Analysis of Risk Factors with Assessment of the Impact of the Microbiome on the Risk of Squamous Cell Carcinoma of the Larynx
Abstract
1. Introduction
2. Materials and Methods
Study Group
3. Microbiome Profiling Workflow
- DNA isolation from cotton swab samples using a commercial kit following the manufacturer’s protocol (GeneMATRIX Swab-Extract DNA Purification Kit, Eurx, Gdańsk, Poland).
- Quality control of isolated DNA—concentration and purity evaluation (Qubit 4 Fluorometer, Invitrogen and DeNovix DS-11 spectrophotometer, Connecticut, US); DNA integrity check by electrophoresis on 1.5% agarose gel.
- Amplicon libraries construction following two rounds of PCR amplification-
- Amplification of specific target DNA region of bacterial 16S ribosomal RNA (V3–V4) using universal primers connected with Illumina sequencing adapters; PCR Clean-Up using AMPure XP beads, Indianapolis, US.
- Index PCR attaching dual indices and Illumina sequencing adapters using the Nextera XT Index Kit. San Diego, US; PCR Clean-Up using AMPure XP beads, Indianapolis, US.
- Library QC, quantification, normalization, and pooling.
- Sequencing on MiSeq–Using paired 300-bp reads.
Control Group
4. Statistical Analysis
5. Results
5.1. Analysis of General Characteristics of Patients in the Study and Control Group
5.2. Analysis of Medical History of the Control Group and the Study Group
5.3. Characteristics of the Study Group
5.4. Analysis of Laboratory Parameters between the Study Group and the Control Group
5.5. Analysis of Bacterial Cultures between the Study Group and the Control Group
5.6. Microbiome Analysis
5.7. Microbiome Analysis in Terms of Phylum
6. Discussion
7. Limitations of the Study
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; Botta, L.; Sánchez, M.J.; Anderson, L.A.; Pierannunzio, D.; Licitra, L. EUROCARE Working Group: Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur. J. Cancer 2015, 51, 2130–2143. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global Cancer Statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef]
- Huang, T.T.; Lai, J.B.; Du, Y.L.; Xu, Y.; Ruan, L.M.; Hu, S.H. Current understanding of gut microbiota in mood disorders: An update of human studies. Front. Genet. 2019, 10, 98. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Annunziata, C.; Tornesello, A.L.; Buonaguro, L.; Buonaguro, F.M. Human oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers 2018, 10, 213. [Google Scholar] [CrossRef]
