Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohorts
2.2. Ethics Approval and Consent to Participate
2.3. The SeptiCyte RAPID Test
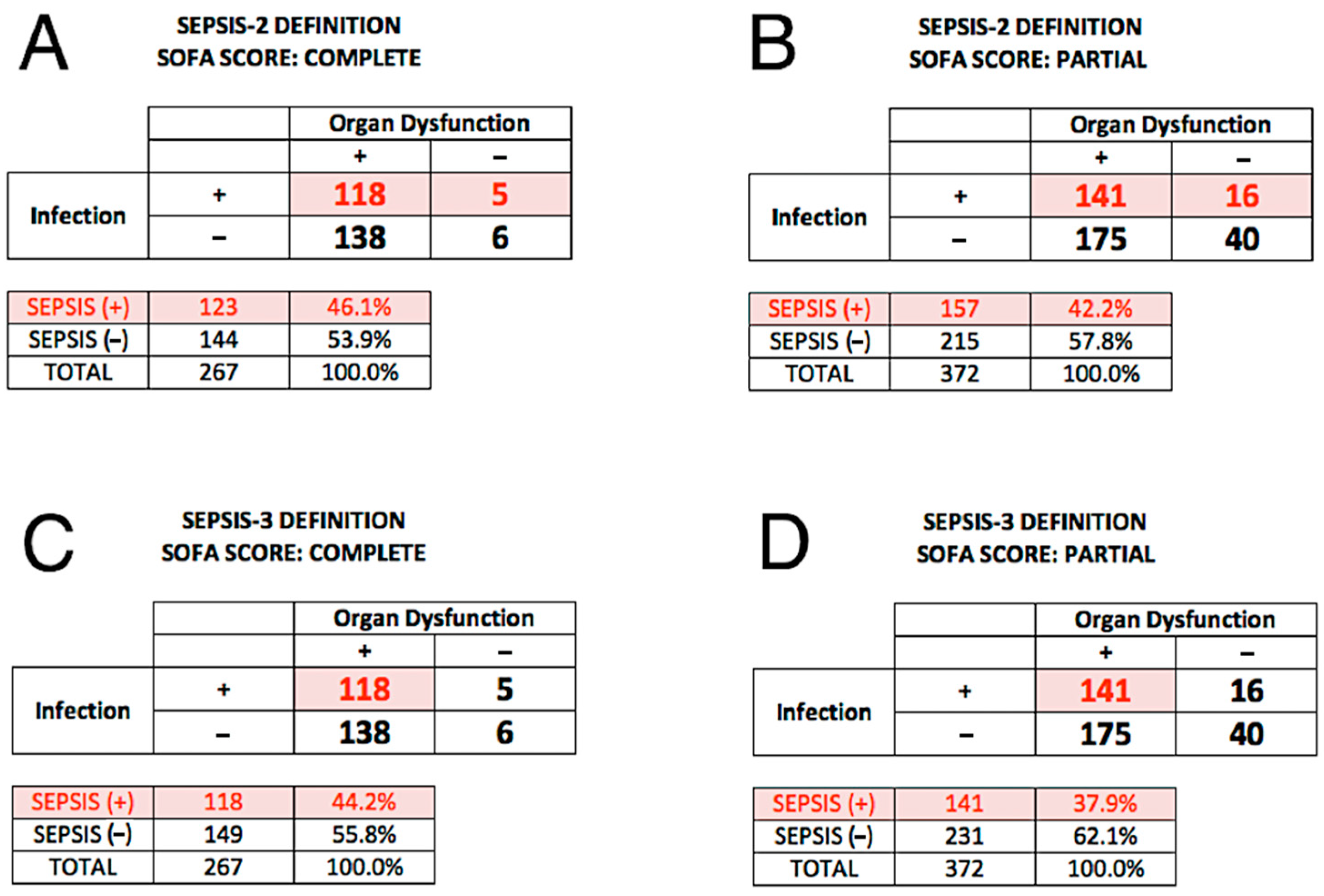
2.4. Reference Diagnosis (Comparator) under Sepsis-2
2.5. Reference Diagnosis (Comparator) under Sepsis-3
2.6. Statistical Calculations
3. Results
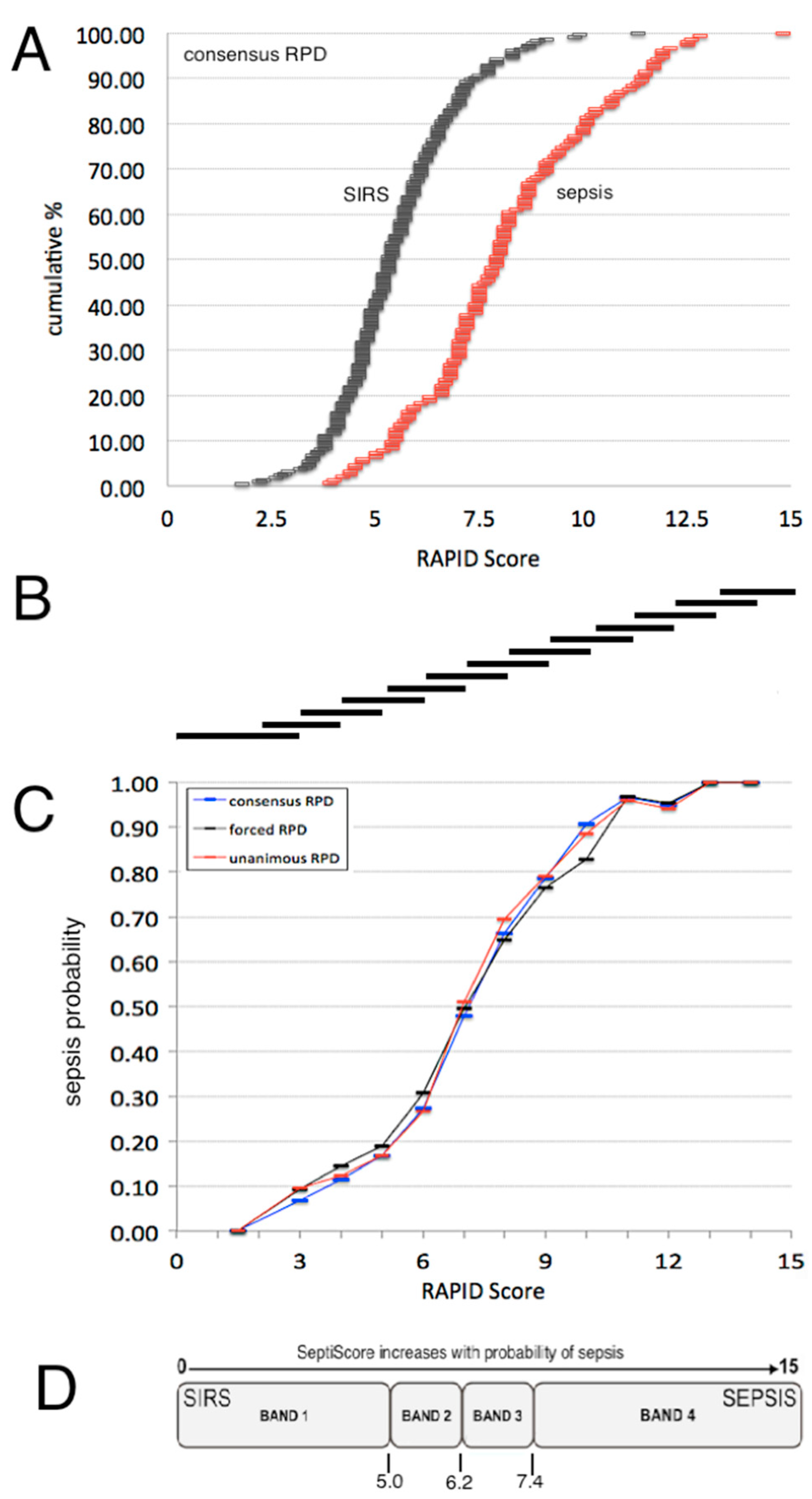
3.1. Description of the SeptiCyte RAPID Test
3.2. Demographic and Clinical Characteristics of the Study Cohorts
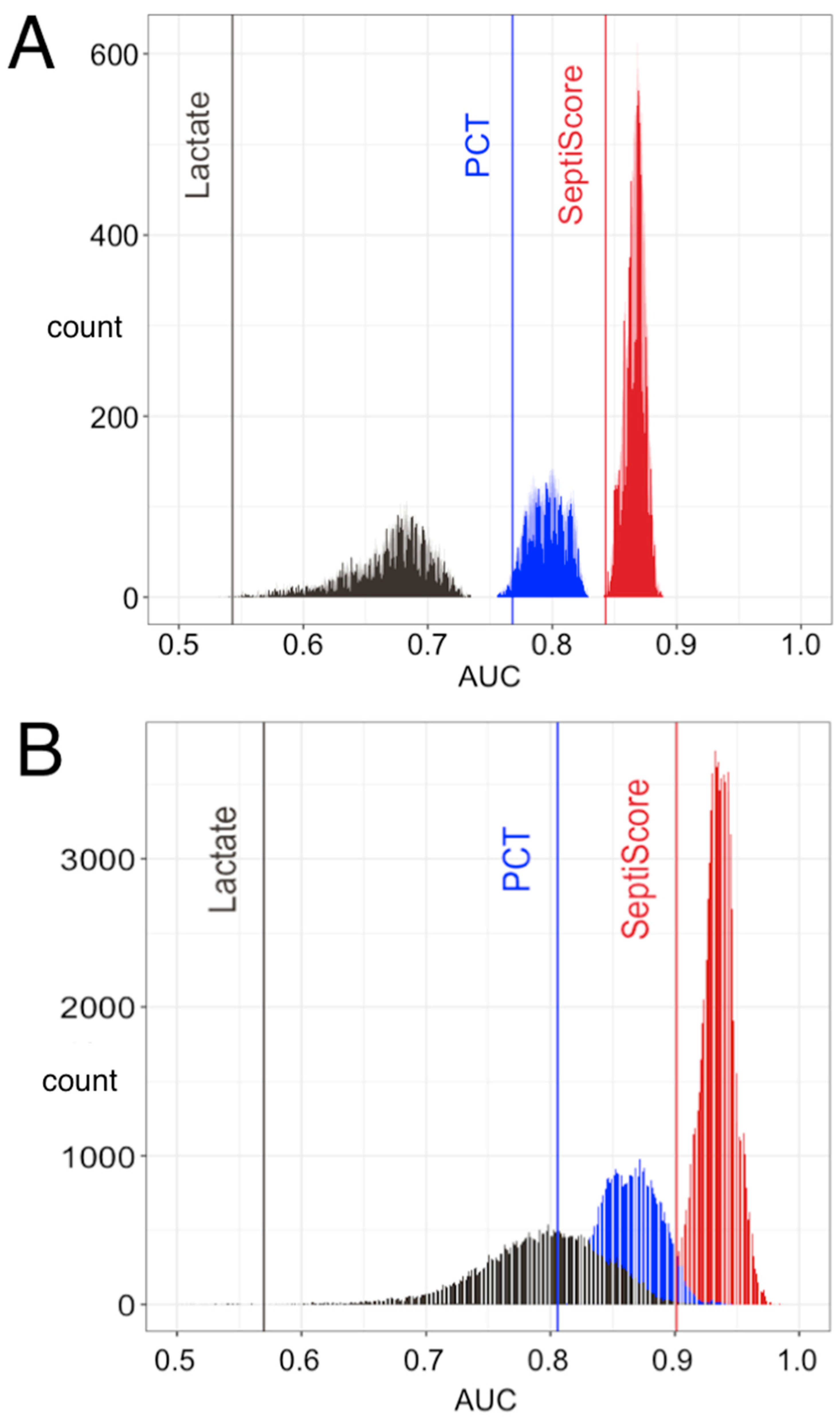
3.3. SeptiCyte RAPID Performance under Sepsis-2
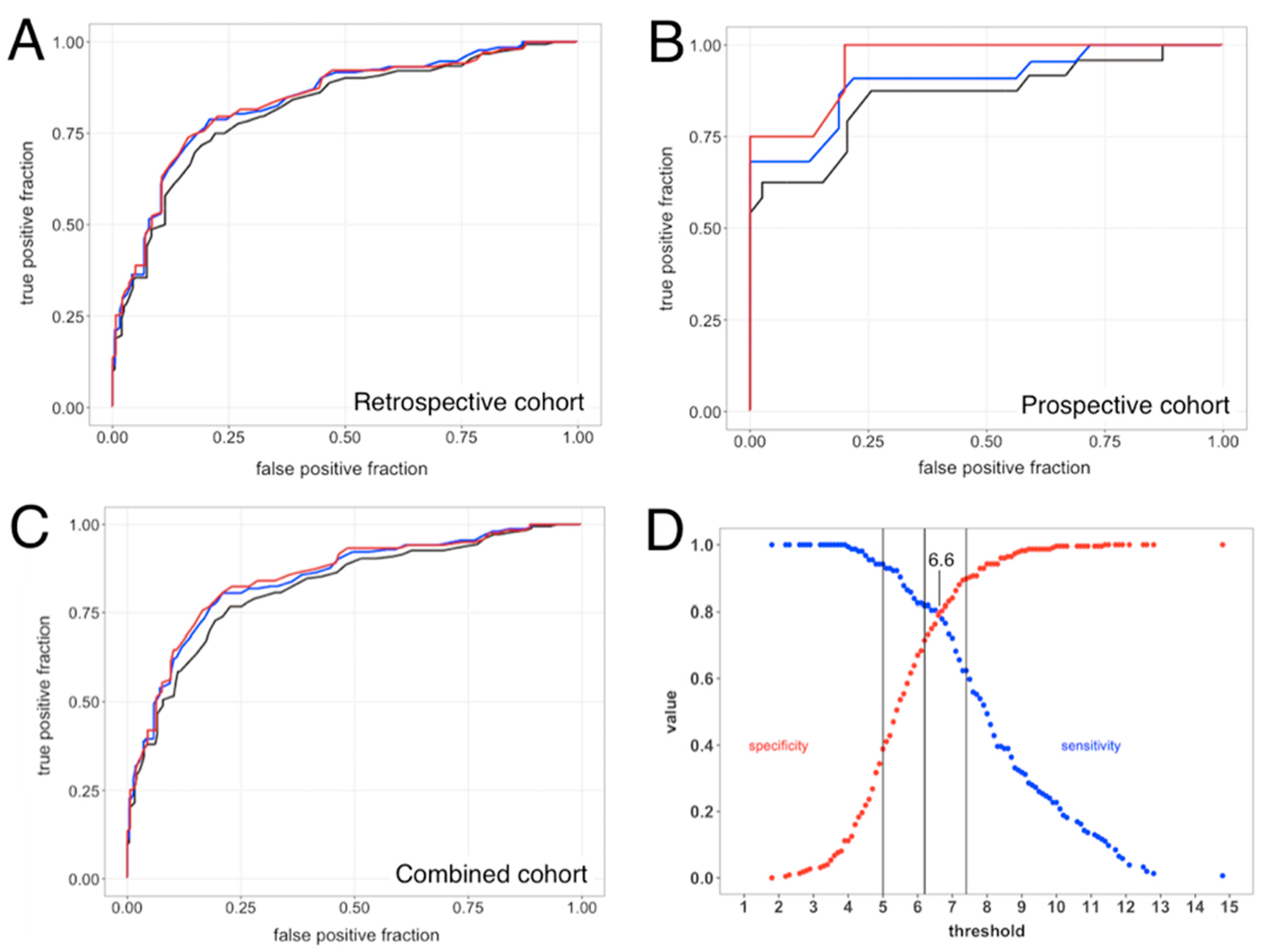
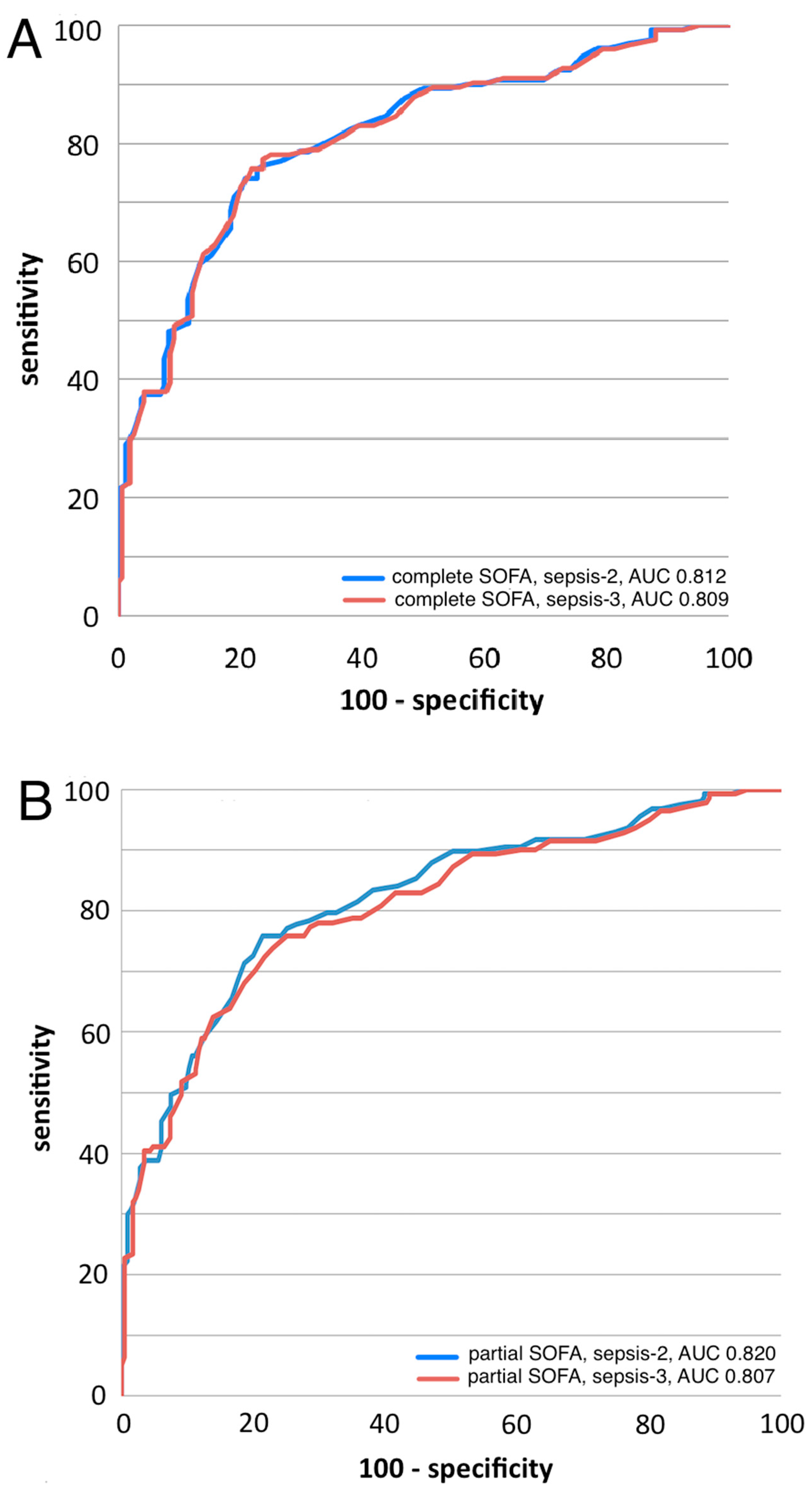
3.3.1. ROC Curve Analyses
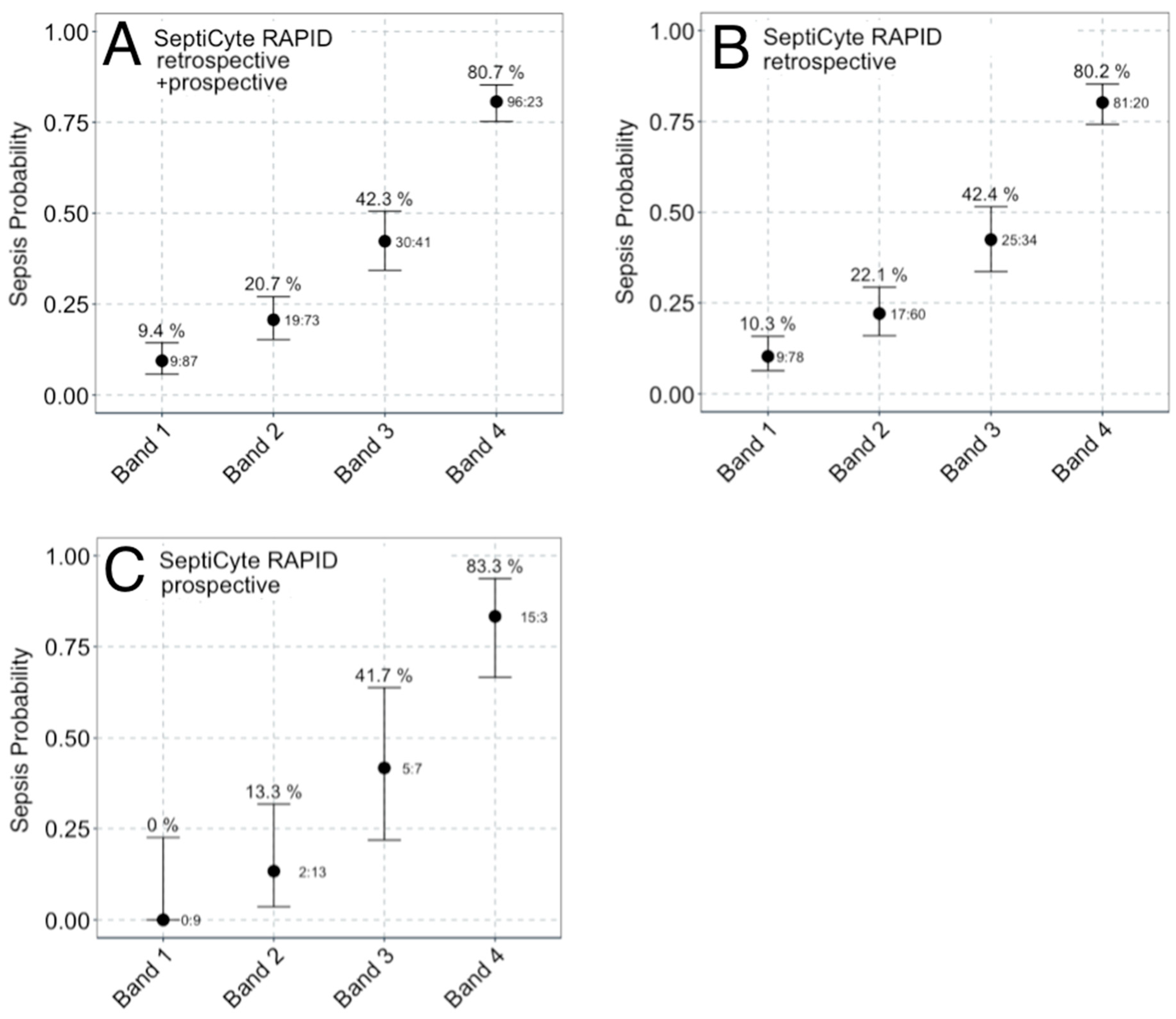
3.3.2. Sepsis Probability Distributions
3.3.3. Multivariable Analysis
3.4. SeptiCyte RAPID Performance under Sepsis-3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vincent, J.L. Highlighting the huge global burden of sepsis. Anaesth. Crit. Care Pain Med. 2020, 39, 171–172. [Google Scholar] [CrossRef]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Torio, C.M.; Moore, B.J. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2013. In Healthcare Cost and Utilization Project (HCUP) Statistical Briefs [Internet]; Statistical Brief #204; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2016. Available online: https://www.ncbi.nlm.nih.gov/books/NBK368492/ (accessed on 17 February 2024).
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States—An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Dantes, R.; Epstein, L.; Murphy, D.J.; Seymour, C.W.; Iwashyna, T.J.; Kadri, S.S.; Angus, D.C.; Danner, R.L.; Fiore, A.E.; et al. CDC Prevention Epicenter Program. Incidence and Trends of Sepsis in US Hospitals Using Clinical vs Claims Data, 2009–2014. JAMA 2017, 318, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Elezkurtaj, S.; Greuel, S.; Ihlow, J.; Michaelis, E.G.; Bischoff, P.; Kunze, C.A.; Sinn, B.V.; Gerhold, M.; Hauptmann, K.; Ingold-Heppner, B.; et al. Causes of death and comorbidities in hospitalized patients with COVID-19. Sci. Rep. 2021, 11, 4263. [Google Scholar] [CrossRef] [PubMed]
- Shappell, C.N.; Klompas, M.; Chan, C.; Chen, T.; Kanjilal, S.; McKenna, C.; Rhee, C.; CDC Prevention Epicenters Program. Use of Electronic Clinical Data to Track Incidence and Mortality for SARS-CoV-2-Associated Sepsis. JAMA Netw. Open 2023, 6, e2335728. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensive Care Med. 2018, 44, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Liu, V.X.; Bhimarao, M.; Greene, J.D.; Manickam, R.N.; Martinez, A.; Schuler, A.; Barreda, F.; Escobar, G.J. The Presentation, Pace, and Profile of Infection and Sepsis Patients Hospitalized Through the Emergency Department: An Exploratory Analysis. Crit. Care Explor. 2021, 3, e0344. [Google Scholar] [CrossRef]
- Lambregts, M.M.C.; Bernards, A.T.; Beek MT van der Visser, L.G.; de Boer, M.G. Time to positivity of blood cultures supports early re-evaluation of empiric broad-spectrum antimicrobial therapy. PLoS ONE 2019, 14, e0208819. [Google Scholar] [CrossRef]
- Panday, R.S.N.; Lammers, E.M.J.; Alam, N.; Nanayakkara, P.W.B. An overview of positive cultures and clinical outcomes in septic patients: A sub-analysis of the Prehospital Antibiotics Against Sepsis (PHANTASi) trial. Crit. Care 2019, 23, 182. [Google Scholar] [CrossRef]
