Role of L1 HPV Protein Expression in the Cytological Diagnosis of Precancerous Cervical Lesions
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
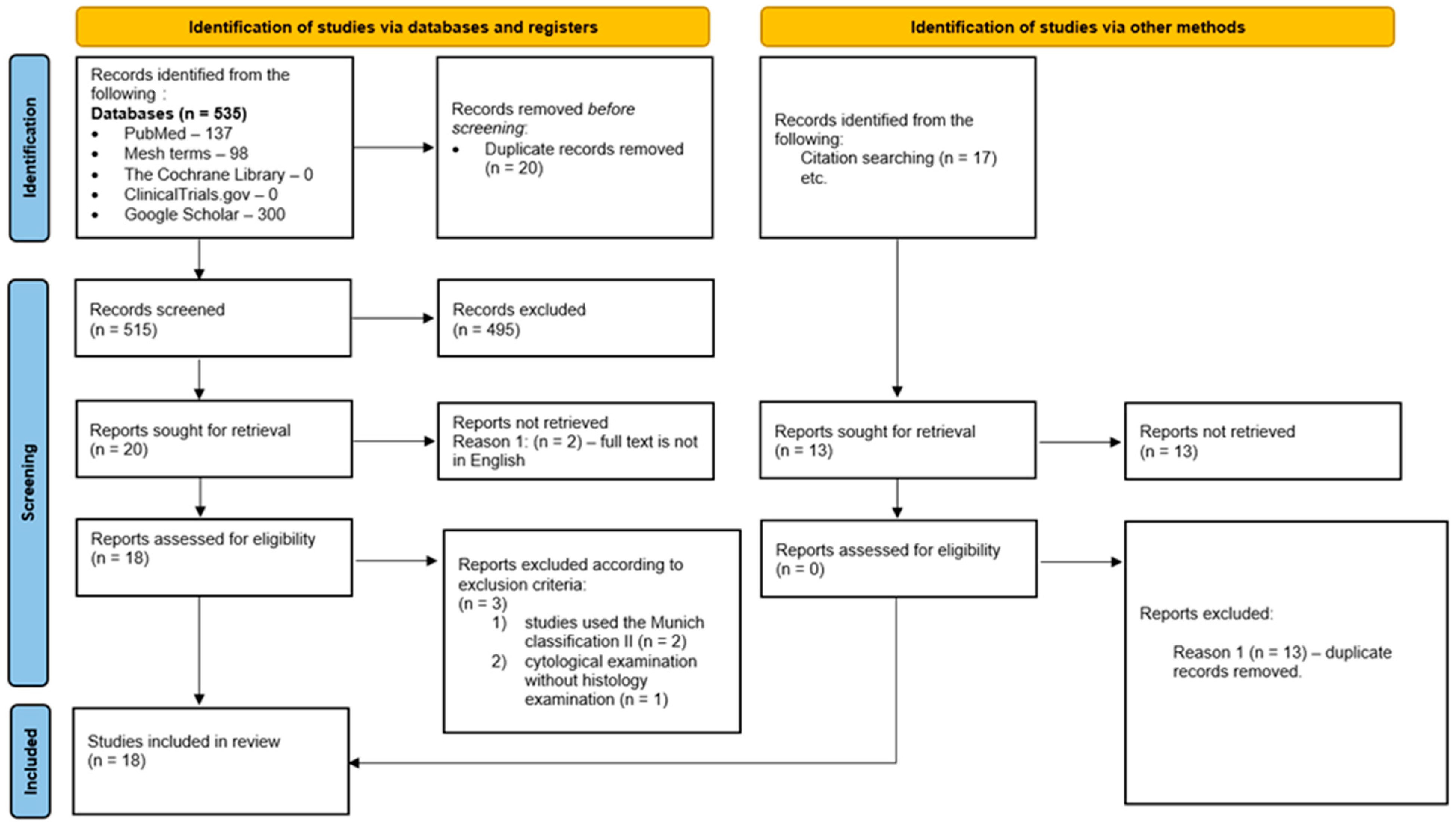
2.3. Study Selection and Reference Standard
2.4. Data Items
2.5. Data Analysis
2.6. Quality Assessment
3. Results
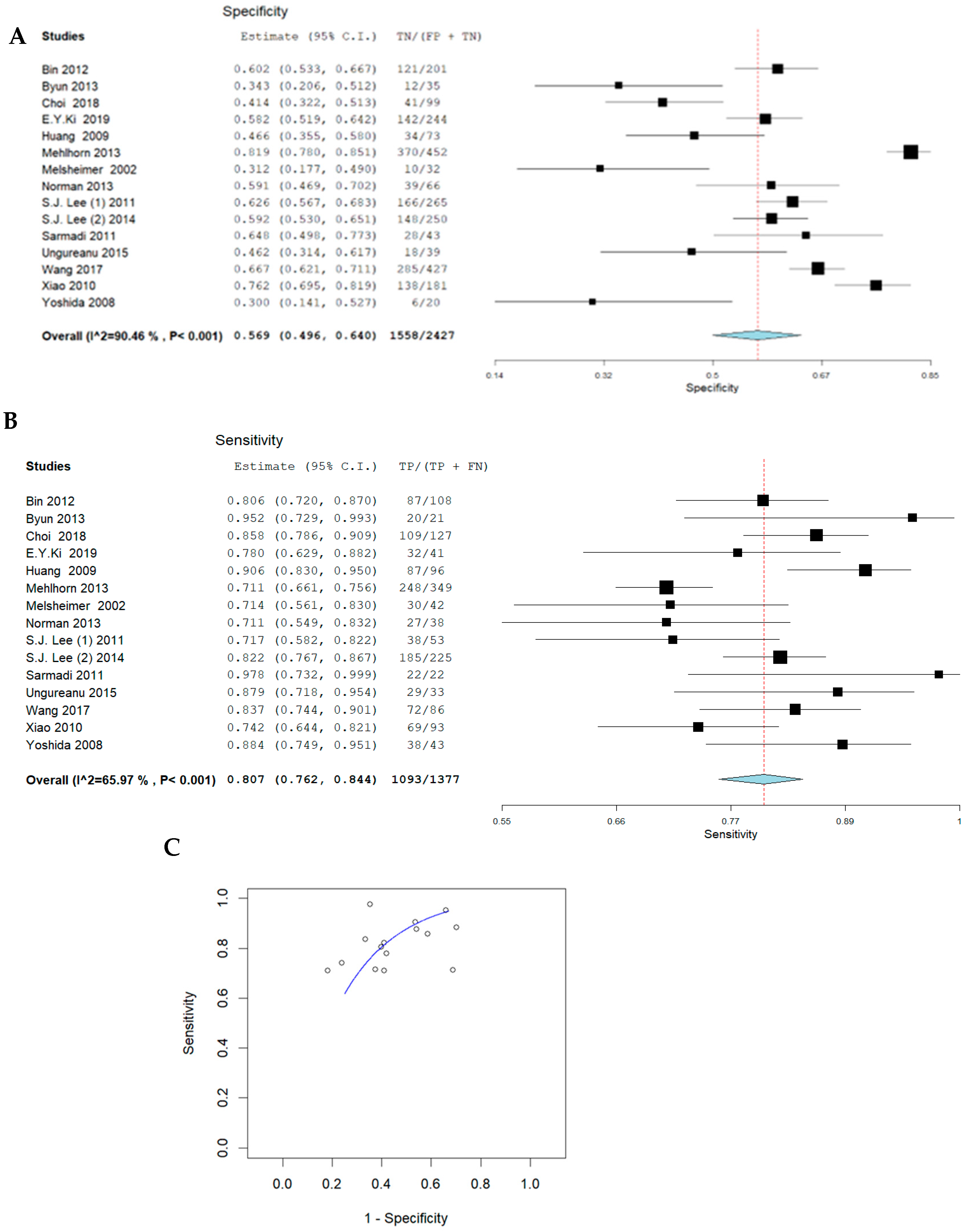
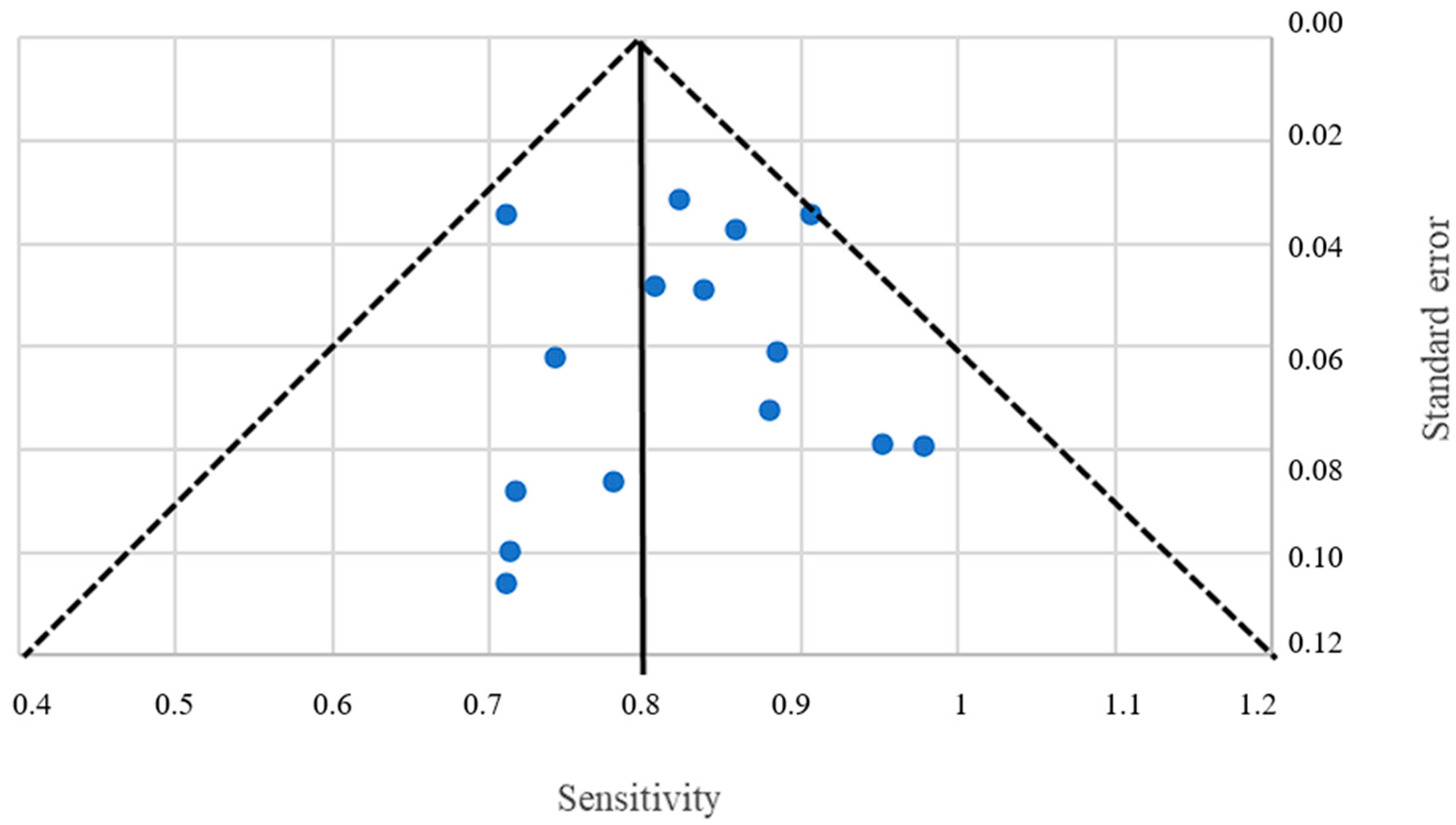
3.1. Pooled Sensitivity and Specificity
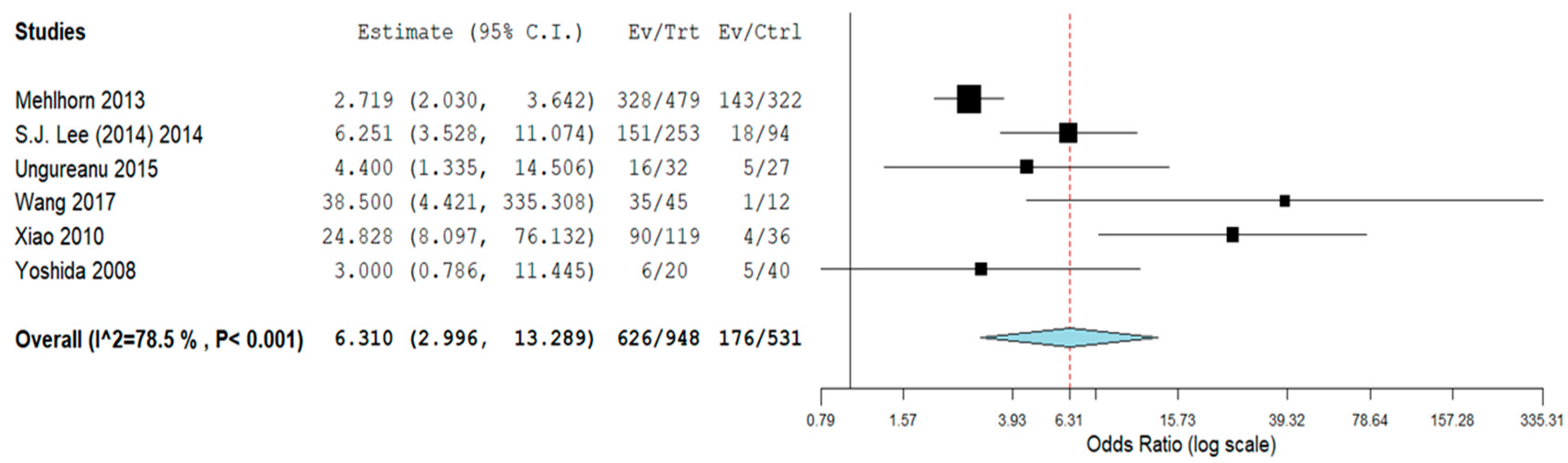
3.2. Subgroup Analyses
4. Discussion
4.1. Translating These Insights into Clinical Practice
4.2. Limitations of the Review
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ASC-H | Atypical squamous cell cannot exclude HSIL |
| ASC-US | Atypical squamous cells of undetermined significance |
| CI | Confidence interval |
| CIN | Cervical intraepithelial neoplasia |
| CIN2+ | High-grade cervical intraepithelial neoplasia |
| HPV | Human papillomavirus |
| HSIL | High-grade squamous intraepithelial lesions |
| ICC | Immunocytochemistry |
| LSIL | Low-grade squamous intraepithelial lesions |
| NILM | Negative for intraepithelial lesion or malignancy |
| NPV | Negative predictive value |
| OR | Odds ratio |
| PCS | Prospective cohort study |
| PPV | Positive predictive value |
| RCS | Retrospective cohort studies |
| RCT | Randomized clinical trials |
| Sn | Sensitivity |
| Sp | Specificity |
| TBS | The Bethesda System |
| WNL | NILM according to TBS |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Alhamlan, F.S.; Alfageeh, M.B.; Al Mushait, M.A.; Al-Badawi, I.A.; Al-Ahdal, M.N. Human Papillomavirus-Associated Cancers. In Microbial Pathogenesis; Springer: Cham, Switzerland, 2021; Volume 1313, pp. 1–14. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Ouh, Y.-T.; Kim, H.Y.; Yi, K.W.; Lee, N.-W.; Kim, H.-J.; Min, K.-J. Enhancing cervical cancer screening: Review of p16/Ki-67 dual staining as a promising triage strategy. Diagnostics 2024, 14, 451. [Google Scholar] [CrossRef]
- Contreras, N.E.; Roldán, J.S.; Castillo, D.S. Novel competitive enzyme-linked immunosorbent assay for the detection of the high-risk Human Papillomavirus 18 E6 oncoprotein. PLoS ONE 2023, 18, e0290088. [Google Scholar] [CrossRef]