- Mesia, R.; Iglesias, L.; Lambea, J.; Martínez-Trufero, J.; Soria, A.; Taberna, M.; Trigo, J.; Chaves, M.; García-Castaño, A.; Cruz, J. SEOM clinical guidelines for the treatment of head and neck cancer (2020). Clin. Transl. Oncol. 2021, 23, 913–921, Erratum in Clin. Transl. Oncol. 2021, 23, 1001. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm. Bowel Dis. 2016, 22, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, C.; Yue, J. Radiotherapy and the gut microbiome: Facts and fiction. Radiat. Oncol. 2021, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Wootla, B.; Anderson, G. Multiple sclerosis, gut microbiota and permeability: Role of tryptophan catabolites, depression and the driving down of local melatonin. Curr. Pharm. Des. 2016, 22, 6134–6141. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, A.; Okuma, Y. Perspective on immune oncology with liquid biopsy, peripheral blood mononuclear cells, and microbiome with non-invasive biomarkers in cancer patients. Clin. Transl. Oncol. 2018, 20, 966–974. [Google Scholar] [CrossRef]
- Floch, P.; Megraud, F.; Lehours, P. Helicobacter pylori strains and gastric MALT lymphoma. Toxins 2017, 9, 132. [Google Scholar] [CrossRef]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1, 24. [Google Scholar] [CrossRef]
- Kareva, I. Metabolism and gut microbiota in cancer immunoediting, CD8/Treg Ratios, immune cell homeostasis, and cancer (immuno) therapy: Concise review. Stem Cells 2019, 37, 1273–1280. [Google Scholar] [CrossRef]
- Irfan, M.; Delgado, R.Z.R.; Frias-Lopez, J. The Oral Microbiome and Cancer. Front. Immunol. 2020, 11, 591088. [Google Scholar] [CrossRef]
- Hooper, S.J.; Wilson, M.J.; Crean, S.J. Exploring the link between microorganisms and oral cancer: A systematic review of the literature. Head. Neck 2009, 31, 1228–1239. [Google Scholar] [CrossRef]
- Baban, C.K.; Cronin, M.; O’Hanlon, D.; O’Sullivan, G.C.; Tangney, M. Bacteria as vectors for gene therapy of cancer. Bioeng. Bugs 2010, 1, 385–394. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Alfano, M.; Canducci, F.; Nebuloni, M.; Clementi, M.; Montorsi, F.; Salonia, A. The interplay of extracellular matrix and microbiome in urothelial bladder cancer. Nat. Rev. Urol. 2016, 13, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Gail, M.H.; Shi, J.; Klepac-Ceraj, V.; Paster, B.J.; Dye, B.A.; Wang, G.Q.; Wei, W.Q.; Fan, J.H.; Qiao, Y.L.; et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol. Biomark. Prev. 2014, 23, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2016, 67, 120–127. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Dong, J.; Li, Y.; Xiao, H.; Zhang, S.; Wang, B.; Wang, H.; Li, Y.; Fan, S.; Cui, M. Oral microbiota affects the efficacy and prognosis of radiotherapy for colorectal cancer in mouse models. Cell Rep. 2021, 37, 109886. [Google Scholar] [CrossRef]
- Miranda-Galvis, M.; Loveless, R.; Kowalski, L.P.; Teng, Y. Impacts of Environmental Factors on Head and Neck Cancer Pathogenesis and Progression. Cells 2021, 10, 389. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Eslami, H.; Yousefi, M.; Asgharzadeh, M.; Aghazadeh, M.; Kafil, H.S. Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 2016, 84, 552–558. [Google Scholar] [CrossRef]
