In-Advance Prediction of Pressure Ulcers via Deep-Learning-Based Robust Missing Value Imputation on Real-Time Intensive Care Variables
Abstract
:1. Introduction
2. Methods
2.1. Study Population
2.2. Data Collection
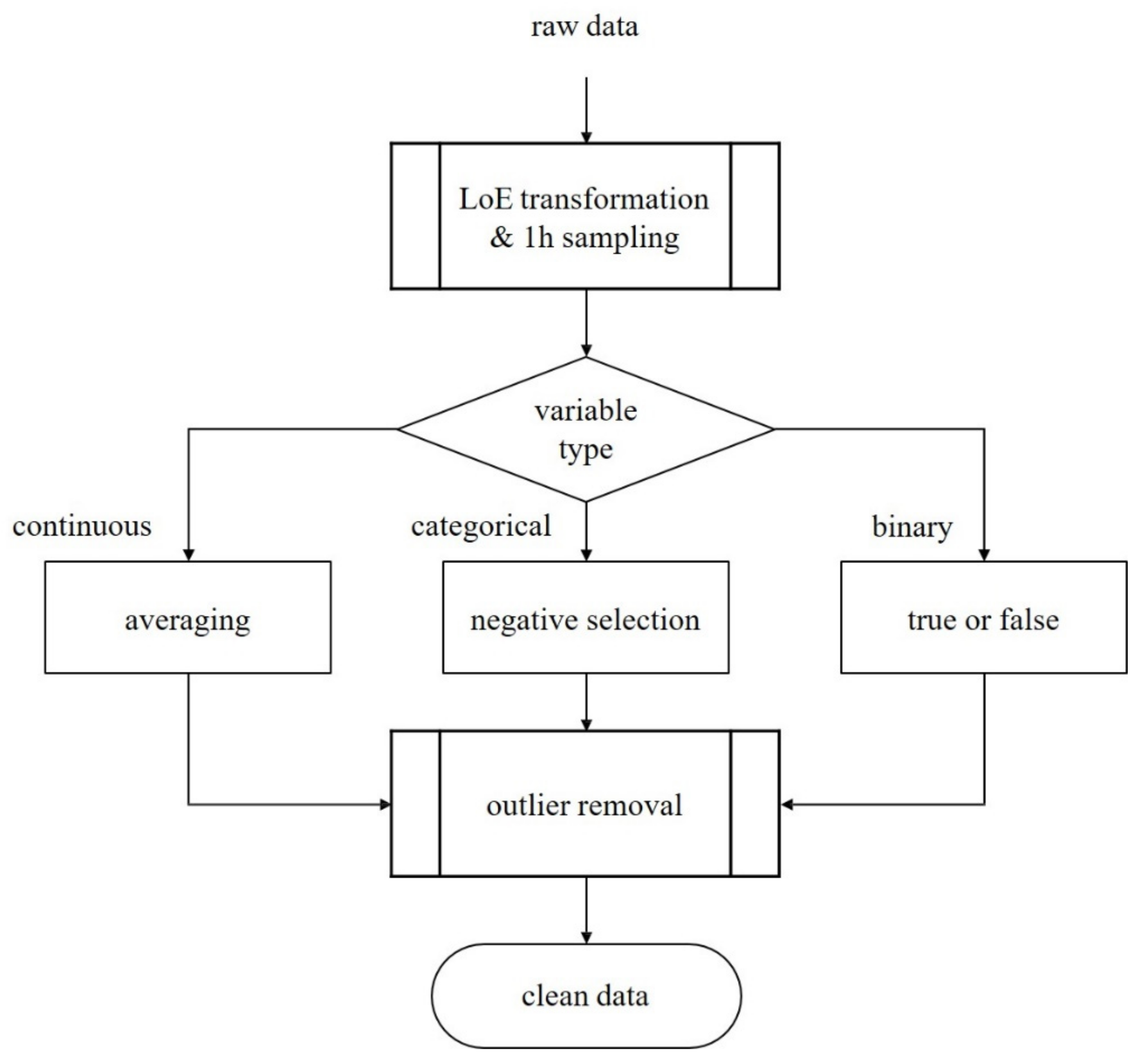
2.3. Data Preprocessing
- We sampled patient data hourly from ICU admission to discharge;
- If multiple sampling data exist in an hour, we selected the average values for continuous variables, the most negative values for categorical variables, and binarized medication information;
- We applied the interquartile range method to discard outliers.