- Panday, R.S.N.; Wang, S.; Ven PM van de Hekker, T.A.M.; Alam, N.; Nanayakkara, P.W.B. Evaluation of blood culture epidemiology and efficiency in a large European teaching hospital. PLoS ONE 2019, 14, e0214052. [Google Scholar] [CrossRef]
- Minderhoud, T.C.; Spruyt, C.; Huisman, S.; Oskam, E.; Schuit, S.C.E.; Levin, M.D. Microbiological outcomes and antibiotic overuse in Emergency Department patients with suspected sepsis. Neth. J. Med. 2017, 75, 196–203. [Google Scholar] [PubMed]
- Shappell, C.N.; Klompas, M.; Ochoa, A.; Rhee, C.; CDC Prevention Epicenters Program. Likelihood of Bacterial Infection in Patients Treated with Broad-Spectrum IV Antibiotics in the Emergency Department. Crit. Care Med. 2021, 49, e1144–e1150. [Google Scholar] [CrossRef]
- Bae, E.Y.; Smith, T.T.; Monogue, M.L. A case-control study evaluating the unnecessary use of intravenous broad-spectrum antibiotics in presumed sepsis and septic-shock patients in the emergency department. Antimicrob. Steward. Healthc. Epidemiol. 2022, 2, e193. [Google Scholar] [CrossRef] [PubMed]
- Lukaszewski, R.A.; Jones, H.E.; Gersuk, V.H.; Russell, P.; Simpson, A.; Brealey, D.; Walker, J.; Thomas, M.; Whitehouse, T.; Ostermann, M.; et al. Presymptomatic diagnosis of postoperative infection and sepsis using gene expression signatures. Intensive Care Med. 2022, 48, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Smischney, N.J.; Zhang, H.; Van Poucke, S.; Tsirigotis, P.; Rello, J.; Honore, P.M.; Kuan, W.S.; Ray, J.J.; Zhou, J.; et al. AME evidence series 001-The Society for Translational Medicine: Clinical practice guidelines for diagnosis and early identification of sepsis in the hospital. J. Thorac. Dis. 2016, 8, 2654–2665. [Google Scholar] [CrossRef] [PubMed]
- Tusgul, S.; Carron, P.N.; Yersin, B.; Calandra, T.; Dami, F. Low sensitivity of qSOFA, SIRS criteria and sepsis definition to identify infected patients at risk of complication in the prehospital setting and at the emergency department triage. Scand. J. Trauma Resusc. Emerg. Med. 2017, 25, 108. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, M.; Kroll, M.; Smith, C.S.; Asaro, P.; Liang, S.Y.; Moy, H.P. qSOFA has poor sensitivity for Prehospital identification of severe sepsis and septic shock. Prehosp. Emerg. Car. 2017, 21, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Sartelli, M.; Kluger, Y.; Ansaloni, L.; Hardcastle, T.C.; Rello, J.; Watkins, R.R.; Bassetti, M.; Giamarellou, E.; Coccolini, F.; Abu-Zidan, F.M.; et al. Raising concerns about the Sepsis-3 definitions. World J. Emerg. Surg. 2018, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Park, S. Sepsis: Early Recognition and Optimized Treatment. Tuberc. Respir. Dis. 2019, 82, 6–14. [Google Scholar] [CrossRef]
- Miller, R.R., 3rd; Lopansri, B.K.; Burke, J.P.; Levy, M.; Opal, S.; Rothman, R.E.; D’Alessio, F.R.; Sidhaye, V.K.; Aggarwal, N.R.; Balk, R.; et al. Validation of a Host Response Assay, SeptiCyte LAB, for Discriminating Sepsis from Systemic Inflammatory Response Syndrome in the ICU. Am. J. Respir. Crit. Care Med. 2018, 198, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.; Yager, T.; Cermelli, S.; Sampson, D.; Brandon, R.; Sillekens, P.; Keuleers, I.; Vanhoey, T. Clinical performance of a rapid sepsis test on a near-patient molecular testing platform. In Proceedings of the ISICEM 2020, Brussels, Belgium, 19–22 March 2020. Abstract # P481. [Google Scholar]