- Koliopoulos, G.; Nyaga, V.N.; Santesso, N.; Bryant, A.; Martin-Hirsch, P.P.; Mustafa, R.A.; Schünemann, H.; Paraskevaidis, E.; Arbyn, M. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 2017, 8, CD008587. [Google Scholar] [CrossRef]
- Abbas, M.; Erduran, I.; De Jonge, J.; Bettendorf, O. Evaluation of P16/Ki67 (CINtecPlus) and L1-capsid compared with HPV-genotyping in cervical cytology in women ≥35 years old focusing on patients with atypical squamous cells of undetermined significance. Oncol. Lett. 2022, 24, 242. [Google Scholar] [CrossRef]
- Przybylski, M.; Pruski, D.; Millert-Kalińska, S.; Krzyżaniak, M.; de Mezer, M.; Frydrychowicz, M.; Jach, R.; Żurawski, J. Expression of E4 Protein and HPV Major Capsid Protein (L1) as A Novel Combination in Squamous Intraepithelial Lesions. Biomedicines 2023, 11, 225. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, A.W.; Kim, T.J.; Kim, J.H.; Bae, J.H.; Lee, C.W.; Song, M.J.; Yoon, J.H.; Hur, S.Y.; Park, J.S. Correlation between immunocytochemistry of human papilloma virus L1 capsid protein and behavior of low-grade cervical cytology in Korean women. J. Obstet. Gynaecol. Res. 2011, 37, 1222–1228. [Google Scholar] [CrossRef]
- Solomon, D.; Davey, D.; Kurman, R.; Moriarty, A.; O’Connor, D.; Prey, M.; Raab, S.; Sherman, M.; Wilbur, D.; Wright, T. The 2001 Bethesda system terminology for reporting results of cervical cytology. JAMA 2002, 287, 2114–2119. [Google Scholar] [CrossRef]
- Nayar, R.; Wilbur, D.C. The Bethesda system for reporting cervical cytology: A historical perspective. Acta Cytol. 2017, 61, 359–372. [Google Scholar] [CrossRef]
- Melsheimer, P.; Kaul, S.; Dobeck, S.; Bastert, G. Immunocytochemical Detection of HPV High-Risk Type L1 Capsid Proteins in LSIL and HSIL as Compared with Detection of HPV L1 DNA The expression of L1 capsid proteins is significantly reduced in HPV 16 DNA-positive HSIL and HPV hr DNA-positive HSIL. Acta Cytol. 