- Stashenko, P.; Yost, S.; Choi, Y.; Danciu, T.; Chen, T.; Yoganathan, S.; Kressirer, C.; Ruiz-Tourrella, M.; Das, B.; Kokaras, A.; et al. The Oral Mouse Microbiome Promotes Tumorigenesis in Oral Squamous Cell Carcinoma. mSystems 2019, 4, e00323-19. [Google Scholar] [CrossRef]
- Frank, D.N.; Qiu, Y.; Cao, Y.; Zhang, S.; Lu, L.; Kofonow, J.M.; Robertson, C.E.; Liu, Y.; Wang, H.; Levens, C.L.; et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene 2022, 41, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.L.; Shi, Y.; Zhou, L.; Wu, C.P.; Cao, P.Y.; Tao, L.; Xu, C.; Hou, D.S.; Wang, Y.Z. The Composition of Microbiome in Larynx and the Throat Biodiversity between Laryngeal Squamous Cell Carcinoma Patients and Control Population. PLoS ONE 2013, 8, e66476. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.B.; Ahn, J.; Fan, X.; Peters, B.A.; Ma, Y.; Yang, L.; Agalliu, I.; Burk, R.D.; Ganly, I.; Purdue, M.P.; et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA Oncol. 2018, 4, 358–365. [Google Scholar] [CrossRef]
- Katz, J.; Onate, M.D.; Pauley, K.M.; Bhattacharyya, I.; Cha, S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int. J. Oral. Sci. 2011, 3, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Segers, S.; Hayes, R.B. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis 2012, 33, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Gleber-Netto, F.O.; Fernandes, G.R.; Amorim, M.; Barbosa, L.F.; Francisco, A.L.; de Andrade, A.G.; Setubal, J.C.; Kowalski, L.P.; Nunes, D.N.; et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 2014, 14, 250. [Google Scholar] [CrossRef] [PubMed]
- Lafuente Ibáñez de Mendoza, I.; Maritxalar Mendia, X.; García de la Fuente, A.M.; Quindós Andrés, G.; Aguirre Urizar, J.M. Role of Porphyromonas gingivalis in oral squamous cell carcinoma development: A systematic review. J. Periodont Res. 2020, 55, 13–22. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Kipanyula, M.J.; Seke Etet, P.F.; Vecchio, L.; Farahna, M.; Nukenine, E.N.; Nwabo Kamdje, A.H. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal 2013, 25, 403–416. [Google Scholar] [CrossRef]
- Atasever Akkas, E.; Yucel, B. Prognostic value of systemic ımmune ınflammation ındex in patients with laryngeal cancer. Eur. Arch. Otorhinolaryngol. 2021, 278, 1945–1955. [Google Scholar] [CrossRef]