- Rand, K.H.; Beal, S.G.; Rivera, K.; Allen, B.; Payton, T.; Lipori, G.P. Hourly Effect of Pretreatment with IV Antibiotics on Blood Culture Positivity Rate in Emergency Department Patients. Open Forum Infect. Dis. 2019, 6, ofz179. [Google Scholar] [CrossRef] [PubMed]
- Scheer, C.S.; Fuchs, C.; Gründling, M.; Vollmer, M.; Bast, J.; Bohnert, J.A.; Zimmermann, K.; Hahnenkamp, K.; Rehberg, S.; Kuhn, S.O. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin. Microbiol. Infect. 2019, 25, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, F.; Blet, A.; Mebazaa, A. The new sepsis definition: Limitations and contribution to research and diagnosis of sepsis. Curr. Opin. Anesthesiol. 2017, 30, 200–204. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Seymour, C.W.; Liu, V.X.; Iwashyna, T.J.; Brunkhorst, F.M.; Rea, T.D.; Scherag, A.; Rubenfeld, G.; Kahn, J.M.; Shankar-Hari, M.; Singer, M.; et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 762–774, Erratum in JAMA 2016, 315, 2237. [Google Scholar] [CrossRef] [PubMed]
- Grissom, C.K.; Brown, S.M.; Kuttler, K.G.; Boltax, J.P.; Jones, J.; Jephson, A.R.; Orme, J.F., Jr. A modified sequential organ failure assessment score for critical care triage. Disaster Med. Public Health Prep. 2010, 4, 277–284. [Google Scholar] [CrossRef]
- Lambden, S.; Laterre, P.F.; Levy, M.M.; Francois, B. The SOFA score-development, utility and challenges of accurate assessment in clinical trials. Crit. Care 2019, 23, 374–382. [Google Scholar] [CrossRef]
- Arasu, A.; Manku, G.S. Approximate Counts and Quantiles over Sliding Windows. In Proceedings of the PODS 2004: Twenty-third ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems, Paris, France, 14–16 June 2004; pp. 286–296. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77–84. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Faust, J.S.; Weingart, S.D. The Past, Present, and Future of the Centers for Medicare and Medicaid Services Quality Measure SEP-1: The Early Management Bundle for Severe Sepsis/Septic Shock. Emerg. Med. Clin. N. Am. 2017, 35, 219–231. [Google Scholar] [CrossRef]
- Rhee, C.; Chiotos, K.; Cosgrove, S.E.; Heil, E.L.; Kadri, S.S.; Kalil, A.C.; Gilbert, D.N.; Masur, H.; Septimus, E.J.; Sweeney, D.A.; et al. Infectious Diseases Society of America Position Paper: Recommended Revisions to the National Severe Sepsis and Septic Shock Early Management Bundle (SEP-1) Sepsis Quality Measure. Clin. Infect. Dis. 2020, 72, ciaa059. [Google Scholar] [CrossRef]
- Pepe, M.S.; Janes, H.; Li, C.I. Net Risk Reclassification P Values: Valid or Misleading? J. Natl. Cancer Inst. 2014, 106, dju041. [Google Scholar] [CrossRef] [PubMed]
- Engoren, M.; Seelhammer, T.; Freundlich, R.E.; Maile, M.D.; Sigakis, M.J.G.; Schwann, T.A. A Comparison of Sepsis-2 (Systemic Inflammatory Response Syndrome Based) to Sepsis-3 (Sequential Organ Failure Assessment Based) Definitions-A Multicenter Retrospective Study. Crit. Care Med. 2020, 48, 1258–1264. [Google Scholar] [CrossRef]
