 L six family member 1 in oral submucous fibrosis. Oncol. Rep. 2015, 33, 2753–2760. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ai, R.; Hao, Y.; Jiang, L.; Dan, H.; Ji, N.; Zeng, X.; Zhou, Y.; Chen, Q. Role of miR-155 in immune regulation and its relevance in oral lichen planus. Exp. Ther. Med. 2019, 17, 575–586. [Google Scholar] [CrossRef]
- Farshbaf, A.; Mohajertehran, F.; Sahebkar, A.; Garmei, Y.; Sabbagh, P.; Mohtasham, N. The role of altered microRNA expression in premalignant and malignant head and neck lesions with epithelial origin. Health Sci. Rep. 2022, 5, e921. [Google Scholar] [CrossRef]
- Deng, Z.Y.; Wang, Y.H.; Quan, H.Z.; Liu, O.S.; Li, Y.P.; Li, Y.; Zhu, W.; Munnee, K.; Tang, Z.G. Investigation of the association between miR-181b, Bcl-2 and LRIG1 in oral verrucous carcinoma. Mol. Med. Rep. 2016, 14, 2991–2996. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, H. miR-451 elevation relieves inflammatory pain by suppressing microglial activation-evoked inflammatory response via targeting TLR4. Cell Tissue Res. 2018, 374, 487–495. [Google Scholar] [CrossRef]
- Ge, X.; Xie, H.; Wang, L.; Li, R.; Zhang, F.; Xu, J.; Zhao, B.; Du, J. MicroRNA-122 promotes apoptosis of keratinocytes in oral lichen planus through suppressing VDR expression. J. Cell. Mol. Med. 2021, 25, 3400–3407. [Google Scholar] [CrossRef]
- Aghbari, S.M.H.; Abushouk, A.I.; Shakir, O.G.; Zayed, S.O.; Attia, A. Correlation between tissue expression of microRNA-137 and CD8 in oral lichen planus. Clin. Oral Investig. 2018, 22, 1463–1467. [Google Scholar] [CrossRef]
- Yakirevich, E.; Matoso, A.; Sabo, E.; Wang, L.J.; Tavares, R.; Meitner, P.; Morris, D.J.; Pareek, G.; Delellis, R.A.; Resnick, M.B. Expression of the glucocorticoid receptor in renal cell neoplasms: An immunohistochemical and quantitative reverse transcriptase polymerase chain reaction study. Hum. Pathol. 2011, 42, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Meijer, O.C.; Koorneef, L.L.; Kroon, J. Glucocorticoid receptor modulators. Ann. Endocrinol. 2018, 79, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.C.; Sun, H.Y.; Zhen, Y.X.; Zhang, H.; Shi, H.; Wang, X.X. Low expression of glucocorticoid receptor a in oral lichen planus correlates with activation of nuclear factor kappaB: A preliminary study. J. Oral Pathol. Med. 2014, 43, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.X.; Yang, X.; Jiang, L.; Zhou, Z.J.; Zhu, Y.Q. Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. BMC Cancer 2013, 13, 129. [Google Scholar] [CrossRef] [PubMed]
| Risk of Bias | Applicability Concerns | ||||||
|---|---|---|---|---|---|---|---|
| Domains | Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard |
| Signaling questions | Could the Selection of Patients Have Introduced Bias? | Could the Conduct or Interpretation of the Index Test Have Introduced Bias? | Could the Reference Standard, Its Conduct, or Its Interpretation Have Introduced Bias? | Could the Patient Flow Have Introduced Bias? | Are There Concerns That the Included Patients and Setting Do Not Match the Review Question? | Are There Concerns That the Index Test, Its Conduct, or Its Interpretation Differ from the Review Question? | Are There Concerns That the Target Condition as Defined by the Reference Standard Does Not Match the Question? |
| No | Title | Author | Year | Number of Participants | Reference | |
|---|---|---|---|---|---|---|
| OPMD or OSCC Group | Control Group | |||||
| 1 | Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer | F. Momen-Heravi | 2014 | 9 patients with OSCC, 8 patients with OSCC in remission, 8 patients with OLP | 9 healthy controls | [26] |
| 2 | Difficulties detecting miRNA-203 in human whole saliva by the use of PCR | Martin Lundegard | 2015 | 7 patients with OLP | 14 healthy controls | [27] |