2003, 47, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Sarmadi, S.; Izadi-Mood, N.; Pourlashkari, M.; Yarandi, F.; Sanii, S. HPV L1 capsid protein expression in squamous intraepithelial lesions of cervix uteri and its relevance to disease outcome. Arch. Gynecol. Obstet. 2012, 285, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, J.B.; Rutjes, A.W.; Whiting, P.; Yang, B.; Leeflang, M.M.; Bossuyt, P.M.; Deeks, J.J. Chapter 8 Assessing Risk of Bias and Alicability. 2023. Available online: https://training.cochrane.org/handbook-diagnostic-test-accuracy/current (accessed on 1 January 2023).
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Duval, S.; Tweedie, R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000, 56, 455–463. [Google Scholar] [CrossRef]
- Byun, S.W.; Lee, A.; Kim, S.; Choi, Y.J.; Lee, Y.S.; Park, J.S. Immunostaining of p16INK4a/Ki-67 and L1 capsid protein on liquid-based cytology specimens obtained from ASC-H and LSIL-H cases. Int. J. Med. Sci. 2013, 10, 1602–1607. [Google Scholar] [CrossRef]
- Mehlhorn, G.; Obermann, E.; Negri, G.; Bubendorf, L.; Mian, C.; Koch, M.; Sander, H.; Simm, B.; Lütge, M.; Bánrévi, Z.; et al. HPV L1 detection discriminates cervical precancer from transient HPV infection: A prospective international multicenter study. Mod. Pathol. 2013, 26, 967–974. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Lee, A.; Kim, T.-J.; Jin, H.-T.; Seo, Y.-B.; Park, J.-S.; Lee, S.-J. E2/E6 ratio and L1 immunoreactivity as biomarkers to determine HPV16-positive high-grade squamous intraepithelial lesions (CIN2 and 3) and cervical squamous cell carcinoma. J. Gynecol. Oncol. 2018, 29, e38. [Google Scholar] [CrossRef]
- Bin, H.; Ruifang, W.; Ruizhen, L.; Yiheng, L.; Zhihong, L.; Juan, L.; Chun, W.; Yanqiu, Z.; Leiming, W. Detention of HPV L1 Capsid Protein and hTERC Gene in Screening of Cervical Cancer. Iran J. Basic Med. Sci. 2013, 16, 797–802. [Google Scholar] [CrossRef]
- Ungureanu, C.; Socolov, D.; Anton, G.; Mihailovici, M.S.; Teleman, S. Immunocytochemical expression of p16INK4a and HPV L1 capsid proteins as predictive markers of the cervical lesions progression risk O OR RI IG GI IN NA Immunocytochemical expression of p16 INK4a and HPV L1 capsid proteins as predictive markers of the cervical lesions progression risk. Rom. J. Morphol. Embryol. 2010, 51, 497–503. Available online: https://www.rjme.ro/RJME/resources/files/510310497503.pdf (accessed on 8 May 2025).
