The Role of Radiomics and Artificial Intelligence Applied to Staging PSMA PET in Assessing Prostate Cancer Aggressiveness
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Bias Analysis
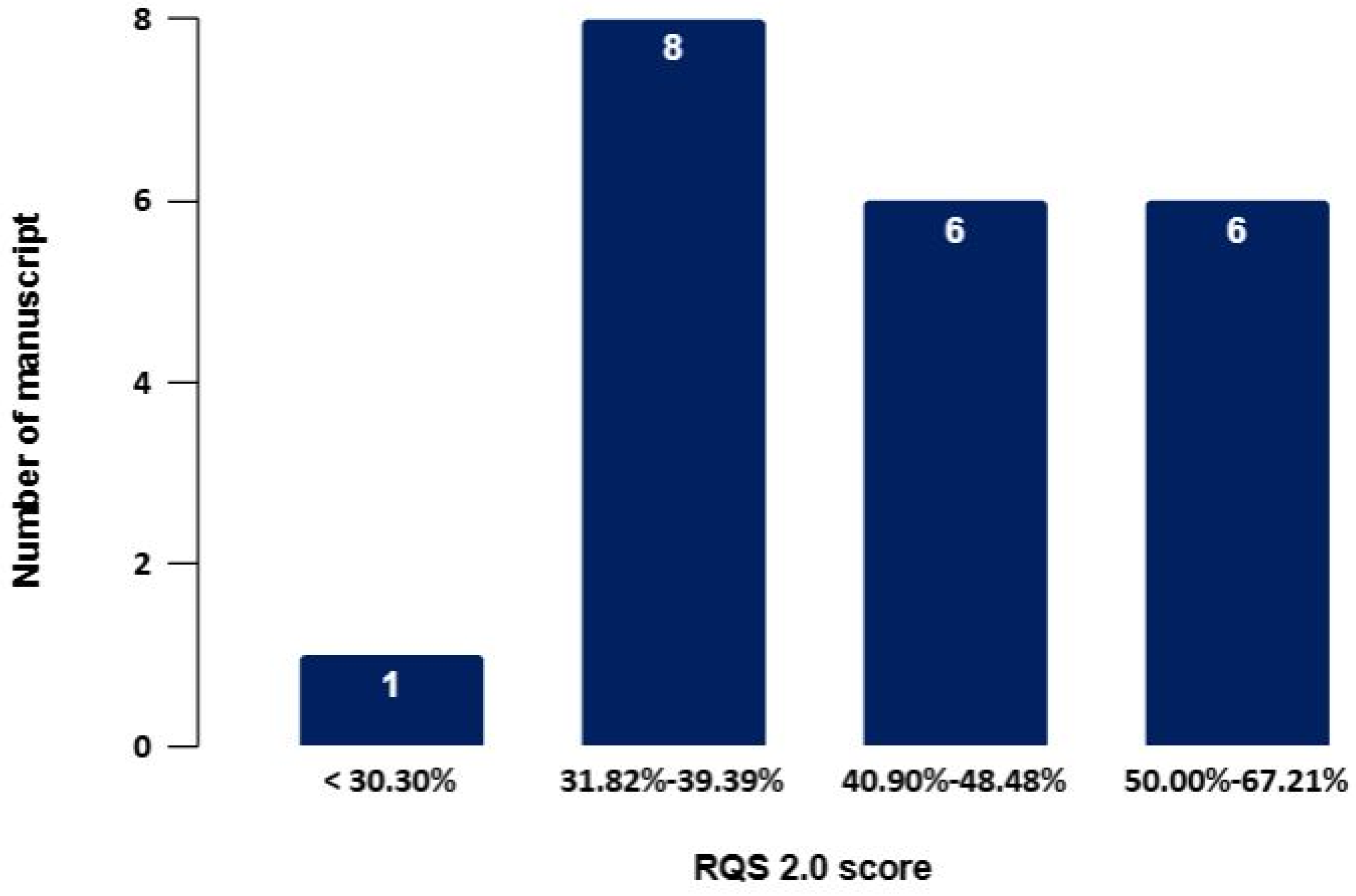
3.2. RQS
3.3. Characterization of the Primary Tumor Prediction of Adverse Pathologic Features of the Primary Tumor
3.4. Prediction of BCR and/or Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Ilic, D.; Djulbegovic, M.; Jung, J.H.; Hwang, E.C.; Zhou, Q.; Cleves, A.; Agoritsas, T.; Dahm, P. Prostate Cancer Screening with Prostate-Specific Antigen (PSA) Test: A Systematic Review and Meta-Analysis. BMJ 2018, 362, k3519. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Brunckhorst, O.; Darraugh, J.; Eberli, D.; De Meerleer, G.; De Santis, M.; Farolfi, A.; et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG Guidelines on Prostate Cancer-2024 Update. Part I: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2024, 86, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Vale, C.L.; Fisher, D.; Kneebone, A.; Parker, C.; Pearse, M.; Richaud, P.; Sargos, P.; Sydes, M.R.; Brawley, C.; Brihoum, M.; et al. Adjuvant or Early Salvage Radiotherapy for the Treatment of Localised and Locally Advanced Prostate Cancer: A Prospectively Planned Systematic Review and Meta-Analysis of Aggregate Data. Lancet 2020, 396, 1422–1431. [Google Scholar] [CrossRef]
- Kupelian, P.A.; Mahadevan, A.; Reddy, C.A.; Reuther, A.M.; Klein, E.A. Use of Different Definitions of Biochemical Failure after External Beam Radiotherapy Changes Conclusions about Relative Treatment Efficacy for Localized Prostate Cancer. Urology 2006, 68, 593–598. [Google Scholar] [CrossRef]
- Cookson, M.S.; Aus, G.; Burnett, A.L.; Canby-Hagino, E.D.; D’Amico, A.V.; Dmochowski, R.R.; Eton, D.T.; Forman, J.D.; Goldenberg, S.L.; Hernandez, J.; et al. Variation in the Definition of Biochemical Recurrence in Patients Treated for Localized Prostate Cancer: The American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel Report and Recommendations for a Standard in the Reporting of Surgical Outcomes. J. Urol. 2007, 177, 540–545. [Google Scholar] [CrossRef]
- Roach, M.; Hanks, G.; Thames, H.; Schellhammer, P.; Shipley, W.U.; Sokol, G.H.; Sandler, H. Defining Biochemical Failure Following Radiotherapy with or without Hormonal Therapy in Men with Clinically Localized Prostate Cancer: Recommendations of the RTOG-ASTRO Phoenix Consensus Conference. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 965–974. [Google Scholar] [CrossRef]
- Bauckneht, M.; Lanfranchi, F.; Albano, D.; Triggiani, L.; Linguanti, F.; Urso, L.; Mazzola, R.; Rizzo, A.; D’Angelo, E.; Dondi, F.; et al. Diverse Imaging Methods May Influence Long-Term Oncologic Outcomes in Oligorecurrent Prostate Cancer Patients Treated with Metastasis-Directed Therapy (the PRECISE-MDT Study). J. Nucl. Med. 2024, 65, 1202–1209. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Filippi, L.; Urso, L.; Bianconi, F.; Palumbo, B.; Marzola, M.C.; Evangelista, L.; Schillaci, O. Radiomics and Theranostics with Molecular and Metabolic Probes in Prostate Cancer: Toward a Personalized Approach. Expert Rev. Mol. Diagn. 2023, 23, 243–255. [Google Scholar] [CrossRef]
- Ibrahim, A.; Primakov, S.; Beuque, M.; Woodruff, H.C.; Halilaj, I.; Wu, G.; Refaee, T.; Granzier, R.; Widaatalla, Y.; Hustinx, R.; et al. Radiomics for Precision Medicine: Current Challenges, Future Prospects, and the Proposal of a New Framework. Methods 2021, 188, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, R.; Treglia, G. Systematic Reviews and Meta-Analyses of Diagnostic Studies: A Practical Guideline. Clin. Transl. Imaging 2017, 5, 83–87. [Google Scholar] [CrossRef]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; The PRISMA-DTA Group; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; et al. Preferred Reporting Items for a Systematic Review and Meta-Analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Papp, L.; Spielvogel, C.P.; Grubmüller, B.; Grahovac, M.; Krajnc, D.; Ecsedi, B.; Sareshgi, R.A.M.; Mohamad, D.; Hamboeck, M.; Rausch, I.; et al. Supervised Machine Learning Enables Non-Invasive Lesion Characterization in Primary Prostate Cancer with [68Ga]Ga-PSMA-11 PET/MRI. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1795–1805. [Google Scholar] [CrossRef]