- Valenstein, P.N. Evaluating diagnostic tests with imperfect standards. Am. J. Clin. Pathol. 1990, 93, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.J.; Sullivan, E.; Yager, T.D.; Cheng, C.; Permut, L.; Cermelli, S.; McHugh, L.; Sampson, D.; Seldon, T.; Brandon, R.B.; et al. Diagnostic Accuracy of a Host Gene Expression Signature That Discriminates Clinical Severe Sepsis Syndrome and Infection-Negative Systemic Inflammation Among Critically Ill Children. Crit. Care Med. 2017, 45, e418–e425. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.; Navalkar, K.; van der Poll, T.; Schultz, M.; Zimmerman, J.; Cremer, O.; Bonton, M. SeptiCyte RAPID in Sepsis Cases with Malignancy or Treated with Antineoplastics/Immunosuppressants. Crit. Care Med. 2021, 49, 643. [Google Scholar] [CrossRef]
- Reyes, M.; Filbin, M.R.; Bhattacharyya, R.P.; Billman, K.; Eisenhaure, T.; Hung, D.T.; Levy, B.D.; Baron, R.M.; Blainey, P.C.; Goldberg, M.B.; et al. An immune-cell signature of bacterial sepsis. Nat. Med. 2020, 26, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Crit. Care Med. 2021, 49, e1063–e1143. [Google Scholar] [CrossRef] [PubMed]
- Deis, A.S.; Whiles, B.B.; Brown, A.R.; Satterwhite, C.L.; Simpson, S.Q. Three-Hour Bundle Compliance and Outcomes in Patients with Undiagnosed Severe Sepsis. Chest 2018, 153, 39–45. [Google Scholar] [CrossRef]
- Leisman, D.E.; Doerfler, M.E.; Ward, M.F.; Masick, K.D.; Wie, B.J.; Gribben, J.L.; Hamilton, E.; Klein, Z.; Bianculli, A.R.; Akerman, M.B.; et al. Survival Benefit and Cost Savings from Compliance with a Simplified 3-Hour Sepsis Bundle in a Series of Prospective, Multisite, Observational Cohorts. Crit. Care Med. 2017, 45, 395–406. [Google Scholar] [CrossRef]
- Balk, R.A. Systemic inflammatory response syndrome (SIRS): Where did it come from and is it still relevant today? Virulence 2014, 5, 20–26. [Google Scholar] [CrossRef]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.H.; Sibbald, W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.N.; Bozza, F.A.; Amâncio, R.T.; Japiassú, A.M.; Vianna, R.C.S.; Larangeira, A.P.; Gouvêa, J.M.; Bastos, M.S.; Zimmerman, G.A.; Stafforini, D.M.; et al. Exogenous platelet- activating factor acetylhydrolase reduces mortality in mice with systemic inflammatory response syndrome and sepsis. Shock 2006, 26, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Gul, F.; Arslantas, M.K.; Cinel, I.; Kumar, A. Changing Definitions of Sepsis. Turk. J. Anesthesia Reanim. 2017, 45, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Honaker, J.; King, G.; Blackwell, M. AmeliaII: A Program for Missing Data. J. Stat. Softw. 2011, 45, 1–47. [Google Scholar] [CrossRef]
- Huang, Z.; Jiang, H.; Cui, X.; Liang, G.; Chen, Y.; Wang, T.; Sun, Z.; Qi, L. Elevated serum levels of lipoprotein-associated phospholipase A2 predict mortality rates in patients with sepsis. Mol. Med. Rep. 2017, 17, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Wang, K.; Shen, J. Lipoprotein-associated phospholipase A2: The story continues. Med. Res. Rev. 2019, 40, 79–134. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.M.; Kerr, M.S.; Slaven, J.E. Plac8-Dependent and Inducible NO Synthase-Dependent Mechanisms Clear Chlamydia muridarum Infections from the Genital Tract. J. Immunol. 2012, 188, 1896–1904. [Google Scholar] [CrossRef]
- Johnson, R.M.; Kerr, M.S.; Slaven, J.E. Perforin Is Detrimental to Controllinγ C. muridarum Replication In Vitro, but Not In Vivo. PLOS ONE 2013, 8, e63340. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Jones, M.E.; Draghi, D.C.; Thornsberry, C.; Sahm, D.F.; Volturo, G.A. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 7. [Google Scholar] [CrossRef][Green Version]