- Ki, E.Y.; Park, J.S.; Lee, A.; Kim, T.J.; Jin, H.T.; Seo, Y.B.; Gen, Y.; Park, M.Y.; Lee, S.J. Utility of human papillomavirus L1 capsid protein and HPV test as prognostic markers for cervical intraepithelial neoplasia 2+ in women with persistent ASCUS /LSIL cervical cytology. Int. J. Med. Sci. 2019, 16, 1096–1101. [Google Scholar] [CrossRef]
- Xiao, W.; Bian, M.; Liu, J.; Chen, Y.; Yang, B.; Wu, Q. Immunochemical analysis of human papillomavirus L1 capsid protein in liquid-based cytology samples from cervical lesions. Acta Cytol. 2010, 54, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Norman, I.; Hjerpe, A.; Andersson, S. High-risk HPV L1 capsid protein as a marker of cervical intraepithelial neoplasia in high-risk HPV-positive women with minor cytological abnormalities. Oncol. Rep. 2013, 30, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Lee, A.-W.; Kang, C.-S.; Park, J.-S.; Park, D.-C.; Ki, E.-Y.; Lee, K.-H.; Yoon, J.-H.; Hur, S.-Y.; Kim, T.-J. Clinicopathological implications of human papilloma virus (HPV) L1 capsid protein immunoreactivity in HPV16-positive cervical cytology. Int. J. Med. Sci. 2013, 11, 80–86. [Google Scholar] [CrossRef]
- Huang, M.; Li, H.; Nie, X.; Wu, X.; Jiang, X. An analysis on the combination expression of HPV L1 capsid protein and p16INK4a in cervical lesions. Diagn. Cytopathol. 2010, 38, 573–578. [Google Scholar] [CrossRef]
- Yoshida, T.; Sano, T.; Kanuma, T.; Owada, N.; Sakurai, S.; Fukuda, T.; Nakajima, T. Immunochemical analysis of HPV L1 capsid protein and p16 protein in liquid-based cytology samples from uterine cervical lesions. Cancer 2008, 114, 83–88. [Google Scholar] [CrossRef]
- Wang, J.-J.; Lyu, L.-P.; Hu, Q.-W.; Wan, Z.-Q.; Dong, J.; Pan, M.; Shen, W.-W.; Zhang, S. A proper triage for detecting women with high-risk human papillomavirus genotypes other than HPV16/18. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 219, 113–118. [Google Scholar] [CrossRef]