| 3 | Salivary microRNAs in oral cancer | F. Zahran | 2015 | 20 patients with OPMD with dysplasia 20 patients with OPMD without dysplasia 20 patients with OSCC | 20 healthy controls | [28] |
| 4 | Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients | Jin-Seok Byun | 2015 | 16 patients with OLP | 8 healthy controls | [29] |
| 5 | MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder | Kai-Feng Hung | 2016 | 46 patients OPMD (hyperkeratosis, epithelial hyperplasia or dysplasia) | 24 healthy controls | [30] |
| 6 | Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: a pilot study | Mehmet Bugrahan Duz | 2016 | 25 Tongue squamous cell carcinoma | 25 healthy subjects | [31] |
| 7 | Predictive value of salivary microRNA-320a, vascular endothelial growth factor receptor 2, CRP and IL-6 in Oral lichen planus progression | Minoo Shahidi | 2017 | 32 patients with OLP, 15 patients with OSCC | 15 healthy subjects | [32] |
| 8 | Study to explore the significance of saliva as a diagnostic tool to detect microRNA in oral potentially malignant disorders | T.N. Uma Maheswari | 2017 | 5 patients with OPMD (oral leukoplakia, oral lichen planus and oral submucous fibrosis) | 5 healthy controls | [33] |
| 9 | Evaluating the accuracy of microRNA27b and microRNA137 as biomarkers of activity and potential malignant transformation in oral lichen planus patients | Sana Maher Hasan Aghbari | 2018 | 20 patients with diagnosed OLP | 20 healthy individuals | [34] |
| 10 | Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma | Chiara Gai | 2018 | 21 patients with OSCC | 11 healthy controls | [35] |
| 11 | Uncovering the potential of CD44v/SYNE1/miR34a axis in salivary fluids of oral cancer patients | Kavan Shah | 2018 | 8 patients with OSCC, 10 patients with leukoplakia | 10 healthy subjects | [36] |
| 12 | Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus—a short report | Masoumeh Mehdipour | 2018 | 30 OLP patients 15 OSCC patients | 15 healthy subjects | [37] |
| 13 | Predicting the Presence of Oral Squamous Cell Carcinoma Using Commonly Dysregulated MicroRNA in Oral Swirls | Tami Yap | 2018 | 30 patients with OSCC | 30 healthy subjects | [38] |
| 14 | Non-invasive screening of a microRNA-based dysregulation signature in oral cancer and oral potentially malignant disorders | Tami Yap | 2019 | 74 patients with OPMD (OLP, OLL, oral leukoplakia, dysplasia, traumatic ulceration) 53 patients with OSCC | 54 healthy subjects | [39] |
| 15 | Expression profile of salivary micro RNA-21 and 31 in oral potentially malignant disorders | Uma Maheswari | 2020 | 12 patients with OSMF, 8 patients with leukoplakia, 9 patients with OLP, 7 patients with OSMF + leukoplakia | 36 healthy subjects | [40] |
| 16 | Expression of Salivary miRNA 21 in Oral Submucous Fibrosis (OSMF): An Observational Study | Shesha R. Prasad | 2020 | 61 patients with chewing habits (chewing gutkha or other forms of areca nut) with OSMF 61 patients with chewing habits without OSMF | 63 healthy subjects | [41] |
| 17 | Systemic investigation identifying salivary mir-196b as a promising biomarker for early detection of head-neck cancer and oral precancer lesions | Cheng Ann-Joy | 2021 | 30 patients with OPMD 86 patients with HNSCC | 52 healthy individuals | [42] |
| 18 | Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma | Chiara Romani | 2021 | 28 patients with OSCC | 14 healthy controls | [43] |
| 19 | Salivary miR-30c-5p as Potential Biomarker for Detection of Oral Squamous Cell Carcinoma | Nikolay Mehterov | 2021 | 33 patients with OSCC | 12 healthy individuals | [44] |
| 20 | Expression of miR-31 in saliva-liquid biopsy in patients with oral squamous cell carcinoma | Parma Kumari | 2021 | 19 patients with OSCC | 2 healthy controls | [45] |