- Zamboglou, C.; Carles, M.; Fechter, T.; Kiefer, S.; Reichel, K.; Fassbender, T.F.; Bronsert, P.; Koeber, G.; Schilling, O.; Ruf, J.; et al. Radiomic Features from PSMA PET for Non-Invasive Intraprostatic Tumor Discrimination and Characterization in Patients with Intermediate- and High-Risk Prostate Cancer—A Comparison Study with Histology Reference. Theranostics 2019, 9, 2595–2605. [Google Scholar] [CrossRef]
- Luo, L.; Wang, X.; Xie, H.; Liang, H.; Gao, J.; Li, Y.; Xia, Y.; Zhao, M.; Shi, F.; Shen, C.; et al. Role of [18F]-PSMA-1007 PET Radiomics for Seminal Vesicle Invasion Prediction in Primary Prostate Cancer. Comput. Biol. Med. 2024, 183, 109249. [Google Scholar] [CrossRef]
- Cysouw, M.C.F.; Jansen, B.H.E.; Van De Brug, T.; Oprea-Lager, D.E.; Pfaehler, E.; De Vries, B.M.; Van Moorselaar, R.J.A.; Hoekstra, O.S.; Vis, A.N.; Boellaard, R. Machine Learning-Based Analysis of [18F]DCFPyL PET Radiomics for Risk Stratification in Primary Prostate Cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 340–349. [Google Scholar] [CrossRef]
- Yao, F.; Bian, S.; Zhu, D.; Yuan, Y.; Pan, K.; Pan, Z.; Feng, X.; Tang, K.; Yang, Y. Machine Learning-Based Radiomics for Multiple Primary Prostate Cancer Biological Characteristics Prediction with 18F-PSMA-1007 PET: Comparison among Different Volume Segmentation Thresholds. Radiol. Med. 2022, 127, 1170–1178. [Google Scholar] [CrossRef]
- Chan, T.H.; Haworth, A.; Wang, A.; Osanlouy, M.; Williams, S.; Mitchell, C.; Hofman, M.S.; Hicks, R.J.; Murphy, D.G.; Reynolds, H.M. Detecting Localised Prostate Cancer Using Radiomic Features in PSMA PET and Multiparametric MRI for Biologically Targeted Radiation Therapy. EJNMMI Res. 2023, 13, 34. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, L.; Zhou, M.; Li, Y.; Cai, Y.; Gan, Y.; Tang, Y.; Hu, S. [68Ga]Ga-PSMA-617 PET-Based Radiomics Model to Identify Candidates for Active Surveillance amongst Patients with GGG 1–2 Prostate Cancer at Biopsy. Cancer Imaging 2024, 24, 86. [Google Scholar] [CrossRef] [PubMed]
- Feliciani, G.; Celli, M.; Ferroni, F.; Menghi, E.; Azzali, I.; Caroli, P.; Matteucci, F.; Barone, D.; Paganelli, G.; Sarnelli, A. Radiomics Analysis on [68Ga]Ga-PSMA-11 PET and MRI-ADC for the Prediction of Prostate Cancer ISUP Grades: Preliminary Results of the BIOPSTAGE Trial. Cancers 2022, 14, 1888. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Spielvogel, C.P.; Haberl, D.; Trachtova, K.; Stoiber, S.; Rasul, S.; Bystry, V.; Wasinger, G.; Baltzer, P.; Gurnhofer, E.; et al. A Novel Assessment of Whole-Mount Gleason Grading in Prostate Cancer to Identify Candidates for Radical Prostatectomy: A Machine Learning-Based Multiomics Study. Theranostics 2024, 14, 4570–4581. [Google Scholar] [CrossRef]
- Luining, W.I.; Oprea-Lager, D.E.; Vis, A.N.; Van Moorselaar, R.J.A.; Knol, R.J.J.; Wondergem, M.; Boellaard, R.; Cysouw, M.C.F. Optimization and Validation of 18F-DCFPyL PET Radiomics-Based Machine Learning Models in Intermediate- to High-Risk Primary Prostate Cancer. PLoS ONE 2023, 18, e0293672. [Google Scholar] [CrossRef]
- Khateri, M.; Babapour Mofrad, F.; Geramifar, P.; Jenabi, E. Machine Learning-Based Analysis of 68Ga-PSMA-11 PET/CT Images for Estimation of Prostate Tumor Grade. Phys. Eng. Sci. Med. 2024, 47, 741–753. [Google Scholar] [CrossRef]
- Yang, F.; Wang, C.; Shen, J.; Ren, Y.; Yu, F.; Luo, W.; Su, X. End-to-End [18F]PSMA-1007 PET/CT Radiomics-Based Pipeline for Predicting ISUP Grade Group in Prostate Cancer. Abdom. Radiol. 2025, 50, 1641–1652. [Google Scholar] [CrossRef]
- Bian, S.; Hong, W.; Su, X.; Yao, F.; Yuan, Y.; Zhang, Y.; Xie, J.; Li, T.; Pan, K.; Xue, Y.; et al. A Dynamic Online Nomogram Predicting Prostate Cancer Short-Term Prognosis Based on 18F-PSMA-1007 PET/CT of Periprostatic Adipose Tissue: A Multicenter Study. Abdom. Radiol. 2024, 49, 3747–3757. [Google Scholar] [CrossRef]