- Rauber, D.; Mehlhorn, G.; Fasching, P.A.; Beckmann, M.W.; Ackermann, S. Prognostic significance of the detection of human papilloma virus L1 protein in smears of mild to moderate cervical intraepithelial lesions. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 140, 258–262. [Google Scholar] [CrossRef]
- Muñoz, N.; Bosch, F.X.; De Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.F.; Meijer, C.J.L.M. Epidemiologic Classification of Human Papillomavirus Types Associated with Cervical Cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Mcmurray, H.R.; Nguyen, D.; Westbrook, T.F.; Mcance, D.J. Biology of human papillomaviruses. Int. J. Exp. Pathol. 2001, 82, 15–33. [Google Scholar] [CrossRef]
- Doorbar, J. The papillomavirus life cycle. J. Clin. Virol. 2005, 32 (Suppl. S1), 7–15. [Google Scholar] [CrossRef]
- Klaes, R.; Woerner, S.M.; Ridder, R.; Wentzensen, N.; Duerst, M.; Schneider, A.; Lotz, B.; Melsheimer, P.; Doeberitz, M.V.K. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999, 59, 6132–6136. [Google Scholar] [PubMed]
- Stanley, M.A. Immune responses to human papilloma viruses. Indian J. Med. Res. 2009, 130, 266. [Google Scholar] [PubMed]
- Zhou, C.; Tuong, Z.K.; Frazer, I.H. Papillomavirus Immune Evasion Strategies Target the Infected Cell and the Local Immune System. Front. Oncol. 2019, 9, 682. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Solomon, D. Findings to date from the ASCUS-LSIL Triage Study (ALTS). Arch. Pathol. Lab. Med. 2003, 127, 946–949. [Google Scholar] [CrossRef]

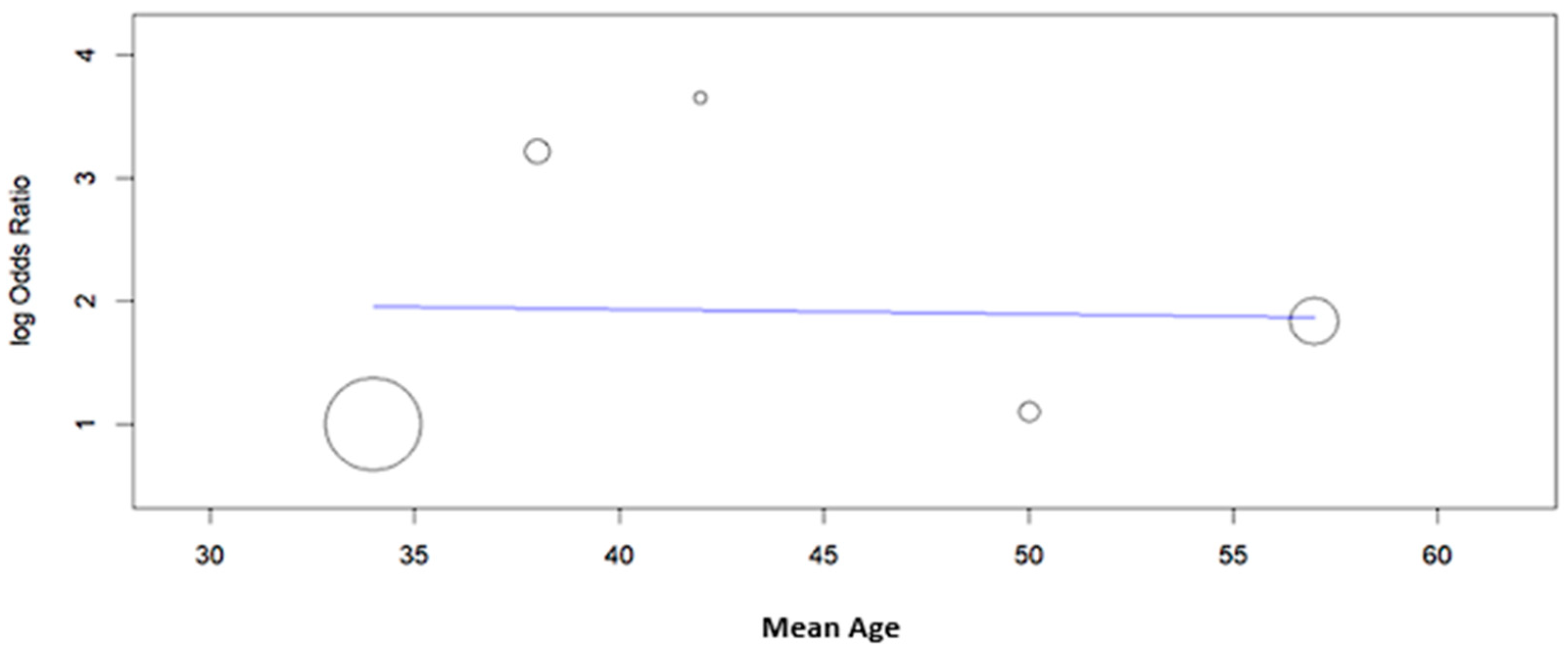
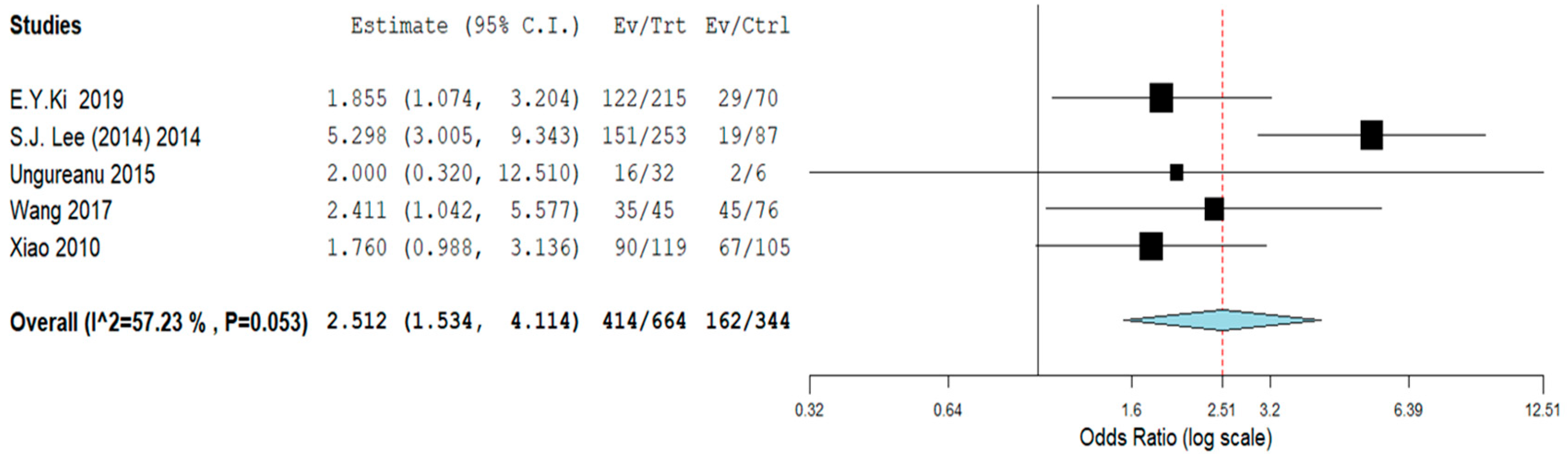
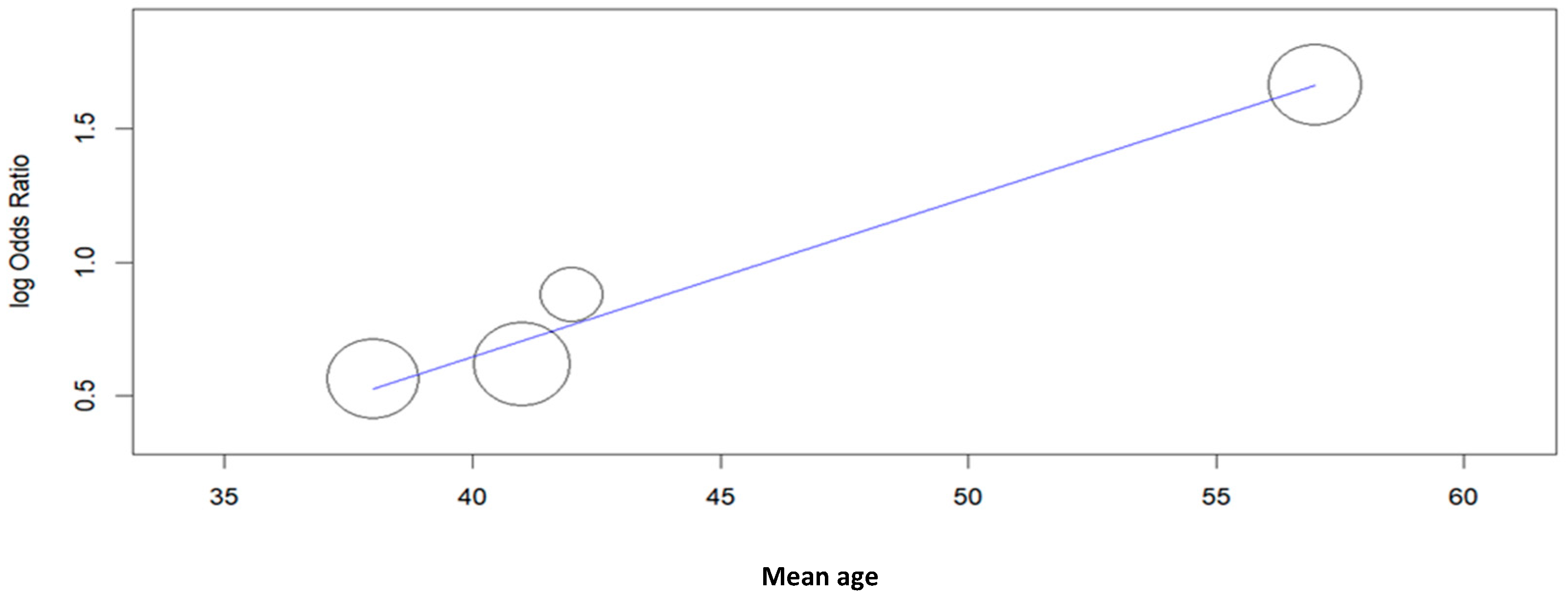
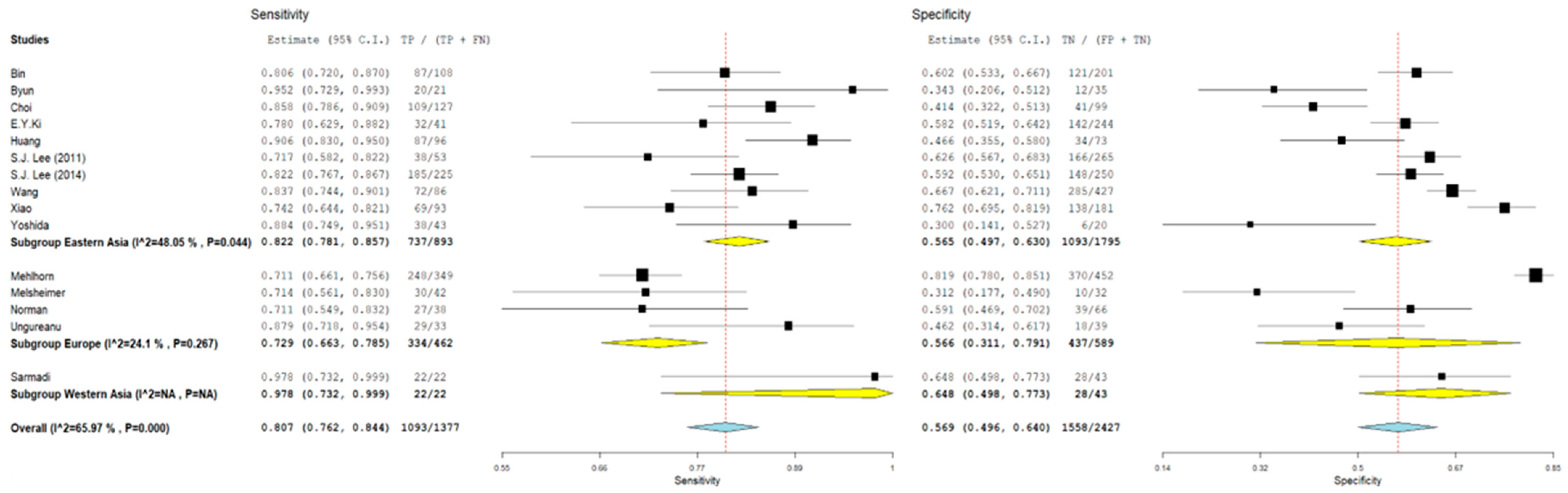
| Study | Type of Study | Mean Age ± SD or (Range), Years | N° of Cases | Cytology (n) | Histology (n) | HPV L1-Positive Cases (n) | Key Findings | |
|---|---|---|---|---|---|---|---|---|
| Cytology (n) | Histology (n) | |||||||
| S. J. Lee, 2011, Korea [9] | PCS | L1+: 41.8 ± 10.1 L1−: 40.9 ± 11.2 | 318 | LSIL (318) | ND | LSIL (181) | ND | Higher HSIL risk in L1-negative; PPV of L1-positive for no progression 91.7%; NPV of L1-negative for HSIL progression 27.7%. |