| 21 | Expression of Salivary miRNA-31 in Oral Submucous Fibrosis | Saba Khan | 2021 | 25 patients with OSMF | 25 healthy controls | [46] |
| 22 | Clinical significance of miR-142-3p in oral lichen planus and its regulatory role in keratinocyte proliferation | Zhichao Meng | 2021 | 56 OLP patients | 44 healthy subjects | [47] |
| 23 | Exploiting salivary miR-375 as a clinical biomarker of oral potentially malignant disorder | Hsi-Feng Tu | 2022 | 41 patients with OPMD | 26 healthy subjects | [48] |
| 24 | Salivary miRNAs Expression in Potentially Malignant Disorders of the Oral Mucosa and Oral Squamous Cell Carcinoma: A Pilot Study on miR-21, miR-27b, and miR-181b | Dario Di Stasio | 2022 | 6 patients with epithelial hyperkeratosis with no dysplasia, 7 patients with low-grade OED 10 patients with high-grade OED 10 patients with OSCC | 10 healthy subjects | [49] |
| 25 | Salivary miR-31-5p, miR-345-3p, and miR-424-3p Are Reliable Biomarkers in Patients with Oral Squamous Cell Carcinoma | Beáta Scholtz | 2022 | 43 patients with OSCC | 44 healthy controls | [50] |
| 26 | Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients | Aditi Patel | 2022 | 12 patients with OSCC | 7 healthy individuals | [51] |
| 27 | Salivary Exosomal MicroRNA-486-5p and MicroRNA-10b-5p in Oral and Oropharyngeal Squamous Cell Carcinoma | Cosmin Ioan Faur | 2022 | 25 patients with OSCC | 25 healthy individuals | [52] |
| 28 | Salivary level of microRNA-146a and microRNA-155 biomarkers in patients with oral lichen planus versus oral squamous cell carcinoma | Masoumeh Mehdipour | 2023 | 15 OLP patients with dysplasia 15 OLP patients without dysplasia 15 OSCC patients | 15 healthy controls | [53] |
| 29 | Evaluation of Diagnostic Significance of Salivary miRNA-184 and miRNA-21 in Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders | Aarushi Garg | 2023 | 30 patients with OPMD 30 patients with OSCC | 30 healthy controls | [54] |
| 30 | Salivary LINC00657 and miRNA-106a as diagnostic biomarkers for oral squamous cell carcinoma, an observational diagnostic study | Nayroz Abdel Fattah Tarrad | 2023 | 12 patients with OLP 12 patients with OSCC | 12 healthy individuals | [55] |
| 31 | Downregulation of salivary miR-3928 as a potential biomarker in patients with oral squamous cell carcinoma and oral lichen planus | Alieh Farshbaf | 2024 | 30 patients with OLP 31 patients with OSCC | 30 healthy individuals | [56] |
| 32 | Expression Analysis of Circulating microRNAs in Saliva and Plasma for the Identification of Clinically Relevant Biomarkers for Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders | Federica Rocchetti | 2024 | 6 patients with OPMD 14 patients with OSCC | 5 healthy individuals | [57] |
| 33 | The oncogenic potential of salivary microRNA-93 and microRNA-412-3p in oral lichen planus: a case–control study | Moataz M ELHefny | 2024 | 20 patients with OLP 20 patients with OSCC | 20 healthy controls | [58] |
| Study | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| F. Momen-Heravi 2014 [26] | − | − | + | + | − | + | + |
| Martin Lundegard 2015 [27] | + | + | + | + | + | + | + |
| F. Zahran 2015 [28] | + | + | + | + | + | + | + |
| Jin-Seok Byun 2015 [29] | + | + | + | + | + | + | + |
| Kai-Feng Hung 2016 [30] | − | − | + | + | + | + | + |
| Mehmet Bugrahan Duz 2016 [31] | − | − | + | + | − | + | + |
| Minoo Shahidi 2017 [32] | − | − | + | + | + | + | + |
| T.N. Uma Maheswari 2017 [33] | − | − | + | ? | − | − | + |
| Sana Maher Hasan Aghbari 2018 [34] | − | − | + | + | + | + | + |
| Chiara Gai 2018 [35] | − | + | + | + | + | + | + |
| Kavan Shah 2018 [36] | − | − | + | + | + | + | + |
| Masoumeh Mehdipour 2018 [37] | + | − | + | + | + | + | + |
| Tami Yap 2018 [38] | + | + | + | + | + | + | + |
| Tami Yap 2019 [39] | + | + | + | + | + | + | + |
| Uma Maheswari 2020 [40] | + | − | + | + | + | + | + |