- Li, T.; Xu, M.; Yang, S.; Wang, G.; Liu, Y.; Liu, K.; Zhao, K.; Su, X. Development and Validation of [18F]-PSMA-1007 PET-Based Radiomics Model to Predict Biochemical Recurrence-Free Survival Following Radical Prostatectomy. Eur. J. Nucl. Med. Mol. Imaging 2024, 51, 2806–2818. [Google Scholar] [CrossRef]
- Öğülmüş, F.E.; Almalıoğlu, Y.; Tamam, M.Ö.; Yıldırım, B.; Uysal, E.; Numanoğlu, Ç.; Özçevik, H.; Tekin, A.F.; Turan, M. Integrating PET/CT, Radiomics and Clinical Data: An Advanced Multi-Modal Approach for Lymph Node Metastasis Prediction in Prostate Cancer. Comput. Biol. Med. 2025, 184, 109339. [Google Scholar] [CrossRef]
- Solari, E.L.; Gafita, A.; Schachoff, S.; Bogdanović, B.; Villagrán Asiares, A.; Amiel, T.; Hui, W.; Rauscher, I.; Visvikis, D.; Maurer, T.; et al. The Added Value of PSMA PET/MR Radiomics for Prostate Cancer Staging. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 527–538. [Google Scholar] [CrossRef]
- Ghezzo, S.; Mapelli, P.; Bezzi, C.; Samanes Gajate, A.M.; Brembilla, G.; Gotuzzo, I.; Russo, T.; Preza, E.; Cucchiara, V.; Ahmed, N.; et al. Role of [68Ga]Ga-PSMA-11 PET Radiomics to Predict Post-Surgical ISUP Grade in Primary Prostate Cancer. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 2548–2560. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Zheng, A.; Gao, J.; Yuan, W.; Shen, C.; Bai, L.; Duan, X. Evaluation of a Radiomics Nomogram Derived from Fluoride-18 PSMA-1007 PET/CT for Risk Stratification in Newly Diagnosed Prostate Cancer. Front. Oncol. 2022, 12, 1018833. [Google Scholar] [CrossRef] [PubMed]
- Aksu, A.; Vural Topuz, Ö.; Yılmaz, G.; Çapa Kaya, G.; Yılmaz, B. Dual Time Point Imaging of Staging PSMA PET/CT Quantification; Spread and Radiomic Analyses. Ann. Nucl. Med. 2022, 36, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Yao, F.; Hong, W.; Xiao, J.; Bian, S.; Zhu, D.; Yuan, Y.; Zhang, Y.; Zhuang, Y.; Yang, Y. Multimodal Radiomics Based on 18F-Prostate-Specific Membrane Antigen-1007 PET/CT and Multiparametric MRI for Prostate Cancer Extracapsular Extension Prediction. Br. J. Radiol. 2024, 97, 408–414. [Google Scholar] [CrossRef]
- Gülbahar Ateş, S.; Demirel, B.B.; Kekilli, E.; Öztürk, E.; Uçmak, G. Primary Tumor Heterogeneity on Pre-Treatment [68Ga]Ga-PSMA PET/CT for the Prediction of Biochemical Recurrence in Prostate Cancer. Rev. Esp. Med. Nucl. Imagen Mol. (Engl. Ed.) 2024, 43, 500032. [Google Scholar] [CrossRef]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.-S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.; Boellaard, R.; Buvat, I. A Guide to ComBat Harmonization of Imaging Biomarkers in Multicenter Studies. J. Nucl. Med. 2022, 63, 172–179. [Google Scholar] [CrossRef]
- Touijer, K.A.; Karnes, R.J.; Passoni, N.; Sjoberg, D.D.; Assel, M.; Fossati, N.; Gandaglia, G.; Eastham, J.A.; Scardino, P.T.; Vickers, A.; et al. Survival Outcomes of Men with Lymph Node-Positive Prostate Cancer After Radical Prostatectomy: A Comparative Analysis of Different Postoperative Management Strategies. Eur. Urol. 2018, 73, 890–896. [Google Scholar] [CrossRef]
- Hofman, M.S.; Lawrentschuk, N.; Francis, R.J.; Tang, C.; Vela, I.; Thomas, P.; Rutherford, N.; Martin, J.M.; Frydenberg, M.; Shakher, R.; et al. Prostate-Specific Membrane Antigen PET-CT in Patients with High-Risk Prostate Cancer before Curative-Intent Surgery or Radiotherapy (proPSMA): A Prospective, Randomised, Multicentre Study. Lancet 2020, 395, 1208–1216. [Google Scholar] [CrossRef]
- Luiting, H.B.; van Leeuwen, P.J.; Busstra, M.B.; Brabander, T.; van der Poel, H.G.; Donswijk, M.L.; Vis, A.N.; Emmett, L.; Stricker, P.D.; Roobol, M.J. Use of Gallium-68 Prostate-Specific Membrane Antigen Positron-Emission Tomography for Detecting Lymph Node Metastases in Primary and Recurrent Prostate Cancer and Location of Recurrence after Radical Prostatectomy: An Overview of the Current Literature. BJU Int. 2020, 125, 206–214. [Google Scholar] [CrossRef]
- Albano, D.; Temponi, A.; Bertagna, F.; Suardi, N.; Talin, A.; Bonù, M.L.; Triggiani, L. The Prognostic Role of Staging [18F]PSMA-1007 PET/CT Volumetric and Dissemination Features in Prostate Cancer. Ann. Nucl. Med. 2025, 39, 518–526. [Google Scholar] [CrossRef]