- Verro, B.; Saraniti, C.; Carlisi, D.; Chiesa-Estomba, C.; Maniaci, A.; Lechien, J.R.; Mayo, M.; Fakhry, N.; Lauricella, M. Biomarkers in Laryngeal Squamous Cell Carcinoma: The Literature Review. Cancers 2023, 15, 5096. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Fu, Y.; Gao, Y.; Yang, A.; Zhang, Q. Platelet-to-lymphocyte ratio predicts long-term survival in laryngeal cancer. Eur. Arch. Otorhinolaryngol. 2018, 275, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Santos, I.C.; Dos Reis, P.F.; Rodrigues, V.D.; Peres, W.A.F. Impact of Nutritional Status on Survival in Head and Neck Cancer Patients After Total Laryngectomy. Nutr. Cancer 2022, 74, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- van Bokhorst-de van der Schueren, M.A.; van Leeuwen, P.A.; Sauerwein, H.P.; Kuik, D.J.; Snow, G.B.; Quak, J.J. Assessment of malnutrition parameters in head and neck cancer and their relation to postoperative complications. Head. Neck 1997, 19, 419–425. [Google Scholar] [CrossRef]

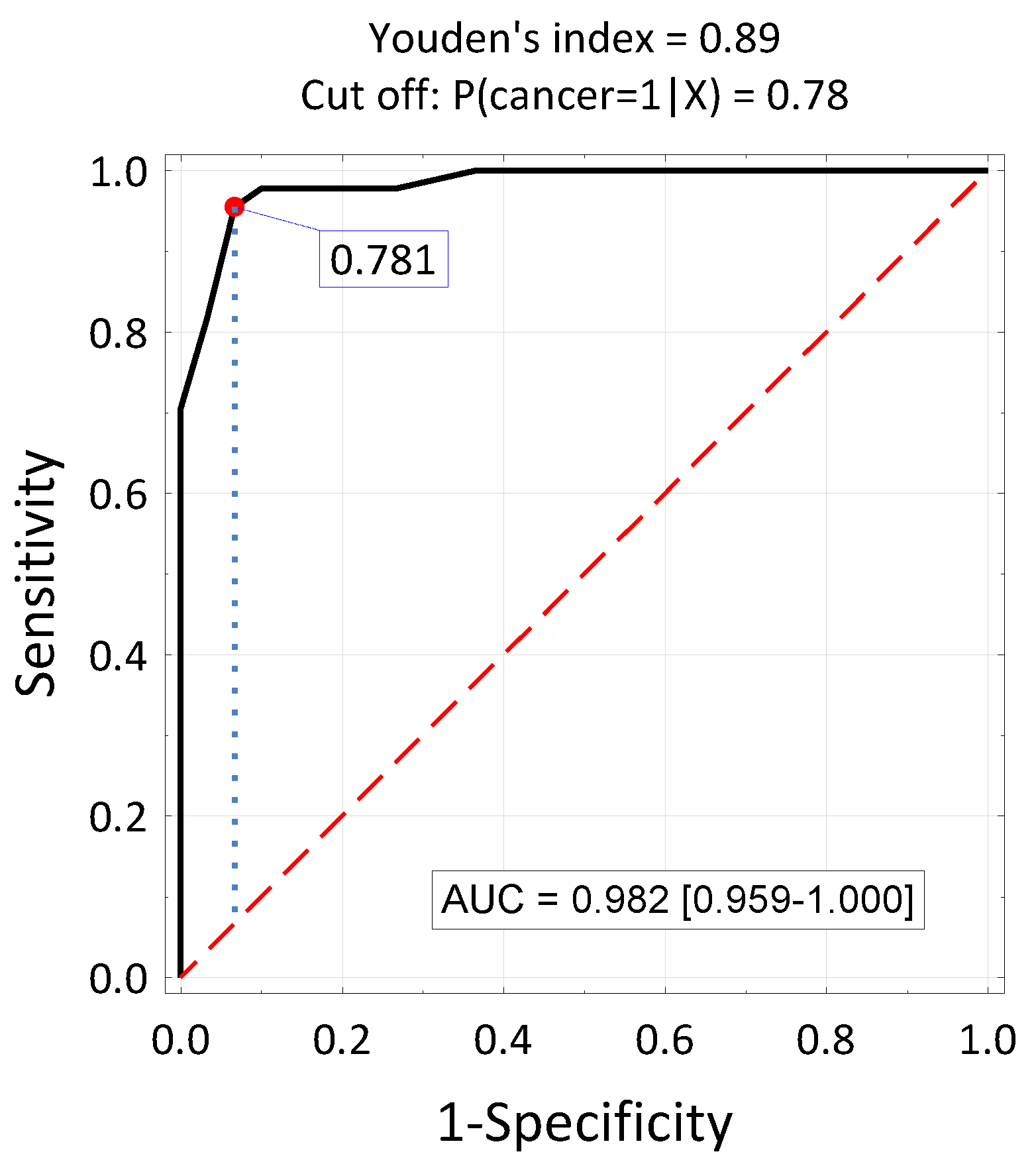
| Variable | Exam Group (E) N = 44 | Control Group (C) N = 30 | E Vs. C p-Value | Post-Hoc Test |
|---|---|---|---|---|
| Gender: | 0.202 a | |||
| Male, n (%) | 35 (79.6%) | 19 (63.3) | ||
| Female, n (%) | 9 (20.4%) | 11 (36.7%) | ||
| Age (years): | 0.703 b | |||
| M ± SD | 63.4 ± 9.0 | 62.6 ± 6.8 | ||
| Education: | 0.001 a | |||
| Primary | 14 (31.8%) | 1 (3.3%) | 0.003 | |
| Secondary | 25 (56.8%) | 16 (53.4%) | 0.773 | |
| Incomplete higher | 1 (2.3%) | 6 (20.0%) | 0.011 | |
| Higher | 4 (9.1%) | 7 (23.3%) | 0.092 | |
| Place of residence: | 0.770 a | |||
| Village | 11 (25.0%) | 8 (26.7%) | ||
| Town up to 20,000 | 6 (13.6%) | 2 (6.7%) | ||
| 21–50,000 inhabitants | 7 (15.9%) | 4 (13.3%) | ||
| Over 50,000 | 20 (45.5%) | 16 (53.3%) | ||
| Economic zone/urban area: | 0.871 a | |||
| Yes | 26 (59.1%) | 18 (60.0%) | ||
| No | 18 (40.9%) | 12 (40.0%) | ||
| Marital status: | 0.004 a | |||
| Single | 17 (38.6%) | 3 (10.0%) | 0.007 | |
| Partner/married relationship | 17 (38.6%) | 23 (76.7%) | 0.001 | |
| With family support | 10 (22.8%) | 4 (13.3%) | 0.306 | |
| BMI (kg/m2): | 0.100 c | |||
| Me [Q1–Q3] | 22.7 [21.3–24.2] | 23.4 [22.4–25.1] |
| Variable | Group E N = 44 | Group C N = 30 | E vs. C p-Value | Post-Hoc Test |
|---|---|---|---|---|
| ECOG scale (score): | <0.001 | |||
| 0—asymptomatic, n (%) | 12 (35.3%) | 22 (64.7%) | 0.013 | |
| 1—symptomatic, but completely ambulatory | 30 (78.9%) | 8 (21.1%) | <0.001 | |
| 2—Symptomatic, <50% in bed during the day | 2 (100.0%) | 0 (0.0%) | <0.001 | |
| Swallowing disorders (yes) | 16 (36.4%) | 0 (0.0%) | <0.001 | |
| Smoking | 43 (97.7%) | 18 (60.0%) | <0.001 | |
| Drinking alcohol regularly | 24 (54.5%) | 0 (0.0%) | <0.001 | |
| Dental condition: | <0.001 | |||
| 1—Normal | 8 (18.2%) | 30 (100.0%) | <0.001 | |
| 2—Cavities, caries, periodontal disease | 30 (68.2%) | 0 (0.0%) | <0.001 | |
| 3—Edentulism | 6 (13.6%) | 0 (0.0%) | 0.035 | |
| Nutritional status: | <0.001 | |||
| 1—Satisfactory | 8 (18.2%) | 30 (100.0%) | <0.001 | |
| 2—Risk of malnutrition | 10 (22.7%) | 0 (0.0%) | 0.005 | |
| 3—Malnutrition | 26 (59.1%) | 0 (0.0%) | <0.001 |
| Study Group | N = 44 (100%) |