| Shesha R. Prasad 2020 [41] | + | + | + | ? | + | + | + |
| Cheng Ann-Joy 2021 [42] | + | + | + | + | + | + | + |
| Chiara Romani 2021 [43] | − | + | + | + | + | + | + |
| Nikolay Mehterov 2021 [44] | + | + | + | + | + | + | + |
| Parma Kumari 2021 [45] | − | − | + | + | − | − | + |
| Saba Khan 2021 [46] | − | + | + | + | + | + | + |
| Zhichao Meng 2021 [47] | + | + | + | + | + | + | + |
| Hsi-Feng Tu 2022 [48] | + | + | + | + | + | + | + |
| Dario Di Stasio 2022 [49] | + | − | + | + | + | + | + |
| Beáta Scholtz 2022 [50] | + | + | + | + | + | + | + |
| Aditi Patel 2022 [51] | − | + | + | + | + | + | + |
| Cosmin Ioan Faur 2022 [52] | + | + | + | + | + | + | + |
| Masoumeh Mehdipour 2023 [53] | + | + | + | + | + | + | + |
| Aarushi Garg 2023 [54] | + | + | + | + | + | + | + |
| Nayroz Abdel Fattah Tarrad 2023 [55] | − | + | + | + | + | + | + |
| Alieh Farshbaf 2024 [56] | + | + | + | + | + | + | + |
| Federica Rocchetti 2024 [57] | − | + | + | + | + | + | + |
| Moataz M ELHefny 2024 [58] | + | + | + | + | + | + | + |
| No | Author | Sample Areas | Type of Saliva | Quantity of Saliva (mL) | Aspects of Collections | RNA Extraction | RNA Method of Analysis |
|---|---|---|---|---|---|---|---|
| 1 | F. Momen-Heravi 2014 [26] | Saliva | Whole unstimulated saliva | 8 | Condition: refrain from eating, drinking and oral hygiene procedures on the day of saliva collection; saliva was collected from 6 to 11 am, after a water rinse Preservation: - Temporary storage: all saliva samples were processed immediately Long-term storage: −80 °C | miRNeasy kit (Qiagen, Valencia, CA, USA) | NanoDrop Spectrophotometer ND2000 (NanoDrop Technologies, Wilmington, DE, USA), Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), NanoString nCounter miRNA expression assay (NanoString Technologies Seattle, WA, USA), TaqMan MicroRNA Assay (Applied Biosystems) using qRT-PCR (MJ Research PTC-200 Thermal Cycler, MJ Research) |
| 2 | Jin-Seok Byun 2015 [29] | Saliva | Whole unstimulated saliva | 5–10 | Condition: saliva was collected from 10 to 12 am, after a mechanical tooth and tongue cleansing, and a rinse with distilled water Preservation: - Temporary storage: - Long-term storage: - | TRIzol reagent (Invitrogen, Carlsbad, CA, USA), miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA) | NanoDrop (Thermo Scientific, Wilmington, DE, USA) Microarray (Human miRNA Microarray 8 × 60K V19.0, Agilent Technologies, Santa Clara, CA, USA) TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA, USA) using qRT-PCR (7500 Real-Time PCR System, Applied Biosystems, Foster City, CA, USA) |
| 3 | F. Zahran 2015 [28] | Saliva | Whole unstimulated saliva | - | Condition: subjects refrained from eating, drinking, using chewing gum for at least one and a half hour prior sampling Preservation: - Temporary storage: - Long-term storage: - | miRNeasy kit (Qiagen, Valencia, CA, USA) | NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA), qRT-PCR (Applied Biosystems, 7500 Real-Time PCR System, Foster City, CA, USA) |
| 4 | Martin Lundegard 2015 [27] | Saliva | Stimulated saliva | 2 | Condition: - Preservation: Saliva Incubation Mix Temporary storage: -room temperature for 1 h Long-term storage: −80 °C | RNeasy Micro Kit (Qiagen, Hilden, Germany) | NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), qRT-PCR (IQ5 Multicolor Real-Time PCR Detection System, Bio-Rad Laboratories Inc., USA) |
| 5 | Kai-Feng Hung 2016 [30] | Saliva | Whole unstimulated saliva | - | Condition: after mouth rinsing Preservation: - Temporary storage: all saliva samples were processed immediately Long-term storage: −80 °C | mirVana PARIS kit (Ambion, Austin, TX, USA) | TaqMan MicroRNA (Applied Biosystems, Foster City, CA, USA) using qRT-PCR |