- Urso, L.; Manco, L.; Cittanti, C.; Adamantiadis, S.; Szilagyi, K.E.; Scribano, G.; Mindicini, N.; Carnevale, A.; Bartolomei, M.; Giganti, M. 18F-FDG PET/CT Radiomic Analysis and Artificial Intelligence to Predict Pathological Complete Response after Neoadjuvant Chemotherapy in Breast Cancer Patients. Radiol. Med. 2025, 130, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, P.; Marturano, F.; Bettinelli, A.; Gregianin, M.; Paiusco, M.; Evangelista, L. Additional Value of PET Radiomic Features for the Initial Staging of Prostate Cancer: A Systematic Review from the Literature. Cancers 2021, 13, 6026. [Google Scholar] [CrossRef] [PubMed]
- Ciarmiello, A.; Giovannini, E.; Tutino, F.; Yosifov, N.; Milano, A.; Florimonte, L.; Bonatto, E.; Bareggi, C.; Dellavedova, L.; Castello, A.; et al. Does FDG PET-Based Radiomics Have an Added Value for Prediction of Overall Survival in Non-Small Cell Lung Cancer? J. Clin. Med. 2024, 13, 2613. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, L.; Fiz, F.; Laudicella, R.; Bianconi, F.; Castello, A.; Guglielmo, P.; Liberini, V.; Manco, L.; Frantellizzi, V.; Giordano, A.; et al. PET Radiomics and Response to Immunotherapy in Lung Cancer: A Systematic Review of the Literature. Cancers 2023, 15, 3258. [Google Scholar] [CrossRef]
- Ulas Babacan, O.; Hasbek, Z.; Seker, K. The Relationship between D’Amico and ISUP Risk Classifications and 68Ga-PSMA PET/CT SUVmax Values in Newly Diagnosed Prostate Cancers. Curr. Oncol. 2024, 31, 5307–5317. [Google Scholar] [CrossRef]
- Pepe, P.; Pepe, L.; Tamburo, M.; Marletta, G.; Savoca, F.; Pennisi, M.; Fraggetta, F. 68Ga-PSMA PET/CT and Prostate Cancer Diagnosis: Which SUVmax Value? In Vivo 2023, 37, 1318–1322. [Google Scholar] [CrossRef]
- Urso, L.; Panareo, S.; Castello, A.; Ambrosio, M.R.; Zatelli, M.C.; Caracciolo, M.; Tonini, E.; Valpiani, G.; Boschi, A.; Uccelli, L.; et al. Glucose Metabolism Modification Induced by Radioligand Therapy with [177Lu]Lu/[90Y]Y-DOTATOC in Advanced Neuroendocrine Neoplasms: A Prospective Pilot Study within FENET-2016 Trial. Pharmaceutics 2022, 14, 2009. [Google Scholar] [CrossRef]
- Goel, S.; Shoag, J.E.; Gross, M.D.; Al Hussein Al Awamlh, B.; Robinson, B.; Khani, F.; Baltich Nelson, B.; Margolis, D.J.; Hu, J.C. Concordance Between Biopsy and Radical Prostatectomy Pathology in the Era of Targeted Biopsy: A Systematic Review and Meta-Analysis. Eur. Urol. Oncol. 2020, 3, 10–20. [Google Scholar] [CrossRef]
- Tilki, D.; Mandel, P.; Schlomm, T.; Chun, F.K.-H.; Tennstedt, P.; Pehrke, D.; Haese, A.; Huland, H.; Graefen, M.; Salomon, G. External Validation of the CAPRA-S Score to Predict Biochemical Recurrence, Metastasis and Mortality after Radical Prostatectomy in a European Cohort. J. Urol. 2015, 193, 1970–1975. [Google Scholar] [CrossRef]
- De Visschere, P.J.L.; Standaert, C.; Fütterer, J.J.; Villeirs, G.M.; Panebianco, V.; Walz, J.; Maurer, T.; Hadaschik, B.A.; Lecouvet, F.E.; Giannarini, G.; et al. A Systematic Review on the Role of Imaging in Early Recurrent Prostate Cancer. Eur. Urol. Oncol. 2019, 2, 47–76. [Google Scholar] [CrossRef]
- Falagario, U.G.; Abbadi, A.; Remmers, S.; Björnebo, L.; Bogdanovic, D.; Martini, A.; Valdman, A.; Carrieri, G.; Menon, M.; Akre, O.; et al. Biochemical Recurrence and Risk of Mortality Following Radiotherapy or Radical Prostatectomy. JAMA Netw. Open 2023, 6, e2332900. [Google Scholar] [CrossRef] [PubMed]
- Van Den Broeck, T.; Van Den Bergh, R.C.N.; Arfi, N.; Gross, T.; Moris, L.; Briers, E.; Cumberbatch, M.; De Santis, M.; Tilki, D.; Fanti, S.; et al. Prognostic Value of Biochemical Recurrence Following Treatment with Curative Intent for Prostate Cancer: A Systematic Review. Eur. Urol. 2019, 75, 967–987. [Google Scholar] [CrossRef] [PubMed]
- Fradin, J.; Kim, F.J.; Lu-Yao, G.L.; Storozynsky, E.; Kelly, W.K. Review of Cardiovascular Risk of Androgen Deprivation Therapy and the Influence of Race in Men with Prostate Cancer. Cancers 2023, 15, 2316. [Google Scholar] [CrossRef] [PubMed]