| P. Melsheimer, 2002, Germany [12] | PCS | 36.5 ± 11.3 | 74 | LSIL (32); HSIL (42) | ND | LSIL (10) HSIL (12) | ND | Reduced HPV L1 expression in HPV 16 DNA-positive HSIL. |
| S. Sarmadi, 2011, Iran [13] | RCS | 38.9 ± 10.7 | 65 | HSIL (22) LSIL (43) | ND | HSIL (22): remission (3) persistence (6) LSIL (28): remission (17) persistent (9) progression (2) | ND | Significant association between HPV L1 positive staining and lack of progression in LSIL. Women under the age of 30 had a significantly higher frequency of positive L1 staining. |
| S. W. Byun, 2013, Korea [17] | PCS | 46 (25–83) | 56 | LSIL (8) ASC-H (26) LSIL-H (30) HSIL (8) | Chronic cervicitis (12) CIN1 (8) CIN2 (5) CIN3 (23) | LSIL (3) HSIL (1) ASC-H LSIL-H (13) | <CIN2 (12) ≥CIN2 (1) | HPV L1 negativity is associated with CIN2+ in follow-up but not with histology; low specificity and PPV for predicting CIN2+. The HPV L1 protein sensitivity and specificity for predicting follow-up CIN2+ were 95.2%, 34.3%, 46.5%, and 92.3%, respectively. |
| G. Mehlhorn, 2013, Germany [18] | PCS | 33.6 (range 15–83) | 908 | LSIL (479) HSIL (322) | CIN1 CIN2 CIN3+ | 471 HPV L1-positive | LSIL (328) HSIL (143) etc. | Significantly higher spontaneous remission in L1-positive LSIL; higher progression risk in L1-negative. |
| Y.-J. Choi, 2018, Korea [19] | RCS |
Normal: 37.8 CIN1: 37.3 CIN2/3: 38.6 SCC: 52.6 | 226 | Normal (21) ASCUS (53) LSIL (66) HSIL (49) SCC (37) | Normal (57) CIN1 (42) CIN 2/3 (69) SCC (48) | ND | Normal (27) CIN1 (14) CIN 2/3 (17) SCC (1) | Significantly lower HPV L1 positivity in CIN2/3 and SCC vs. normal tissue; poor discrimination between CIN1 and CIN2/3 (p < 0.001; 95% l CI = 0.071–0.445). The sensitivity and specificity of the combination of the HPV E2/E6 ratio and HPV L1 were 99.2% and 31.3%, respectively. |
| H. Bin, 2012, China [20] | RCS | 37.3 ± 8.9 | 309 | NILM (72) ASCUS (71) LSIL (80) ASC-H (19) HSIL (49) SCC (18) | Normal (33) CIN 1 (168) CIN 2/3 (84) SCC (24) | ND | Normal (9) CIN 1 (112) CIN 2/3 (21) SCC (0) | HPV L1 negativity correlated with higher-grade cervical lesions. (rs = −0.272, p < 0.001). |