| 6 | Mehmet Bugrahan Duz 2016 [31] | Saliva | Whole saliva | - | Condition: in the morning, stop eating, drinking and oral hygiene 1 h before Preservation: - Temporary storage: all saliva samples were processed immediately Long-term storage: −80 °C | mirVana PARIS kit (Ambion, Darmstadt, Germany) | NanoDrop ND-2000c (Thermo Fisher Scientific Inc., Wilmington, DE, USA), Microarray (Agilent Technologies, Santa Clara, CA, USA), TaqMan MicroRNA (Applied Biosystems, Foster City, CA, USA) using qRT-PCR (LightCycler 480-II Real-Time PCR System, Roche, Switzerland) |
| 7 | Minoo Shahidi 2017 [32] | Saliva and tissue | Whole unstimulated saliva | 5 | Condition: refrain from consuming food, drinking and oral hygiene procedures for at least 1 h prior to sampling Preservation: RNA Stabilization Reagent Temporary storage: - Long-term storage: −80 °C | mirVana PARIS Kit (Ambion, USA) | Nanodrop ND-1000 (Thermo Scientific, Worcester, MA, USA), qRT-PCR (Rotor-Gene 6000 Real-Time PCR System, Corbett Life Science) |
| 8 | T.N. Uma Maheswari 2017 [33] | Saliva | Whole saliva | 8–10 | Condition: saliva was collected after a rinse with water, between 6 and 11 am, refrain from eating, drinking or applying oral hygiene in that day Preservation: - Temporary storage: dry ice Long-term storage: −80 °C | RNeasy Kit (Qiagen) | Nanodrop spectrophotometer, TaqMan MicroRNA (Applied Biosystems) using qRT-PCR |
| 9 | Kavan Shah 2018 [36] | Saliva and serum | Whole saliva | - | Condition: - Preservation: - Temporary storage: - Long-term storage: - | TRIzol reagent (Invitrogen, Carlsbad, CA, USA) | qRT-PCR (AriaMx Real-Time PCR System, Agilent Technologies) |
| 10 | Masoumeh Mehdipour 2018 [37] | Saliva | Whole unstimulated saliva | 3–5 | Condition: saliva was collected after a rinse with water; subjects refrained from eating, drinking or using oral hygiene procedures for at least 1h prior sampling; Preservation: RNAlater Temporary storage: - Long-term storage: −80 °C | mirVana PARIS kit (Ambion, USA) | qRT-PCR |
| 11 | Sana Maher Hasan Aghbari 2018 [34] | Saliva and tissue | Whole unstimulated saliva | - | Condition: - Preservation: - Temporary storage: - Long-term storage: −80 °C | miRNeasy extraction kit (Qiagen, Valencia, CA, USA) | qRT-PCR (Rotor-Gene Q Real-time PCR System, Qiagen, USA) |
| 12 | Chiara Gai 2018 [35] | Saliva | Whole unstimulated saliva | - | Condition: saliva was collected after a rinse with water, between 9 and 11 am, refrain from eating, drinking or applying oral hygiene Preservation: - Temporary storage: - Long-term storage: −80 °C | mirVana Isolation kit (Thermo Fisher Scientific, Waltham, MA, USA) | Nanodrop ND-1000 (Thermo Fisher Scientific), TaqMan MicroRNA (Thermo Fisher) using qRT-PCR (QuantStudio 12k Flex, Thermo Fisher) |
| 13 | Tami Yap 2018 [38] | Saliva and tissue | Oral swirl | 10 | Condition: patients were asked to swirl deionized water 50–60 s Preservation: - Temporary storage: Long-term storage: −20 °C | mirVana Isolation Kit kit (Life Technologies, USA) | NanoDrop 1000 (NanoDrop Technologies Wilmington, DE, USA), qRT-PCR (Eco Real-Time PCR System, Illumina) |
| 14 | Tami Yap 2019 [39] | Saliva | Oral swirl | 10 | Condition: patients were asked to swirl deionized water 50–60 s Preservation: - Temporary storage: - Long-term storage: - | mirVana Isolation Kit (Life Technologies) | NanoDrop 1000 (NanoDrop Technologies Wilmington, DE, USA), qRT-PCR (Eco Real-Time PCR System, Illumina) |
| 15 | Shesha R. Prasad 2020 [41] | Saliva | Whole unstimulated saliva | 0.5–1.0 | Condition: saliva was collected from 9 to 11 am, after a rinse with distilled water for 1 min; subjects were refrained from eating, drinking, smoking, or using oral hygiene procedures for at 1 h prior sampling; Preservation: RNAlater Temporary storage: - Long-term storage: −20 °C | miRNeasy kit (Qiagen) | qRT-PCR (Applied Biosystems StepOne Real-Time PCR System) |