|---|---|
| Tumor location: | |
| Glottis | 33 (75.0%) |
| Epiglottis | 11 (25.0%) |
| Tumor: | |
| T1 | 11 (25%) |
| T2 | 17 (38.7%) |
| T3 | 13 (29.5%) |
| T4a | 3 (6.8%) |
| Node: | |
| N0 | 25 (56.8%) |
| N1 | 5 (11.3%) |
| N2a | 1 (2.3%) |
| N2b | 7 (15.9%) |
| N2c | 5 (11.4%) |
| N3a | 1 (2.3%) |
| M0 | 44 (100.0%) |
| Stage: | |
| AND | 11 (25.0%) |
| II | 9 (20.4%) |
| III | 10 (22.8%) |
| Iva | 5 (11.4%) |
| IVb | 9 (20.4%) |
| Treatment *: | |
| Surgery | 20 (45.5%) |
| Radiotherapy | 40 (90.9%) |
| Chemotherapy | 17 (38.6%) |
| Only surgery | 3 (6.8%) |
| Surgery + radiotherapy | 8 (18.2%) |
| Only radiotherapy | 21 (47.8%) |
| Surgery + radiotherapy + chemotherapy | 6 (13.6%) |
| Radiotherapy + chemotherapy | 6 (13.6%) |
| Variable, Me [Q1–Q3] | Study Group (S) N = 44 | Control Group (C) N = 30 | S vs. C p-Value |
|---|---|---|---|
| Hemoglobin (g/dL) | 12.3 [11.1–13.3] | 13.5 (12.8–14.6) | <0.001 |
| Leukocytes (×103/mm) | 9.1 [8.1–11.3] | 5.1 [4.3–6.3] | <0.001 |
| APTT(s) | 28.5 [26.3–31.7] | 27.5 [26.0–29.3] | 0.050 |
| INR (-) | 1.05 [0.98–1.15] | 1.00 [0.96–1.04] | 0.007 |
| Total protein (g/dL) | 6.3 [5.9–6.8] | 7.3 [7.1–7.8] | <0.001 |
| CRP (mg/L) | 12.0 [6.3–25.9] | 3.2 [2.1–4.1] | <0.001 |
| Total cholesterol [mg/dL] | 144 [112–168] | 184 [175–192] | <0.001 |
| LDL cholesterol [mg/dL] | 105 [66–125] | 124 [115–135] | <0.001 |
| HDL cholesterol [mg/dL] | 34 [27–41] | 45 [34–48] | <0.001 |
| Triglycerides [mg/dL] | 109 [98–135] | 128 [117–140] | 0.009 |
| Iron level (mcg/dL) | 45 [32–57] | 75 [73–87] | <0.001 |
| TSH (mIU/L) | 1.42 [1.03–2.22] | 2.56 [1.98–2.98] | <0.001 |
| Cultureresult | Group S N = 44 | Group C N = 30 | S vs. C p-Value |
|---|---|---|---|
| Streptococcus oralis | 16 (36.4%) | 21 (70.0%) | 0.009 |
| Staphylococcus aureus | 4 (9.1%) | 0 (0.0%) | 0.142 |
| Streptococcus pneumoniae, pneumococcus | 0 (0.0%) | 0 (0.0%) | 1.00 |
| Candida albicans | 17 (38.6%) | 5 (16.7%) | 0.069 |
| Neisseria | 4 (9.1%) | 4 (13.3%) | 0.707 |
| Pseudomonas | 5 (11.4%) | 0 (0.0%) | 0.076 |
| Serratia mercescens | 3 (6.8%) | 0 (0.0%) | 0.276 |
| Bifidobacterium longum | 2 (4.6%) | 0 (0.0%) | 0.511 |
| Corynebacterium | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Enterococcus faecalis | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Klebsiella, Enterobacter and Serratia | 3 (6.8%) | 0 (0.0%) | 0.276 |
| Citrobacter freundii | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Lacticaseibicillus paracasei | 2 (4.6%) | 0 (0.0%) | 0.511 |
| Morganella morganii | 2 (4.6%) | 0 (0.0%) | 0.511 |
| Streptococcus dysgalactiae | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Proteus | 0 (0.0%) | 0 (0.0%) | 1.000 |
| Enterobacter cloacae | 0 (0.0%) | 0 (0.0%) | 1.000 |
| Veillonella parvula | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Escherichia coli | 1 (2.3%) | 0 (0.0%) | 1.000 |