- Dale, W.; Hemmerich, J.; Bylow, K.; Mohile, S.; Mullaney, M.; Stadler, W.M. Patient Anxiety About Prostate Cancer Independently Predicts Early Initiation of Androgen Deprivation Therapy for Biochemical Cancer Recurrence in Older Men: A Prospective Cohort Study. J. Clin. Oncol. 2009, 27, 1557–1563. [Google Scholar] [CrossRef]
- Karpinski, M.J.; Hüsing, J.; Claassen, K.; Möller, L.; Kajüter, H.; Oesterling, F.; Grünwald, V.; Umutlu, L.; Kleesiek, J.; Telli, T.; et al. Combining PSMA-PET and PROMISE to Re-Define Disease Stage and Risk in Patients with Prostate Cancer: A Multicentre Retrospective Study. Lancet Oncol. 2024, 25, 1188–1201. [Google Scholar] [CrossRef]


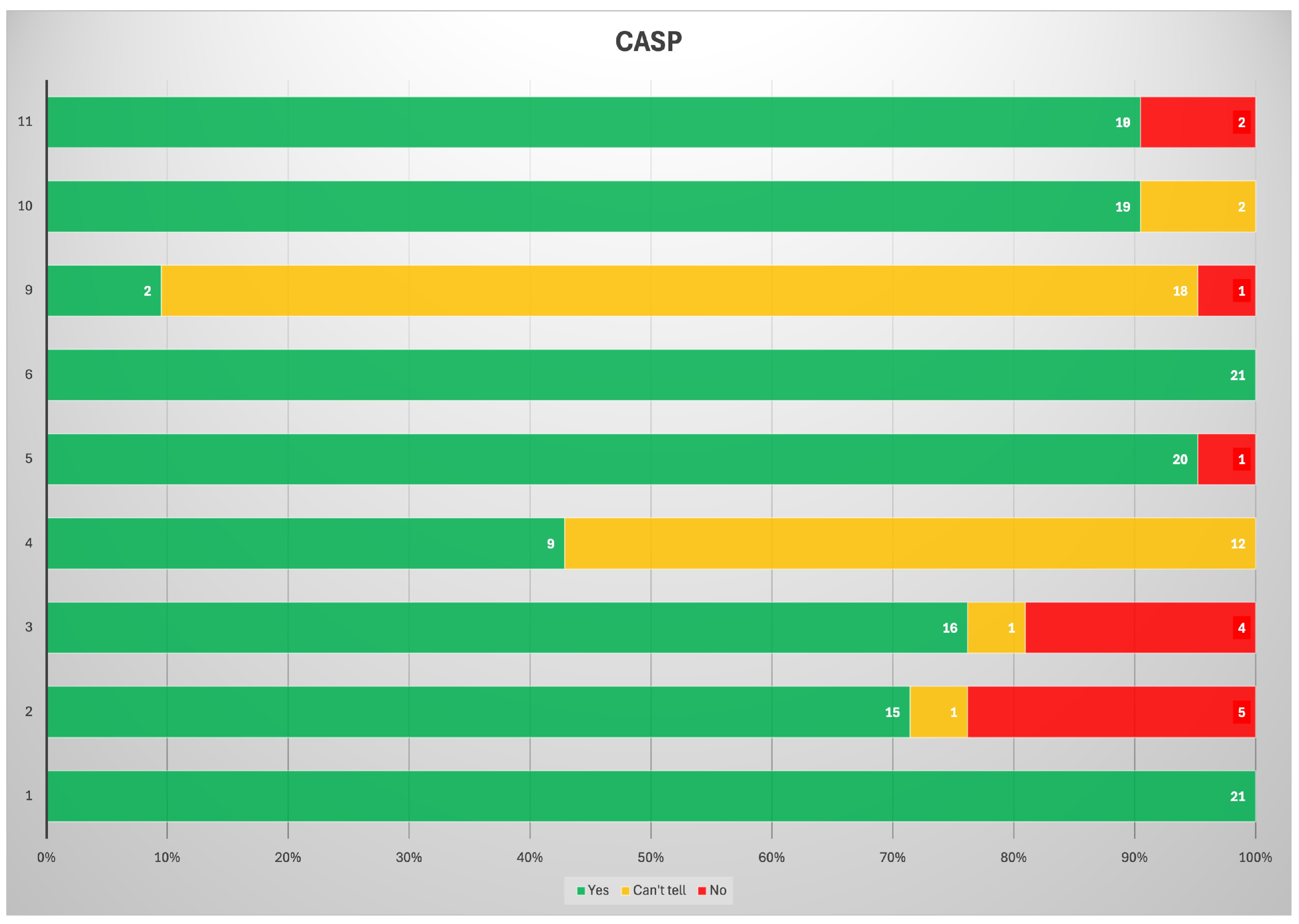
| Author | Year | Study Design | N. pts | Radiotracer Imaging Modality of Radiomic Analysis | Aim | Standard of Reference | Main Results |
|---|---|---|---|---|---|---|---|
| Papp et al. [15] | 2020 | P | 52 | [18F]F-choline and [68Ga]Ga-PSMA-11PET/MRI | Predict GS (≥4 vs. <4), BCR after RP, and OPR | Histopathology after RP | ML models showed better accuracy than conventional PET parameters used in daily clinical practice to predict ISUP grade group, BCR, and OPR |
| Zamboglou et al. [16] | 2019 | P | 20 | [68Ga]Ga-PSMA-11PET | Predict ISUP grade group (≥ 3 vs. <3) and lymph node involvement | Histopathology after RP | GLSZM was able to accurately predict ISUP grade group and pN0 vs. pN1. |
| Luo et al. [17] | 2024 | R | 140 | [18F]F-PSMA-1007PET/CT | Predict seminal vesicle invasion and BCR after RP | Histopathology after RP | The ML models accurately predicted seminal vesicle involvement and stratified PCa patients according to their risk of BCR after RP. |
| Cysouw et al. [18] | 2021 | P | 76 | [18F]DCFPyLPET/CT | Predict ISUP grade group (≥3 vs. <3), ECE, lymph node involvement, and metastatic disease | Histopathology after RP | ML models outperformed conventional PET parameters for risk stratification of PCa patients |
| Yao et al. [19] | 2022 | R | 173 | [18F]F-PSMA-1007PET | Predict ISUP grade group, ECE, and VI | Histopathology after RP | [18F]F-PSMA-1007 PET-based radiomics features at 40–50% SUVmax were able to predict multiple PCa biological features |
| Chan et al. [20] | 2023 | P | 19 | [68Ga]Ga-PSMA-11PET and mpMRI | Predict tumor location and grade | Histopathology after RP | ML models, based on PET and mpMRI, differentiated well between low- and high-risk PCa |
| Yang et al. [21] | 2024 | P | 75 | [68Ga]Ga-PSMA-117PET/CT | Predict adverse pathology in patients with biopsy GS 1–2 | Histopathology after RP | The combined model, radiomics + PSA ratio, was superior to the single model for stratifying GS 1–2 patients. |
| Feliciani et al. [22] | 2022 | R | 28 | [68Ga]Ga-PSMA-11PET and mpMRI | Predict ISUP grade group (1 vs. ≥2) | Histopathology after RP | Radiomics extracted from both MRI-ADC and [68Ga]Ga-PSMA-11 PET could predict ISUP grade group. |
| Ning et al. [23] | 2024 | P | 65 | [68Ga]Ga-PSMA-11PET/MRI | Assess total GS grading compared to biopsy-derived GS | Histopathology after RP | Random forest was superior to biopsy alone GS in terms of AUC, accuracy, specificity, and NPV. |
| Luining et al. [24] | 2023 | R | 72 | [18F]DCFPyLPET | Validate the results obtained in the previous original article [Cysouw et al.] [18] through a multicenter experience | Histopathology after RP | The models maintained a high prediction accuracy at external validation to discriminate high-risk vs. low-risk PCa |
| Khateri et al. [25] | 2024 | R | 90 | [68Ga]Ga-PSMA-11PET | Build ML models to predict GS (≤7 vs. >7) using imaging acquired in three different institutions | Histopathology after RP | ML models could accurately predict post-surgical ISUP grade group. ComBat harmonization algorithm enhanced the models’ performances and enables inter-center generalizability. |
| Yang et al. [26] | 2024 | R | 356 | [18F]F-PSMA-1007PET/CT | Build ML models to predict ISUP grade group (<4 vs. ≥4) | Histopathology at prostate biopsy or after RP | ML models built on radiomic analysis outperformed the clinical model to predict ISUP grade group. |