| 16 | Uma Maheswari 2020 [40] | Saliva | Whole saliva | - | Condition: refrain from consuming food for 2 h prior to sampling, and avoid the use of chewing tobacco or smoking for 1 day before the collection of samples; Preservation: - Temporary storage: DNAse/RNAse-free 50 mL falcon containers at room temperature for maximum 3 h; Long-term storage: −4 °C | NucleoSpin® miRNA (Macherey-Nagel, Germany) | qRT-PCR (Rotor-Gene Q 5-plex, Qiagen) |
| 17 | Zhichao Meng 2021 [47] | Saliva, tissue and serum | Whole saliva | - | Condition: - Preservation: - Temporary storage: - Long-term storage: −80 °C | TRIzol kit (Thermo Fisher) | qRT-PCR |
| 18 | Cheng Ann-Joy 2021 [42] | Saliva | Oral swabbing | - | Condition: oral swabbing was soaked in 1 mL of normal saline for over 30 min Preservation: - Temporary storage: - Long-term storage: - | miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) | qRT-PCR (Bio-Rad CFX96 Real-Time PCR System, Bio-Rad, Hercules, CA, USA) |
| 19 | Chiara Romani 2021 [43] | Saliva | Whole unstimulated saliva | - | Condition: saliva was collected after a rinse with PBS, refrain from eating, drinking or applying oral hygiene Preservation: - Temporary storage: immediately processed Long-term storage: −80 °C | miRNeasy Mini Kit (Qiagen, Hilden, Germany) | Microarray (Agilent Technologies, Santa Clara, CA, USA) and qRT-PCR (Bio-Rad CFX96 Real-Time PCR System, Bio-Rad, Hercules, CA, USA) |
| 20 | Nikolay Mehterov 2021 [44] | Saliva Tissue | Whole unstimulated saliva | 4–5 mL | Condition: saliva was collected after a rinse with distilled water, between 8 and 9 am, refrain from eating, drinking, smoking or applying oral hygiene Preservation: - Temporary storage: immediately pro-cessed Long-term storage: −80 °C | TRI Reagent (Invitrogen, ThermoFisher Scientific, Massachusetts, MA, USA) | NanoDrop 2000 (Thermo Scientific, Massachusetts, MA, USA) TaqMan MicroRNA (Thermo Fisher Scientific, Massachusetts, MA, USA) using qRT-PCR (Rotor-Gene Q Real-Time PCR System, Qiagen, Hilden, Germany) |
| 21 | Parma Kumari 2021 [45] | Saliva | Whole unstimulated saliva | - | Condition: fasting saliva via PureSAL device Preservation: - Temporary storage: - Long-term storage: −80 °C | mirVANA PARIS kit (Invitrogen, Thermo Fisher Scientific, USA) | TaqMan MicroRNA Assay Kit (Thermo Fisher Scientific, USA) using qRT-PCR (QuantStudio™ 7 Flex Real-Time PCR System, Applied Biosystems, USA) |
| 22 | Saba Khan 2021 [46] | Saliva | Whole saliva | - | Condition: - Preservation: - Temporary storage: - Long-term storage: −80 °C | - | qRT-PCR |
| 23 | Beáta Scholtz 2022 [50] | Saliva | Whole unstimulated saliva | 5 mL | Condition: saliva was collected between 9 and 11 am, refrain from eating, drinking, smoking, gum chewing or applying oral hygiene for at least 60 min before sampling Preservation: PAXgene reagent Temporary storage: - Long-term storage: −70 °C | TRIzol Reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) | TaqMan Mi-croRNA (Applied Biosystems) using qRT-PCR (ABI PRISM® 7000 Sequence Detection System, Applied Biosystems) |
| 24 | Dario Di Stasio 2022 [49] | Saliva | Whole unstimulated saliva | - | Condition: saliva was collected from 8 to 12 am Preservation: - Temporary storage: all saliva samples were processed within the shortest time Long-term storage: - | mirVana PARIS KIT (Thermo Fisher Scientific, Waltham, MA, USA) | TaqMan MicroRNA Assay (Life Technologies, Carlsbad, CA, USA) using qRT-PCR (ViiA™ 7 Real-Time PCR System, Applied Biosystems, Foster City, CA, USA) |