| Absentee | 10 (22.7%) | 9 (30.0%) | 0.590 |
| Culture Result—Genus (%) | Group S | Group C | p-Value |
|---|---|---|---|
| Streptococcus | 7.4 [4.4–10.2] | 29.6 [20.0–36.3] | <0.001 |
| Prevotella melaninogenica | 13.2 [4.0–18.7] | 5.5 [1.0–11.6] | 0.005 |
| Prevotella | 15.7 [8.1–24.9] | 5.0 [1.9–7.3] | <0.001 |
| Rothia micilaginosa | 3.2 [1.6–8.1] | 9.2 [5.1–14.7] | 0.004 |
| Aggregatibacter | 0.0 [0.0–0.3] | 0.1 [0.0–0.4] | 0.273 |
| Gemella | 0.4 [0.0–1.2] | 1.7 [0.8–3.0] | <0.001 |
| Porphyromonas | 2.1 [0.3–5.7] | 1.5 [0.0–4.9] | 0.982 |
| Fusobacterium | 1.3 [0.1–2.7] | 0.8 [0.3–2.1] | 0.582 |
| Corynebacterium | 0.0 [0.0–0.5] | 0.1 [0.0–2.0] | 0.202 |
| Neisseria | 2.0 [0.2–6.8] | 4.4 [0.0–10.7] | 0.347 |
| Kingella | 0.0 [0.0–0.0] | 0.0 [0.0–0.6] | 0.316 |
| Lactobacillales | 2.7 [1.5–4.6] | 2.6 [1.5–3.9] | 0.451 |
| Actinobacteria | 1.9 [0.4–4.7] | 1.5 [0.4–3.1] | 0.447 |
| Actinomyces | 3.7 [1.0–6.6] | 3.0 [1.4–5.6] | 0.541 |
| Haemophilus | 4.0 [0.2–10.9] | 6.6 [2.2–13.1] | 0.102 |
| Capnocytophaga granulosa/gingivalis | 0.3 [0.0–1.5] | 0.8 [0.0–2.8] | 0.148 |
| Treponema | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.947 |
| Candidadus Sacharimonas aalborgensis | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.438 |
| Clostridiales | 0.5 [0.0–0.8] | 0.0 [0.0–0.2] | 0.004 |
| Veilonella | 0.0 [0.0–0.2] | 0.0 [0.0–0.2] | 0.688 |
| Campylobacter | 1.8 [0.5–3.7] | 1.1 [0.0–2.1] | 0.087 |
| Granulicatella | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.425 |
| Cloacibacterium | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.804 |
| Lautropia | 0.0 [0.0–1.2] | 0.0 [0.0–1.0] | 0.978 |
| Abiotrophia defective | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.813 |
| Eikenella corrodens | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.406 |
| Shaalia odontolytica | 1.1 [0.0–2.7] | 0.0 [0.0–1.6] | 0.098 |
| Leptotricha | 0.7 [0.0–1.8] | 0.2 [0.0–0.8] | 0.074 |
| Stomatobaculum longum | 0.3 [0.0–0.5] | 0.0 [0.0–0.0] | 0.005 |
| Granucicatella elegans | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Tannerell | 0.0 [0.0–0.3] | 0.0 [0.0–0.0] | 0.438 |
| Pasteurellaceae | 0.2 [0.0–0.7] | 0.5 [0.0–1.2] | 0.198 |
| Bifidobacteriaceae | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.241 |
| Atopobium | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.218 |
| Culture Result—Genus (%) | Group S | Group C | p-Value |
|---|---|---|---|
| Oribacterium | 0.0 [0.0–0.4] | 0.0 [0.0–0.0] | 0.005 |
| Enterobacteriaceae | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.624 |
| Parascardovia denticolens | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Butyrivibrio | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Cardiobacterium hominis | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Bergeyella cardium | 0.0 [0.0–0.2] | 0.0 [0.0–0.0] | 0.013 |