- Klouwenberg, P.M.C.K.; Cremer, O.L.; van Vught, L.A.; Ong, D.S.Y.; Frencken, J.F.; Schultz, M.J.; Bonten, M.J.; van der Poll, T. Likelihood of infection in patients with presumed sepsis at the time of intensive care unit admission: A cohort study. Crit. Care 2015, 19, 319. [Google Scholar] [CrossRef] [PubMed]
- Klein Klouwenberg, P.M.; Ong, D.S.; Bonten, M.J.; Cremer, O.L. Classification of sepsis, severe sepsis and septic shock: The impact of minor variations in data capture and definition of SIRS criteria. Intensive Care Med. 2012, 38, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Klouwenberg, P.M.C.K.; Ong, D.S.Y.; Bos, L.D.J.; de Beer, F.M.; van Hooijdonk, R.T.M.; Huson, M.A.; Straat, M.; van Vught, L.A.; Wieske, L.; Horn, J.; et al. Interobserver Agreement of Centers for Disease Control and Prevention Criteria for Classifying Infections in Critically Ill Patients. Crit. Care Med. 2013, 41, 2373–2378. [Google Scholar] [CrossRef]
- Ledford, J.G.; Kovarova, M.; Koller, B.H. Impaired Host Defense in Mice Lacking ONZIN. J. Immunol. 2007, 178, 5132–5143. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.-L.; Ramsay, G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef]
- Levy, M.M.; Dellinger, R.P.; Townsend, S.R.; Linde-Zwirble, W.T.; Marshall, J.C.; Bion, J.; Schorr, C.; Artigas, A.; Ramsay, G.; Beale, R.; et al. The Surviving Sepsis Campaign: Results of an international guideline-based performance improvement program targeting severe sepsis. Intensive Care Med. 2010, 38, 367–374. [Google Scholar] [CrossRef]
- Lopansri, B.K.; Iii, R.R.M.; Burke, J.P.; Levy, M.; Opal, S.; Rothman, R.E.; D’alessio, F.R.; Sidhaye, V.K.; Balk, R.; Greenberg, J.A.; et al. Physician agreement on the diagnosis of sepsis in the intensive care unit: Estimation of concordance and analysis of underlying factors in a multicenter cohort. J. Intensive Care. 2019, 7, 13. [Google Scholar] [CrossRef]
- McHugh, L.; Seldon, T.A.; Brandon, R.A.; Kirk, J.T.; Rapisarda, A.; Sutherland, A.J.; Presneill, J.J.; Venter, D.J.; Lipman, J.; Thomas, M.R.; et al. A Molecular Host Response Assay to Discriminate between Sepsis and Infection-Negative Systemic Inflammation in Critically Ill Patients: Discovery and Validation in Independent Cohorts. PLOS Med. 2015, 12, e1001916. [Google Scholar] [CrossRef]
- Pankla, R.; Buddhisa, S.; Berry, M.; Blankenship, D.M.; Bancroft, G.J.; Banchereau, J.; Lertmemongkolchai, G.; Chaussabel, D. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009, 10, R127. [Google Scholar] [CrossRef]
- Velásquez, S.Y.; Coulibaly, A.; Sticht, C.; Schulte, J.; Hahn, B.; Sturm, T.; Schefzik, R.; Thiel, M.; Lindner, H.A. Key Signature Genes of Early Terminal Granulocytic Differentiation Distinguish Sepsis from Systemic Inflammatory Response Syndrome on Intensive Care Unit Admission. Front. Immunol. 2022, 13, 864835. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Opal, S.M.; Marshall, J.C.; Tracey, K.J. Sepsis definitions: Time for change. Lancet 2013, 381, 774–775. [Google Scholar] [CrossRef]
- Wieland, S.; Thimme, R.; Purcell, R.H.; Chisari, F.V. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 2004, 101, 6669–6674. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, J.; Chen, X.; Zhang, Y.; Jiang, X.; Guo, X.; Zhao, G. Decrease of plasma platelet-activating factor acetylhydrolase activity in lipopolysaccharide induced Mongolian gerbil sepsis model. PLoS ONE 2010, 5, e9190. [Google Scholar] [CrossRef] [PubMed]