| Bian et al. [27] | 2024 | R | 268 | [18F]F-PSMA-1007PET/CT | Predict short term prognosis, based on periprostatic adipose tissue assessment | Histopathology at prostate biopsy or after RP | The radiomics-clinical combined model demonstrated an optimal performance in terms of AUC. |
| Li et al. [28] | 2024 | R | 236 | [18F]F-PSMA-1007PET | Predict BCR after RP | BCR after RP | PET-based clinical-radiomics model showed high predictive performance. |
| Öğülmüş et al. [29] | 2025 | R | 229 | [68Ga]Ga-PSMA-11PET/CT | Predict lymph nodes metastases | PET visual analysis | The AI model outperformed the reader’s analysis. |
| Solari et al. [30] | 2021 | R | 101 | [68Ga]Ga-PSMA-11PET/MRI | Predict ISUP grade group (≥3 vs. <3) | Histopathology after RP | The combination of PET + ADC radiomic analysis outperformed the prostate biopsy in terms of prediction accuracy of post-surgical ISUP grade group. |
| Ghezzo et al. [31] | 2023 | R | 47 | [68Ga]Ga-PSMA-11PET | Build ML models to predict ISUP grade group (<4 vs. ≥4) | Histopathology after RP | All radiomics-based ML models trained with at least two RFs outperformed the control models. |
| Wang et al. [32] | 2022 | R | 161 | [18F]F-PSMA-1007PET/CT | Combine clinical and PSMA PET/CT radiomic features to build a nomogram for prognostic stratification of PCa patients | Histopathology at prostate biopsy or after RP | A radiomic signature identified was significantly correlated to both PSA values and ISUP grade group. The radiomics nomogram demonstrated a higher specificity (81.3%) than the radiomics features alone (78.1%). |
| Aksu et al. [33] | 2022 | R | 41 | [68Ga]Ga-PSMA-11PET | Predict ISUP grade group (≥3 vs. <3) according volumetric and radiomic analysis of early and late PSMA PET; investigate the relationship between Dmax obtained in early PET images and histopathology and PSA. | Grading at prostate biopsy | Some radiomic features extracted from both early and late PSMA PET images accurately predicted the ISUP grade group. Dmax was strongly correlated with higher values of PSA and PSMA PET volumetric parameters and was higher in patients with ISUP grade group ≥ 3 |
| Pan et al. [34] | 2024 | R | 197 | [18F]F-PSMA-1007PET/CT and mpMRI | Predict ECE with multimodal radiomic analysis | Histopathology after RP | The mpMRI radiomic model was the most accurate for predicting ECE. The multimodal radiomic model outperformed the PET/CT model but did not improve the accuracy of the mpMRI model. |
| Gülbahar Ates et al. [35] | 2024 | R | 51 | [68Ga]Ga-PSMA-11PET | Predict BCR in patients who underwent RT or RP | BCR after RP or RT | INTENSITY-BASED-minimum grey level and GLCM-sum variance were independent predictors of BCR. |
| Author | Segmentation Method (Algorithm) | Segmentation SW (Class) | Radiomics FTs Type (n) | Selected FTs | Radiomic SW (Class) | Statistical Analysis to Reduce Redundant Variables | RQS 2.0 (%) |
|---|---|---|---|---|---|---|---|
| Papp et al. [15] | Semi-automatic (standard three-dimensional iso-count VOIs) | Hybrid 3D V4.0.0 (C) | shaped-based first-order second or higher order (GLCM, GLSZM, GLRLM, NGLDM, GLDZM) (FTs n = 446) | 80 | MUW Radiomics Engine V2.0 (IH) | Covariance matrix analysis, Pearson correlation coefficient | 27 (40.91%) |
| Zamboglou et al. [16] | Semi-automatic (WL 0–5 SUV, threshold of 40% of SUVmax, coregistration of the histopathology with PET image) | MITK V2016.11 (OS) 3D-Slicer V4 (OS) | first order second or higher order (GLCM, GLRLM, GLSZM, NGTDM, WBFP) (FTs n = 133) | 131 | MATLAB (C) | Wilcoxon Rank test, Spearman correlation coefficient | 30 (45.45%) |
| Luo et al. [17] | Manual and semi-automatic (threshold 40% SUVmax) | uAI Research Portal (C) | shaped-based first-order second or higher order (GLCM, GLSZM, GLRLM, GLDM, NGTDM) (FTs n = 2264) | PET:20 CT:11 | uAI Research Portal (C) | Relief, SelectKBest and LASSO | 21 (31.82%) |
| Cysouw et al. [18] | Semi-automatic (region-growing algorithm with a background adapted peak threshold) | N.A. | shaped-based first-order second or higher order (GLCM, GLRLM, GLSZM, GLDZM, NGTDM, NGLDM) (FTs n = 480) | 48 | RaCaT (OS) | PCA, RF, ANOVA | 23 (34.85%) |