| Catonella | 0.0 [0.0–0.2] | 0.0 [0.0–0.0] | 0.048 |
| Vallitalae okinawensis | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.624 |
| Olsenella | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Mogibacterium | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.250 |
| Eubacterium | 0.0 [0.0–0.2] | 0.0 [0.0–0.0] | 0.032 |
| Sulfurihydrogenibium | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.324 |
| Oceanivirga miroungae | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Peptostreptococcus anaerobius | 0.0 [0.0–0.5] | 0.0 [0.0–0.0] | 0.013 |
| Mycoplasma salivarius/faucium | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.250 |
| Dialister pneumosintes | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Pseudoramibacter alactolyticus | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Slackia | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Filifactor alocis | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.250 |
| Gordonibacter | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Lachnoanaerobaculum saburreum | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.624 |
| Cryptobacterium curtum | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Scardovia wiggsiae | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.324 |
| Dysgonomonas | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Parvimonas | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.512 |
| Ihubacter | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.624 |
| Phocaeicola abscessus | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Marinifilum | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.996 |
| Klebsiella | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Staphylococcus | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Anaerococcus | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Eschericia coli | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Salmonella | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Shigella | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Finegoldia | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.996 |
| Lawsonella clevelandensis | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.873 |
| Moryella indoligens | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.745 |
| Bacillales | 0.0 [0.0–0.0] | 0.0 [0.0–0.0] | 0.996 |
| Other genus | 0.7 [0.3–2.8] | 1.5 [0.7–3.1] | 0.055 |
| Culture Result—Phylum (%) | Group S | Group C | p-Value |
|---|---|---|---|
| Bacteroidetes | 33.2 [21.2–44.6] | 10.0 [4.2–18.9] | <0.001 |
| Bacteroidota | 3.2 [1.2–7.6] | 3.8 [0.6–10.1] | 0.830 |
| Fusobacteriota | 2.5 [0.3–4.2] | 1.3 [0.5–3.2] | 0.259 |
| Proteobacteria | 15.4 [5.9–21.4] | 19.4 [9.6–27.2] | 0.269 |