| Parameter | SIRS (N = 224) (Consensus) | Sepsis (N = 154) (Consensus) | p-Value | |
|---|---|---|---|---|
| N (%) | N (%) | |||
| Age | Median (Interquartile range, IQR)) | 57 (42–69) | 62 (49–71) | p = 0.016 |
| Sex | Female | 98 (58%) | 71 (42%) | female vs. male p = 0.65 |
| Male | 126 (60%) | 83 (40%) | ||
| Race/Ethnicity | Asian | 10 (53%) | 9 (47%) | white vs. non-white 1 p = 0.75 white vs. black p = 0.39 |
| Black (American or European of African descent) | 65 (64%) | 37 (36%) | ||
| Hispanic | 11 (52%) | 10 (48%) | ||
| Other | 1 (50%) | 1 (50%) | ||
| Unknown | 2 (50%) | 2 (50%) | ||
| White | 135 (59%) | 95 (41%) | ||
| Study Site | Europe (1 site) | 76 (60%) | 50 (40%) | Europe vs. USA p = 0.76 |
| United States (9 sites) | 148 (59%) | 104 (41%) | ||
| Source of Admission | Emergency department | 138 (59%) | 95 (41%) | ED vs. ward p = 0.0026 (post-anesthesia or post-op) vs. ward p < 0.00001 ICU or CCF vs. ward p = 0.024 |
| Post Anesthesia Unit or Post-Operating Room | 34 (83%) | 7 (17%) | ||
| ICU | 7 (58%) | 5 (42%) | ||
| ICU, other hospital | 14 (54%) | 12 (46%) | ||
| Coronary care facility (CCF) | 4 (80%) | 1 (20%) | ||
| Wards | 13 (33%) | 26 (67%) | ||
| Other | 13 (62%) | 8 (38%) | ||
| Category | SIRS (n = 224; 59.3%) | Sepsis (n = 154; 40.7%) | p-Value | |
|---|---|---|---|---|
| N (%) or Median (IQR) | N (%) or Median (IQR) | |||
| Culture results | ||||
| Blood (+/− Other) | 3 (1.34%) | 38 (24.7%) | <0.0001 | |
| Urine (+/− Other) | 2 (0.89%) | 21 (13.6%) | <0.0001 | |
| Blood + Urine (+/− Other)(double positive) | 0 (0%) | 10 (6.49%) | <0.0001 | |
| Sputum (+/− Other) | 13 (5.8%) | 19 (12.3%) | 0.03 | |
| Other culture * | 19 (8.5%) | 20 (13.0%) | 0.2 | |
| Viral positives/coinfections (by PCR) | 9 (4.0%) | 16 (10.4%) | 0.02 | |
| No culture data recorded ** | 178 (79.5%) | 30 (19.5%) | <0.0001 | |
| Presumed initial site of infection | ||||
| Pulmonary (N = 58) | 8 (3.6%) | 50 (32.5%) | <0.0001 | |
| Abdominal (N = 28) | 1 (0.45%) | 27 (17.5%) | <0.0001 | |
| Blood (N = 16) | 0 (0%) | 16 (10.4%) | <0.0001 | |
| Urinary Tract (N = 25) | 2 (0.89%) | 23 (14.9%) | <0.0001 | |
| Central Nervous System (N = 9) | 4 (1.8%) | 5 (3.25%) | 0.4 | |
| Other (N = 16) | 2 (0.89%) | 14 (9.1%) | 0.0001 | |
| Multiple (N = 5) | 0 (0%) | 5 (3.25%) | 0.007 | |
| Not Identified at Initial Evaluation (N = 226) | 207 (92.4%) | 19 (12.3%) | <0.0001 | |
| Clinical parameters (Median, IQR) | ||||
| Minimum Temperature (°C) | 36 (35.5–36.4) | 36.1 (35.2–36.7) | 0.9 | |
| Maximum Temperature (°C) | 37.2 (36.9–37.8) | 37.8 (37.2–38.6) | <0.001 | |
| SOFA | 6.0 (4.0–8.0) | 7.0 (5.0–9.0) | 0.004 | |
| Procalcitonin (ng/mL) | 0.28 (0.05–1.33) | 5.19 (0.85–25.72) | <0.0001 | |
| WBC max (cells/uL × 10−3) | 12.65 (8.3–17.20) | 14.9 (9.94–21.15) | 0.0002 | |
| Lactate (mmol/L) | 2.05 (1.40–3.27) | 2.40 (1.55–4.05) | 0.5 | |
| Interventions (Median, IQR) | Interventions (Median, IQR) | |||
| Invasive mechanical ventilation | 86 (38.4%) | 52 (33.8%) | 0.4 | |
| Antibiotic administration | 134 (59.8%) | 149 (96.8%) | <0.0001 | |
| Vasopressor Use | 59 (26.3%) | 68 (44.2%) | 0.003 | |
| Outcomes (Median, IQR) | Outcomes (Median, IQR) | |||
| Hospital length of stay (days) | 4 (3–7) | 8 (4–15) | <0.001 | |
| ICU length of stay (days) | 1.8 (1.2–3.3) | 3.2 (1.7–7.0) | <0.001 | |
| Death | 19 (8.5%) | 22 (14.3%) | 0.08 | |