| 25 | Hsi-Feng Tu 2022 [48] | Saliva | Whole unstimulated saliva | 2 | Condition: saliva was collected from 9 to 10 am with prior mouth rinsing with water and the patients refrained from eating, drinking, smoking or using oral hygiene applying oral hygiene for at least 60 min before sampling Preservation: protease inhibitors Temporary storage: all saliva samples were processed within the shortest time Long-term storage: −80 °C | mirVana PARIS KIT (Ambion, Thermo Fisher Scientific, Austin, TX, USA) | TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA, USA), using qRT-PCR (ABI Prism 7700 Sequence Detection System, Applied Biosystems, Foster City, CA, USA) |
| 26 | Aditi Patel 2022 [51] | Saliva Tissue | Whole unstimulated saliva | - | Condition: saliva was collected between 8 and 13 pm, refrain from eating or applying oral hygiene for at least 60 min before sampling Preservation: - Temporary storage: - Long-term storage: −80 °C | TRIzol LS reagent (Thermo Fisher Scientific, Waltham, MA, USA) | TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, USA), using RT-PCR (ABI Prism 7700 qPCR System, Applied Biosystems, Foster City, CA, USA) |
| 27 | Cosmin Ioan Faur 2022 [52] | Saliva | Whole unstimulated saliva | 0.8–1.6 | Condition: saliva was collected between 7 and 10 am, refrain from eating, drinking or applying oral hygiene for at least 60 min before sampling, after a rinse with water 15 min before sampling Preservation: - Temporary storage: −20 °C Long-term storage: −80 °C | Plasma/Serum Circulating and Exosomal RNA Purification Kit (Norgen Biotek Corp., Thorold, ON, Canada) | TaqMan Mi-croRNA (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA) using qRT-PCR (ViiA7 Real-Time PCR System, Applied Biosystems, Waltham, MA, USA) |
| 28 | Masoumeh Mehdipour 2023 [53] | Saliva | Whole unstimulated saliva | 3–5 | Condition: saliva was collected between 8 and 13 pm, refrain from eating, drinking or applying oral hygiene for at least 60 min before sampling Preservation: RNAlater Temporary storage: - Long-term storage: −80 °C | TRIzol Reagent (Anacell Teb, Tehran, Iran) | NanoDrop One (Thermo Scientific/Thermo Fisher, Waltham, MA, USA), qRT-PCR (Roche, Switzerland) |
| 29 | Aarushi Garg 2023 [54] | Saliva | Whole unstimulated saliva | 3–5 | Condition: - Preservation: RNAlater Temporary storage: - Long-term storage: −80 °C | QIAzol Lysis Reagent and miR-Neasy Mini Kit (Qiagen, Germany) | NanoPhotometer P330 (Implen / Biochrom International, Delhi, India), qRT-PCR (Rotor-Gene Q Real-Time PCR System, Qiagen, Germany) |
| 30 | Nayroz Abdel Fattah Tarrad 2023 [55] | Saliva | Whole unstimulated saliva | - | Condition: saliva was collected in the morning Preservation: - Temporary storage: −20 °C Long-term storage: - | - | qRT-PCR (Rotor-Gene Q real-time PCR system, Qiagen, USA) |
| 31 | Alieh Farshbaf 2024 [56] | Saliva | Whole saliva | - | Condition: saliva was collected in the morning, without eating or drinking Preservation: - Temporary storage: - Long-term storage: −80 °C | RNX-Plus (SinaClon, Tehran, Iran) | NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, SUA), qRT-PCR (LightCycler 96, Roche Diagnostics, Basel, Switzerland) |
| 32 | Federica Rocchetti 2024 [57] | Saliva Plasma | Whole saliva | 2 | Condition: saliva was collected and patients refrained from eating, drinking or consuming chewing gum for at least 90 min before sampling Preservation: RNA preservation Temporary storage: 16–24 °C Long-term storage: −80 °C | Saliva RNA purification kit (Norgen Biotek Corp., Thorold, ON, Canada) | TaqMan Mi-croRNA (Thermo Fisher Scientific, Waltham, MA, USA) using qRT-PCR (7500 Fast Real-Time PCR System, Thermo Fisher Scientific, Waltham, MA, USA) |
| 33 | Moataz M ELHefny 2024 [58] | Saliva | Whole unstimulated saliva | 0.4 | Condition: saliva was collected in the morning, refrain from eating, drinking or smoking for at least 30 min before sampling Preservation: - Temporary storage: on ice Long-term storage: −80 °C | MiRNeasy extraction kit (Qiagen Valencia, CA, USA) | qRT-PCR (Rotor-Gene Q PCR system, Qiagen Hilden/USA office, USA) |