| C. Ungureanu, 2015, Romania [21] | PCS | ND | 76 | NILM (8) ASC-US (6) LSIL (32) HSIL (27) SCC (3) | Normal (12) CIN1 (31) CIN2 (17) CIN3 (13) SCC (3) | NILM (1) ASC-US (2) LSIL (16) HSIL (5) SCC (0) | ND | L1-/p16-pattern frequent in NILM; moderate diagnostic accuracy. The diagnostic test results were Sn = 87.88%, Sp = 46.51%, PPV = 55.8%, and NPV = 83.3%. |
| E. Y. Ki, 2019, Korea [22] | RCS | 40.7 (20–78) | 285 | ASC-US (70) LSIL (215) | CIN1 or cervicitis (244), CIN2+ (41) | ASC-US (29) LSIL (122) | ND | HPV L1 (−) ASC-US/LSIL and HPV 16/18 had a higher risk of CIN2+ than HPV L1 (+). HPV L1 (−) ASCUS progressed to CIN2+ more frequently than LSIL. HPV L1 (−) was more common with other HPV types. |
| W. Xiao, 2010, China [23] | PCS | 37.99 ± 9.40 | 274 | ASC-US (105) LSIL (119) ASC-H (9) HSIL (36) SCC (6) | Cervicitis (96) CIN 1 (85) CIN 2 (55) CIN 3 (32) SCC (6) | LSIL (90) ASC-US (67) ASC-H (1) HSIL + and SCC (4) | Cervicitis (67) CIN 1 (71) CIN 2 (23) CIN 3 (1) SCC (0) | HPV L1 (+) was more frequent in CIN 1 than in CIN 2 and higher in the ASCUS group. Sensitivity of L1 (−) for ≥CIN2 was 74.19%, specificity was 76.24%, and NPV was 85.18%. Regression at 1 year was more frequent with HPV L1 (+). |
| I. Norman, 2013, Sweden [24] | RCS | 32 (23–57) | 104 | WNL (ND) LSIL (ND) | Normal (23) CIN1 (43) CIN2 (23) CIN3+ (15) | ND | Normal (13) CIN1 (26) CIN2+ (11) | L1-negative cases are significantly associated with higher CIN grade and progression to CIN2+ (OR 3.2 [95% CI, 1.081–9.417]). |
| S. J. Lee, 2014, Korea [25] | RCS | 56.6 | 475 | ASC-US (87) LSIL (253) HSIL (94) SCC (41) | ≤CIN1 ≥CIN2 ≤CIN2 ≥CIN3 | ASC-US (19) LSIL (151) HSIL (18) SCC (0) | CIN 1 (148) ≤CIN2 (164) ≥CIN2 (40) ≥CIN3 (24) | Highest HPV L1 positivity in LSIL (p < 0.0001); high percentage of L1-negative cases in ASC-US; higher risk of ≥CIN2 progression in L1-negative ASCUS and LSIL |
| M.-Z. Huang, 2009, China [26] | RCS | 35.09 ± 8.9 | 169 | LSIL HSIL SCC | CIN1 (73) CIN2 (41) CIN3 (28) SCC(27) | ND | CIN1 (34) CIN2 (7) CIN3 (2) SCCs (0) | HPV L1 expression decreased with histological grade; combining L1 with p16 improved specificity. (x2 = 32.86, p < 0.001). |