2.4. Prediction Models
3. Results
3.1. Baseline Characteristics
3.2. Predictive Performances
4. Discussion
4.1. Analysis of Predictive Performances
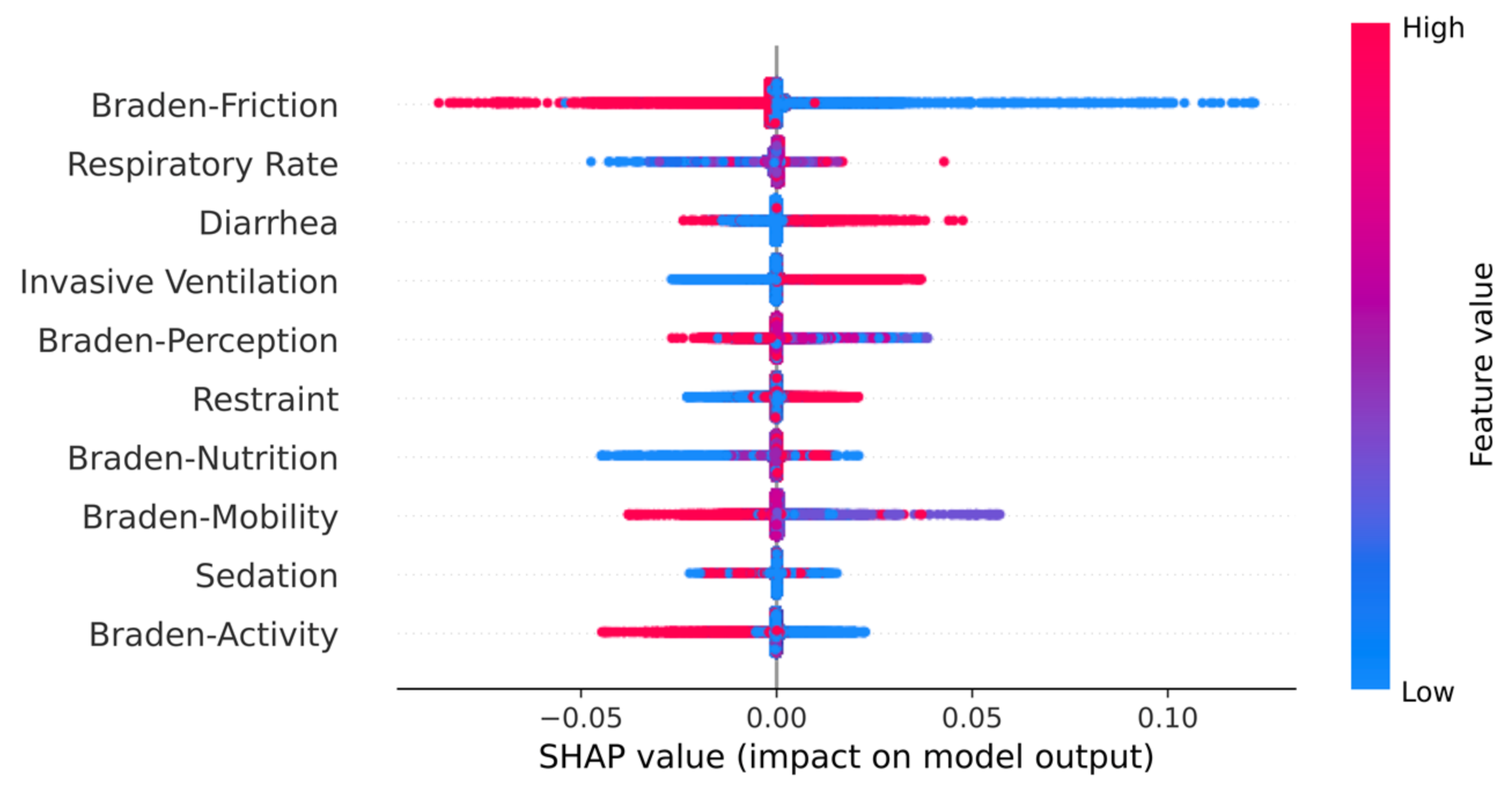
4.2. Clinical Findings
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Details of GRU-D++
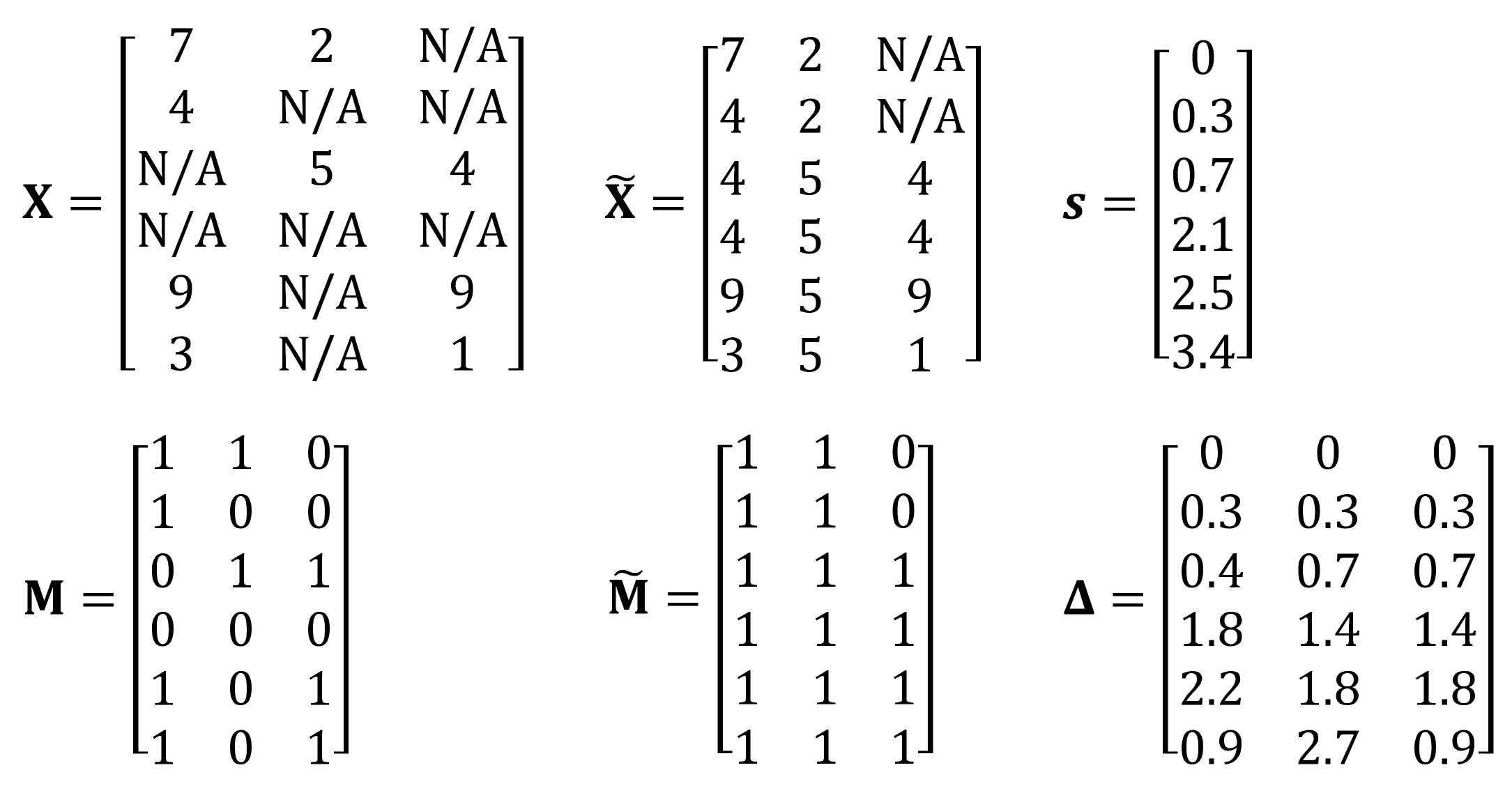
Appendix A.2. Further Experiments
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| LR | 0.653 (±0.008) | 0.212 (±0.002) | 0.655 (±0.008) | 0.220 (±0.002) | 0.656 (±0.008) | 0.229 (±0.003) | 0.667 (±0.008) | 0.249 (±0.003) |
| DT | 0.504 (±0.015) | 0.125 (±0.005) | 0.502 (±0.037) | 0.131 (±0.010) | 0.519 (±0.012) | 0.141 (±0.006) | 0.520 (±0.014) | 0.153 (±0.009) |
| RF | 0.652 (±0.011) | 0.195 (±0.010) | 0.631 (±0.038) | 0.203 (±0.016) | 0.645 (±0.029) | 0.215 (±0.012) | 0.662 (±0.033) | 0.237 (±0.016) |
| XGBoost | 0.603 (±0.025) | 0.189 (±0.010) | 0.605 (±0.022) | 0.199 (±0.015) | 0.627 (±0.026) | 0.221 (±0.018) | 0.635 (±0.016) | 0.233 (±0.015) |