| Author | OPMD Analyzed | Reference Probe | Salivary miRNA | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Uma Maheswari [40] | Leukoplakia OLP OSMF OSMF + leukoplakia | Healthy controls | miR-21 | 66% | 69% |
| Healthy controls | miR-31 | 36% | 40% | ||
| F. Momen-Heravi [26] | OLP | OSCC | miR-27b | 85.71% | 100% |
| Dario Di Stasio [49] | Dysplastic lesions | Healthy controls | miR-181b | 94.1% | 81.2% |
| F. Zahran [28] | OPMD with or without dysplasia | Healthy controls | miR-21 | 90% | 60% |
| Healthy controls | miR-184 | 75% | 80% | ||
| Healthy controls | miR-145 | 70% | 60% | ||
| Sana Maher Hasan Aghbari [41] | OLP | Healthy controls | miR-27b | 75% | 100% |
| miR-137 | 100% | 80% | |||
| Zhichao Meng [47] | OLP | Healthy controls | miR-142-3p | 75% | 86.4% |
| Cheng Ann-Joy 2021 [42] | OPMD | Healthy controls | miR-196b | 90.0% | 98.1% |
| Chiara Romani 2021 [43] | OSCC | Healthy controls | miR-423-5p + miR-106b-5p, miR-193b-3p | 85.4% | 85.1% |
| Nikolay Mehterov 2021 [44] | OSCC | Healthy controls | miR-30c-5p | 86% | 74% |
| Hsi-Feng Tu 2022 [48] | OPMD | Healthy controls | miR-375 | 82% | 80% |
| OPMD in dysplasia state | OPMD in non-dysplasia state | 71% | 83% | ||
| Beáta Scholtz 2022 [50] | OSCC | Healthy controls | miR-31-5p, miR-345-3p, miR-424-3p | 77% | 86% |
| Aditi Patel 2022 [51] | OSCC | Healthy controls | miR-1307-5p | 99.99% | 99.99% |
| Cosmin Ioan Faur 2022 [52] | OSCC | Healthy controls | miR-486-5p | 72% | 59% |
| Aarushi Garg 2023 [54] | OSCC | Healthy controls | miR-21 | 80% | 70% |
| OPMD | Healthy controls | 70% | 53% | ||
| OPMD | OSCC | 70% | 60% | ||
| OSCC | Healthy controls | miR-184 | 80% | 74% | |
| OPMD | Healthy controls | 70% | 60% | ||
| OPMD | OSCC | 70% | 60% | ||
| Nayroz Abdel Fattah Tarrad 2023 [55] | OSCC | Healthy controls | miR-106a | 100% | 70.8% |
| OLP | Healthy controls | 83.5% | 50% | ||
| OLP | OSCC | 66.7% | 83.3% | ||
| Moataz M ELHefny 2024 [58] | OSCC | Healthy controls | miR-93 | 100% | 100% |
| OLP | Healthy controls | 95% | 95% | ||
| OSCC | Healthy controls | miR-412-3p | 100% | 100% | |
| OLP | Healthy controls | 100% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osan, C.; Berindan-Neagoe, I.; Dinu, C.; Armencea, G.; Bran, S.; Kretschmer, W.; Baciut, G.; Onisor, F.; Baciut, M. Salivary MicroRNAs as Innovative Biomarkers for Diagnosis and Prediction of the Oral Potentially Malignant Disorders Transition Towards Oral Cancer: A Systematic Review. J. Clin. Med. 2025, 14, 8128. https://doi.org/10.3390/jcm14228128
Osan C, Berindan-Neagoe I, Dinu C, Armencea G, Bran S, Kretschmer W, Baciut G, Onisor F, Baciut M. Salivary MicroRNAs as Innovative Biomarkers for Diagnosis and Prediction of the Oral Potentially Malignant Disorders Transition Towards Oral Cancer: A Systematic Review. Journal of Clinical Medicine. 2025; 14(22):8128. https://doi.org/10.3390/jcm14228128
Chicago/Turabian StyleOsan, Ciprian, Ioana Berindan-Neagoe, Cristian Dinu, Gabriel Armencea, Simion Bran, Winfried Kretschmer, Grigore Baciut, Florin Onisor, and Mihaela Baciut. 2025. "Salivary MicroRNAs as Innovative Biomarkers for Diagnosis and Prediction of the Oral Potentially Malignant Disorders Transition Towards Oral Cancer: A Systematic Review" Journal of Clinical Medicine 14, no. 22: 8128. https://doi.org/10.3390/jcm14228128
APA StyleOsan, C., Berindan-Neagoe, I., Dinu, C., Armencea, G., Bran, S., Kretschmer, W., Baciut, G., Onisor, F., & Baciut, M. (2025). Salivary MicroRNAs as Innovative Biomarkers for Diagnosis and Prediction of the Oral Potentially Malignant Disorders Transition Towards Oral Cancer: A Systematic Review. Journal of Clinical Medicine, 14(22), 8128. https://doi.org/10.3390/jcm14228128