| SeptiCyte RAPID Score *** | (Median, IQR) | 5.3 (4.6–6.3) | 7.9 (6.8–9.6) | <0.0001 |
| Category | Subcategory | SeptiScore 0–4.9 (Band B1) (N = 104; 24.8%) | SeptiScore 5–6.1 (Band B2) (N = 103; 24.6%) | SeptiScore 6.2–7.3 (Band B3) (N = 82; 19.6%) | SeptiScore 7.4–15 (Band B4) (N = 130; 31.0%) | Significance | |
|---|---|---|---|---|---|---|---|
| RPD Process | |||||||
| RPD: Consensus (sepsis + SIRS = 378) | Indeterminate (N = 41; 9.8%) | 8 (7.7%) | 11 (10.7%) | 11 (13.4%) | 11 (8.5%) | p(B1,B2,B3,B4) = 1.6 × 10−9, 4.7 × 10−4, 0.53, <1.0 × 10−15 | |
| Sepsis (N = 154; 36.8%) | 9 (8.6%) | 19 (18.4%) | 30 (36.6%) | 96 (73.8%) | |||
| SIRS (N = 224; 53.5%) | 87 (83.6%) | 73 (70.9%) | 41 (50.0%) | 23 (17.7%) | |||
| RPD: Forced (sepsis + SIRS = 419) | Sepsis (N = 176; 42.0%) | 13 (12.5%) | 23 (22.3%) | 37 (45.1%) | 103 (79.2%) | p(B1,B2,B3,B4) = 1.1 × 10−9, 5.2 × 10−5, 0.57, <1.0 × 10−15 | |
| SIRS (N = 243; 58.0%) | 91 (87.5%) | 80 (77.7%) | 45 (54.9%) | 27 (20.8%) | |||
| RPD: Unanimous (sepsis + SIRS = 276) | 7 (6.7%) | 12 (11.6%) | 23 (28.0%) | 77 (59.2%) | p(B1,B2,B3,B4) = 6.3 × 10−7, 6.0 × 10−4, 0.97, 3.2 × 10−15 | ||
| SIRS (N = 157; 37.5%) | 60 (57.7%) | 51 (49.5%) | 30 (36.6%) | 16 (12.3%) | |||
| N/A (N = 143; 34.1%) | 37 (35.6%) | 40 (38.8%) | 29 (35.4%) | 37 (28.5%) | |||
| Demographics | |||||||
| Age | Median, (Q1–Q3) | 56.5 (42.8–68.2) | 59 (44.5–69) | 58 (41.2–74) | 62 (48.2–70) | p = 0.6 | |
| Sex | female (N = 187; 44.6%) | 46 (44.2%) | 41 (39.8%) | 37 (45.1%) | 63 (48.5%) | p(B1,B2,B3,B4) = 0.93, 0.33, 0.92, 0.38 | |
| male (N = 232; 55.4%) | 58 (55.8%) | 62 (60.2%) | 45 (54.9%) | 67 (51.5%) | |||
| Race | Black (N = 115; 27.4%) | 25 (24.0%) | 33 (32.0%) | 21 (25.6%) | 36 (27.7%) | p(B1,B2,B3,B4) = 0.70, 0.22, 0.20, 0.92 | |
| White (N = 254; 60.6%) | 65 (62.5%) | 63 (61.2%) | 46 (56.1%) | 80 (61.5%) | |||
| Other (N = 50; 11.9%) | 14 (13.5%) | 7 (6.8%) | 15 (18.3%) | 14 (10.8%) | |||
| Culture Results * | |||||||
| Blood (+/− secondary) | Number (%) positive (out of 42) | 1 (2.4%) | 7 (16.6%) | 8 (19.5%) | 26 (63.4%) | p < 0.0001 | |
| Urine (+/− secondary) | Number (%) positive (out of 24) | 4 (16.6%) | 4 (16.6%) | 3 (12.5%) | 13 (54.2%) | p = 0.1 | |
| Blood + Urine (+/− tertiary) | Number (%) double positive (out of 10) | 0 (0.0%) | 1 (10%) | 1 (10%) | 8 (80%) | p = 0.008 | |
| Sputum (+/− secondary) | Number (%) positive (out of 35) | 8 (22.9%) | 1 (2.9%) | 9 (25.7%) | 17 (48.5%) | p = 0.02 | |
| Other culture/results ** | Number (%) positive (out of 42) | 11 (26%) | 10 (24%) | 8 (19%) | 13 (31%) | p = 0.99 | |
| Viral acute (by PCR) | Number (%) positive (out of 24) | 1 (4%) | 3 (12%) | 8 (32%) | 12 (52%) | p = 0.01 | |
| No positive culture or PCR data recorded *** | Number (%) without positive culture or PCR data recorded (out of 242) | 79 (32.8%) | 77 (32%) | 45 (18.7%) | 41 (16.5%) | p < 0.0001 | |
| Presumed Initial Site of Infection * | |||||||
| Pulmonary | Number (%) out of 74 | 6 (8%) | 11 (15%) | 17 (23%) | 40 (54%) | p < 0.0001 | |
| Abdominal | Number (%) out of 34 | 2 (5.9%) | 7 (20.6%) | 6 (17.6%) | 19 (55.9%) | p = 0.01 | |
| Blood | Number (%) out of 17 | 1 (5.9%) | 2 (11.8%) | 3 (17.6%) | 11 (64.7%) | p = 0.02 | |
| Urinary Tract | Number (%) out of 26 | 3 (11.5%) | 5 (19.2%) | 2 (7.7%) | 16 (61.5%) | p = 0.008 | |
| Central Nervous System | Number (%) out of 11 | 2 (18.2%) | 2 (18.2%) | 3 (27.3%) | 4 (36.4%) | p = 0.8 | |