| Firmicutes | 13.9 [8.5–18.5] | 35.9 [24.8–41.6] | <0.001 |
| Bacillota | 2.2 [0.7–4.9] | 0.4 [0.2–1.6] | 0.002 |
| Actinomycetota | 7.1 [4.0–12.3] | 9.6 [5.5–17.5] | 0.169 |
| Pseudomonadota | 0.7 [0.0–2.9] | 1.5 [0.5–2.4] | 0.226 |
| Actinobacteria | 6.7 [2.4–12.2] | 5.4 [3.0–9.3] | 0.509 |
| Other phylum | 0.7 [0.2–3.2] | 1.4 [0.6–3.1] | 0.127 |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| B | Mr | OR | Beta | Mr | OR (95% CI) | |
| Education ≤ 2 | 1.86 | 0.004 | 5.96 | 0 | >0.05 | 1.00 |
| Singles | 1.735 | 0.013 | 5.67 | 0 | >0.05 | 1.00 |
| Hemoglobin ≤ 13.9 | 2.034 | 0.002 | 7.65 | 0 | >0.05 | 1.00 |
| Leukocytes ≥ 6.52 | 4.234 | <0.001 | 69.0 | 4.319 | 0.006 | 75.1 (3.51–1608) |
| APTT ≥ 29.76 | 2.457 | 0.003 | 11.7 | 0 | >0.05 | 1.00 |
| INR ≥ 1.05 | 1.281 | 0.018 | 3.60 | 0 | >0.05 | 1.00 |
| Total protein ≤ 6.9 | 3.718 | <0.001 | 41.2 | 3.663 | 0.007 | 39.0 (2.82–539) |
| CRP ≥ 4.76 | 5.213 | <0.001 | 184 | 0 | >0.05 | 1.00 |
| Total cholesterol ≤ 167 | 2.708 | <0.001 | 15.0 | 0 | >0.05 | 1.00 |
| LDL ≤ 110 | 2.472 | 0.001 | 11.8 | 0 | >0.05 | 1.00 |
| HDL ≤ 42 | 2.251 | <0.001 | 9.50 | 0 | >0.05 | 1.00 |
| Triglycerides ≤ 113 | 2.072 | 0.001 | 7/94 | 0 | >0.05 | 1.00 |
| Iron level ≤ 65 | 5.254 | <0.001 | 191 | 0 | >0.05 | 1.00 |
| TSH ≤ 1.73 | 2.959 | <0.001 | 19.3 | 0 | >0.05 | 1.00 |
| ECOG ≥ 1 | 1.992 | <0.001 | 7.33 | 0 | >0.05 | 1.00 |
| Smoking | 3.356 | 0.003 | 28/7 | 0 | >0.05 | 1.00 |
| Bacteroidetes ≥ 24.7% | 2.853 | <0.001 | 17.3 | 0 | >0.05 | 1.00 |
| Firmicutes ≤ 22.1% | 3.455 | <0.001 | 31.7 | 4.303 | 0.005 | 74.0 (3.94–1388) |
| Bacillota ≥ 1.7% | 2.046 | <0.001 | 7.73 | 0 | >0.05 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorobisz, K.; Dorobisz, T.; Pazdro-Zastawny, K. Analysis of Risk Factors with Assessment of the Impact of the Microbiome on the Risk of Squamous Cell Carcinoma of the Larynx. J. Clin. Med. 2024, 13, 6101. https://doi.org/10.3390/jcm13206101
Dorobisz K, Dorobisz T, Pazdro-Zastawny K. Analysis of Risk Factors with Assessment of the Impact of the Microbiome on the Risk of Squamous Cell Carcinoma of the Larynx. Journal of Clinical Medicine. 2024; 13(20):6101. https://doi.org/10.3390/jcm13206101
Chicago/Turabian StyleDorobisz, Karolina, Tadeusz Dorobisz, and Katarzyna Pazdro-Zastawny. 2024. "Analysis of Risk Factors with Assessment of the Impact of the Microbiome on the Risk of Squamous Cell Carcinoma of the Larynx" Journal of Clinical Medicine 13, no. 20: 6101. https://doi.org/10.3390/jcm13206101
APA StyleDorobisz, K., Dorobisz, T., & Pazdro-Zastawny, K. (2024). Analysis of Risk Factors with Assessment of the Impact of the Microbiome on the Risk of Squamous Cell Carcinoma of the Larynx. Journal of Clinical Medicine, 13(20), 6101. https://doi.org/10.3390/jcm13206101