| Yao et al. [19] | Semi-automatic (thresholds 30%, 40%, 50%, and 60% SUVmax) | LIFEx V6.30 (OS) | shaped-based first-order second or higher order (GLCM, GLRM, NGLDM, GLZLM) (FTs n = 70) | 10 | LIFEx V6.30 (OS) | ICC, mRMR | 23 (34.85%) |
| Chan et al. [20] | Semi-automatic (guided by histology) | 3D-Slicer | Shape features first-order second or higher order (GLCM, GLRLM, GLSZM, NGTDM, GLDM) Wavelet, GM, LoG, LBP (FTs n = n.d.) | 30 | Python (OS) | -reject all highly correlated features -retain the top 10% of the features based on the ANOVA test -retain the top 50 features based on the mean decrease in random forest Gini impurity. | 26 (39.39%) |
| Yang et al. [21] | Manual | 3D-Slicer V5.3.0 | shaped-based first-order second or higher order (GLCM, GLDM, GLRLM, GLSZM, NGTDM) (FTs n = 107) | 6 | Python V3.7.4 (OS) | mRMR, LASSO | 30 (45.45%) |
| Feliciani et al. [22] | Manual for MR imaging; semi-automatic for PET imaging (threshold SUV(bw)max of 3 g/mL) | Watson Elementary for MR imaging (C) MIM Maestro for PET imaging (C) | first order, second or higher order (GLCM, GLRM) (FTs n = 218) | PET: 29 MRI-ADC:87 | SOPHiA DDM™ (C) | ICC, LASSO | 29 (43.94%) |
| Ning et al. [23] | Manual | N.A. | Shape-based first-order second or higher order (GLCM, GLRLM, GLSZM, GLDM, NGTDM) (FTs n = 203) | 57 | Python (OS) | mRMR | 29 (43.94) |
| Luining et al. [24] | Semi-automatic (region growing with threshold 50%, 55%, 60%, 65%, and 70% SUVpeak) | ACCURATE tool (OS) | shaped-based first-order second-order or higher order (FTs n = 480) | 184 | RaCaT (OS) | PCA, RFE, univariate feature selection, LASSO | 33 (50.00%) |
| Khateri et al. [25] | Manual | LIFEx V7.0.0 | first order second or higher order (GLCM, GLRLM, NGLDM, GLZLM) (FTs n = 69) | 30 | Python (OS) | mRMR, ANOVA, KW, Relief | 42 (63.64%) |
| Yang et al. [26] | Manual and automatic (DL tool Total- Segment) | LIFEx V7.3.0 | shaped-based first-order second or higher order (GLCM, GLRLM, NGTDM, GLSZM) (FTs n = 215) | 134 | Python (OS) | LASSO, RFE, REIF, MUIF, mRMR, IFGN | 41 (62.12%) |
| Bian et al. [27] | Semi-automatic | 3D Slicer V4.11 | shaped-based first-order (GLCM, NGLDM, GLZLM, GLRLM) FTs n = n.d. | 25 | LIFEx V6.30 | mRMR, LASSO | 33 (50.00%) |
| Li et al. [28] | Manual | LIFEx V7.3.0 | shaped-based first-order second or higher order (GLCM, GLSZM, GLRLM, NGTDM) (FTs n = 124) | 3 | LIFEx V7.3.0 | ICC, LASSO | 33 (50.00%) |
| Öğülmüş et al. [29] | Automatic (DL) | Python | shape Features first-order second or higher order (GLCM, GLRLM, GLSZM, GLDM, NGTDM) (FTs n = 105) | n.d. | Python (OS) | DL model | * 41 (67.21%) |
| Solari et al. [30] | Automatic on PET images (FLAB); manual on MR images | FLAB segmentation tool (IH) | shaped-based first-order second or higher order (GLCM, GLSZM, LRLM, NGTDM, and GLDM) (FTs n = 107) | T1w: 9 T2w: 7 ADC: 7 PET: 9 PET + T1w: 10 PET + T2w: 7 PET + ADC: 9 | N.A. | RFE | 22 (33.33%) |
| Ghezzo et al. [31] | Manual | 3D-Slicer V29 | shaped-based first-order second or higher order (GLCM, GLSZM, GLRLM, NGTDM, GLDM) (FTs n = 103) | 4 | ComBat SW | mRMR | 25 (37.88%) |
| Wang et al. [32] | Semi-automatic (threshold 40% of SUVmax) | ITKSNAP V3.8 (OS) | shaped-based first-order second or higher order (GLCM, GLSZM, GLRLM, NGTDM, GLDM) Wavelet, LoG, GFF (FTs n = 944) | 30 | Philips Radiomics Tool (C) | LASSO | 29 (43.94%) |
| Aksu et al. [33] | Semi-automatic (PSMA uptake above 2.5 SUV) | LIFEx (OS) | shaped-based second order or higher order (GLCM, NGLDM, GLRLM, GLZLM) (FTs n = 41) | 36 | LIFEx (OS) | Spearman correlation, Mann-Whitney U test | 20 (30.30%) |
| Pan et al. [34] | Manual and semi-automatic (threshold 40% SUVmax) | ITKSNAP V3.6 (OS) | shape features first-Order second or higher order (GLCM, GLDM, GLRLM, GLSZM, NGTDM) (FTs n = 1316) ** shape features first-Order second or higher order (GLCM, GLRLM, NGLDM, GLZLM) (FTs n = 140) *** | 20 | LIFEx V6.3 | mRMR, LASSO | 24 (36.36%) |
| Gülbahar Ates et al. [35] | Semi-automatic (threshold 40% SUVmax) | LIFEx | shaped-based first-order second or higher order (GLCM, GLRLM, NGTDM, GLSZM) (FTs n = n.d.) | 3 | LIFEx V7.3 | univariate and multivariate analysis | 24 (36.36%) |