| RNN | 0.768 (±0.033) | 0.290 (±0.027) | 0.753 (±0.028) | 0.295 (±0.024) | 0.759 (±0.023) | 0.309 (±0.025) | 0.751 (±0.008) | 0.315 (±0.006) |
| GRU | 0.880 (±0.015) | 0.469 (±0.035) | 0.875 (±0.006) | 0.471 (±0.025) | 0.873 (±0.010) | 0.480 (±0.031) | 0.867 (±0.008) | 0.490 (±0.024) |
| LSTM | 0.816 (±0.042) | 0.370 (±0.054) | 0.833 (±0.013) | 0.390 (±0.027) | 0.829 (±0.017) | 0.405 (±0.029) | 0.815 (±0.027) | 0.409 (±0.041) |
| GRU-D | 0.762 (±0.019) | 0.267 (±0.015) | 0.768 (±0.015) | 0.285 (±0.009) | 0.777 (±0.022) | 0.307 (±0.021) | 0.759 (±0.026) | 0.291 (±0.029) |
| GRU-D++ | 0.807 (±0.015) | 0.350 (±0.026) | 0.827 (±0.018) | 0.380 (±0.031) | 0.826 (±0.035) | 0.398 (±0.054) | 0.848 (±0.016) | 0.445 (±0.030) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| LR | 0.643 (±0.010) | 0.217 (±0.004) | 0.644 (±0.009) | 0.226 (±0.004) | 0.646 (±0.010) | 0.235 (±0.004) | 0.657 (±0.007) | 0.255 (±0.004) |
| DT | 0.542 (±0.022) | 0.139 (±0.011) | 0.503 (±0.060) | 0.133 (±0.013) | 0.539 (±0.024) | 0.151 (±0.011) | 0.541 (±0.009) | 0.161 (±0.005) |