| T. Yoshida, 2008, Japan [27] | RCS | 50 (24–74) | 63 | LSIL (20) HSIL (40) SCC (3) | CIN2 SCC | LSIL (6) HSIL (5) SCC (0) | ND | L1(−)/p16(+) is highly prevalent in HSIL and SCC; L1(+)/p16(−) is rare. |
| J.-J. Wang, 2017, China [28] | PCS | 42.19 ± 8.06 | 513 | NILM (368) ASCUS (76) LSIL (45) ASC-H (12) HSIL (12) | Normal (312) CIN1 (15) CIN2 (57) CIN3 (28) ICC (1) | NILM (216) ASCUS (45) LSIL (35) ASC-H (2) HSIL (1) | <CIN2 (285) CIN2+ (14) | No correlation between HPV L1 positivity and viral load; higher sensitivity and NPV but lower specificity in the CIN2+ group for HPV L1 detection. (p > 0.05). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrovolskaya, D.; Asaturova, A.; Badlaeva, A.; Tregubova, A.; Mogirevskaya, O.; Dzharullaeva, Z.; Davydova, Y.; Palicelli, A.; Bayramova, G.; Sukhikh, G. Role of L1 HPV Protein Expression in the Cytological Diagnosis of Precancerous Cervical Lesions. J. Clin. Med. 2025, 14, 3376. https://doi.org/10.3390/jcm14103376
Dobrovolskaya D, Asaturova A, Badlaeva A, Tregubova A, Mogirevskaya O, Dzharullaeva Z, Davydova Y, Palicelli A, Bayramova G, Sukhikh G. Role of L1 HPV Protein Expression in the Cytological Diagnosis of Precancerous Cervical Lesions. Journal of Clinical Medicine. 2025; 14(10):3376. https://doi.org/10.3390/jcm14103376
Chicago/Turabian StyleDobrovolskaya, Darya, Aleksandra Asaturova, Alina Badlaeva, Anna Tregubova, Olga Mogirevskaya, Zaira Dzharullaeva, Yulia Davydova, Andrea Palicelli, Guldana Bayramova, and Gennady Sukhikh. 2025. "Role of L1 HPV Protein Expression in the Cytological Diagnosis of Precancerous Cervical Lesions" Journal of Clinical Medicine 14, no. 10: 3376. https://doi.org/10.3390/jcm14103376
APA StyleDobrovolskaya, D., Asaturova, A., Badlaeva, A., Tregubova, A., Mogirevskaya, O., Dzharullaeva, Z., Davydova, Y., Palicelli, A., Bayramova, G., & Sukhikh, G. (2025). Role of L1 HPV Protein Expression in the Cytological Diagnosis of Precancerous Cervical Lesions. Journal of Clinical Medicine, 14(10), 3376. https://doi.org/10.3390/jcm14103376