| Authors | AI SW (Class) | Data-Mining Methods | Training/Test Set (n. of Patients) | Validation (n. of Patients) | Performance Score | Clinical Application |
|---|---|---|---|---|---|---|
| Papp et al. [15] | N.A. | RF | 52 | Internal (52) | AUC | N.A. |
| Zamboglou et al. [16] | R V.3.4.4 (OS) SPSS V24 (C) | LR | 20 | Internal (40) | AUC, ROC | N.A. |
| Luo et al. [17] | uAI Research Portal IBM SPSS V25.0 (C) | LR, RF, SVM | 112/28 | N.A. | AUC, SEN, SPEC, ACC | N.A. |
| Cysouw et al. [18] | Python V3.6 (OS) | RF | 61 | Internal (15) | AUC | N.A. |
| Yao et al. [19] | IBM SPSS V25.0 (C) R V4.0.2 (OS) | SVM with RBF kernel | 122/51 | N.A. | AUC, ACC, SEN, SPE, F1Score, NRI | N.A. |
| Chan et al. [20] | Python (OS) | RFC, SVC | 19 | N.A. | AUC, ROC, SEN, SPE | N.A. |
| Yang et al. [21] | IBM SPSS V26.0 (C) R (OS) | LR | 52 | Internal (23) | AUC, SEN, SPE, PPV, NPV, Radscore | N.A. |
| Feliciani et al. [22] | R,RStudio (OS) | LR | 19/9 | N.A. | AUC, ROC | N.A. |
| Ning et al. [23] | Python (OS) | kNN, RF, XGBoost, SVM, LR | 45 | Internal (20) | AUC, SPE, ACC, PPV, NPV, SEN | N.A. |
| Luining et al. [24] | Python V3.7 (OS) | RF, LR | 72/24 | External (27) | AUC, ROC, SEN, SPE | nomogram |
| Khateri et al. [25] | Python (OS) | LR, KNN, ET, LDA, RF | 62/16 | External (12) | AUC, ROC, PREC, ACC, REC, F1Score | N.A. |
| Yang et al. [26] | Python (OS) | LR, RF, SVM, GBDT, and XGBoost | 241 | External (115) | AUC, ROC, bACC | DC |
| Bian et al. [27] | IBM SPSS V25.0 (C) R V4.0.2 (OS) | LR | 156/65 | External (47) | AUC, ROC, Radscore, NRI | Nomogram, DC |
| Li et al. [28] | R V4.1.1 (OS) | univariate and multivariate Cox regression analysis | 236 | External (98) | AUC, C-index | Nomogram, DC |
| Öğülmüş et al. [29] | Python V3.9 (OS) | DL | 181/48 | N.A. | AUC, ROC, ACC, PREC, REC, F1 Score, MCC | N.A. |
| Solari et al. [30] | Python (OS) | SVM with RBF and a “one-vs-rest” multi-class approach | 67 + 53 * | Internal (34) | bACC, SEN, SPE | N.A. |
| Ghezzo et al. [31] | Python V3.7 (OS) | LR, SVM, KNN | 50 | Internal (10) | bACC, SEN, SPE, PPV, NPV | N.A. |
| Wang et al. [32] | R V4.0.2 (OS) IBM SPSS V13.0 (C) | LR | 112 | Internal (49) | AUC, ROC, NPV, PPV | nomogram |
| Aksu et al. [33] | IBM SPSS V28.0 (C) | LR | N.A. | N.A. | AUC, ROC | N.A. |
| Pan et al. [34] | IBM SPSS V25.0 © R V4.0.2 (OS) | LR | 139/58 | N.A. | AUC, ACC, SEN, SPE, NPV, PPV, NRI | N.A. |
| Gülbahar Ates et al. [35] | IBM SPSS V25.0 (C) | Cox regression analysis | 51 | N.A. | Youden’s index, ROC | Kaplan-Meier |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urso, L.; Badrane, I.; Manco, L.; Castello, A.; Lancia, F.; Collavino, J.; Crestani, A.; Castellani, M.; Cittanti, C.; Bartolomei, M.; et al. The Role of Radiomics and Artificial Intelligence Applied to Staging PSMA PET in Assessing Prostate Cancer Aggressiveness. J. Clin. Med. 2025, 14, 3318. https://doi.org/10.3390/jcm14103318
Urso L, Badrane I, Manco L, Castello A, Lancia F, Collavino J, Crestani A, Castellani M, Cittanti C, Bartolomei M, et al. The Role of Radiomics and Artificial Intelligence Applied to Staging PSMA PET in Assessing Prostate Cancer Aggressiveness. Journal of Clinical Medicine. 2025; 14(10):3318. https://doi.org/10.3390/jcm14103318
Chicago/Turabian StyleUrso, Luca, Ilham Badrane, Luigi Manco, Angelo Castello, Federica Lancia, Jeanlou Collavino, Alessandro Crestani, Massimo Castellani, Corrado Cittanti, Mirco Bartolomei, and et al. 2025. "The Role of Radiomics and Artificial Intelligence Applied to Staging PSMA PET in Assessing Prostate Cancer Aggressiveness" Journal of Clinical Medicine 14, no. 10: 3318. https://doi.org/10.3390/jcm14103318
APA StyleUrso, L., Badrane, I., Manco, L., Castello, A., Lancia, F., Collavino, J., Crestani, A., Castellani, M., Cittanti, C., Bartolomei, M., & Giannarini, G. (2025). The Role of Radiomics and Artificial Intelligence Applied to Staging PSMA PET in Assessing Prostate Cancer Aggressiveness. Journal of Clinical Medicine, 14(10), 3318. https://doi.org/10.3390/jcm14103318