| Other | Number (%) out of 16 | 1 (6.3%) | 1 (6.3%) | 6 (37.5%) | 8 (50%) | p = 0.03 | |
| Multiple | Number (%) out of 5 | 0 (0.0%) | 2 (40.0%) | 0 (0.0%) | 3 (60.0%) | p = 0.3 | |
| Not Identified at Initial Evaluation | Number (%) out of 241 | 89 (36.9%) | 75 (31.1%) | 45 (18.7%) | 32 (13.3%) | p < 0.0001 | |
| Clinical Parameters | |||||||
| Temperature (Min) | Median, (Q1–Q3) | 36.0 (35.2–36.4) | 36.2 (35.6–36.6) | 36.2 (35.5–36.7) | 36.0 (35.4–36.7) | p = 0.2 | |
| Temperature (Max) | Median, (Q1–Q3) | 37.2 (37.0–37.8) | 37.3 (36.8–37.9) | 37.3 (36.9–38.2) | 37.8 (37.2–38.4) | p < 0.001 | |
| Lactate | Median, (Q1–Q3) | 1.9 (1.3–3.3) | 2.1 (1.6–3.3) | 2.2 (1.5–3.3) | 2.6 (1.6–4.1) | p = 0.7 | |
| WBC.Max | Median, (Q1–Q3) | 13.5 (8.5–17.2) | 12.2 (8.4–17.4) | 13.9 (10.2–17.9) | 14.6 (9.1–21.1) | p = 0.02 | |
| Procalcitonin | Median, (Q1, Q3) | 0.3 (0.1–0.9) | 0.3 (0.1–1.1) | 0.6 (0.2–2.6) | 5.6 (1.7–25.8) | p < 0.0001 | |
| SeptiScore | Median, (Q1–Q3) | 4.3 (3.8–4.7) | 5.6 (5.3–5.8) | 6.8 (6.5–7.0) | 8.7 (8.0–10.1) | p < 0.0001 | |
| SeptiScore Band | Cohort | Average Probability | Sepsis Likelihood Ratio | Percentage of Cohort | |
|---|---|---|---|---|---|
| SIRS | Sepsis | ||||
| Band 4 (7.4–15) High Risk of Sepsis | Retrospective | 20 (20%) | 81 (80%) | 5.89 | 31% |
| Prospective | 3 (17%) | 15 (83%) | 7.27 | 33% | |
| Combined | 23 (19%) | 96 (81%) | 6.07 | 32% | |
| Band 3 (6.2–7.3) | Retrospective | 34 (58%) | 25 (42%) | 1.07 | 18% |
| Prospective | 7 (58%) | 5 (42%) | 1.04 | 22% | |
| Combined | 41 (58%) | 30 (42%) | 1.06 | 19% | |
| Band 2 (5.0–6.1) | Retrospective | 60 (78%) | 17 (22%) | 0.41 | 24% |
| Prospective | 13 (87%) | 2 (13%) | 0.22 | 28% | |
| Combined | 73 (79%) | 19 (21%) | 0.38 | 24% | |
| Band 1 (0–4.9) Low Risk of Sepsis | Retrospective | 78 (90%) | 9 (10%) | 0.17 | 27% |
| Prospective | 9 (100%) | 0 (0%) | 0.00 | 17% | |
| Combined | 87 (91%) | 9 (9%) | 0.15 | 25% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balk, R.; Esper, A.M.; Martin, G.S.; Miller, R.R., III; Lopansri, B.K.; Burke, J.P.; Levy, M.; Opal, S.; Rothman, R.E.; D’Alessio, F.R.; et al. Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation. J. Clin. Med. 2024, 13, 1194. https://doi.org/10.3390/jcm13051194
Balk R, Esper AM, Martin GS, Miller RR III, Lopansri BK, Burke JP, Levy M, Opal S, Rothman RE, D’Alessio FR, et al. Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation. Journal of Clinical Medicine. 2024; 13(5):1194. https://doi.org/10.3390/jcm13051194
Chicago/Turabian StyleBalk, Robert, Annette M. Esper, Greg S. Martin, Russell R. Miller, III, Bert K. Lopansri, John P. Burke, Mitchell Levy, Steven Opal, Richard E. Rothman, Franco R. D’Alessio, and et al. 2024. "Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation" Journal of Clinical Medicine 13, no. 5: 1194. https://doi.org/10.3390/jcm13051194
APA StyleBalk, R., Esper, A. M., Martin, G. S., Miller, R. R., III, Lopansri, B. K., Burke, J. P., Levy, M., Opal, S., Rothman, R. E., D’Alessio, F. R., Sidhaye, V. K., Aggarwal, N. R., Greenberg, J. A., Yoder, M., Patel, G., Gilbert, E., Parada, J. P., Afshar, M., Kempker, J. A., ... Brandon, R. B. (2024). Validation of SeptiCyte RAPID to Discriminate Sepsis from Non-Infectious Systemic Inflammation. Journal of Clinical Medicine, 13(5), 1194. https://doi.org/10.3390/jcm13051194