| RF | 0.513 (±0.051) | 0.162 (±0.026) | 0.590 (±0.044) | 0.197 (±0.023) | 0.581 (±0.067) | 0.202 (±0.029) | 0.555 (±0.073) | 0.202 (±0.029) |
| XGBoost | 0.565 (±0.033) | 0.179 (±0.016) | 0.569 (±0.059) | 0.187 (±0.022) | 0.555 (±0.039) | 0.192 (±0.019) | 0.587 (±0.048) | 0.218 (±0.016) |
| RNN | 0.755 (±0.043) | 0.278 (±0.028) | 0.774 (±0.029) | 0.321 (±0.025) | 0.779 (±0.031) | 0.336 (±0.029) | 0.749 (±0.011) | 0.324 (±0.010) |
| GRU | 0.890 (±0.007) | 0.513 (±0.024) | 0.882 (±0.003) | 0.503 (±0.017) | 0.880 (±0.006) | 0.515 (±0.015) | 0.874 (±0.005) | 0.521 (±0.006) |
| LSTM | 0.834 (±0.014) | 0.393 (±0.027) | 0.838 (±0.031) | 0.394 (±0.048) | 0.819 (±0.017) | 0.390 (±0.026) | 0.811 (±0.028) | 0.398 (±0.045) |
| GRU-D | 0.824 (±0.045) | 0.370 (±0.088) | 0.840 (±0.039) | 0.407 (±0.070) | 0.810 (±0.035) | 0.378 (±0.050) | 0.726 (±0.083) | 0.289 (±0.103) |
| GRU-D++ | 0.895 (±0.032) | 0.534 (±0.078) | 0.892 (±0.022) | 0.538 (±0.044) | 0.889 (±0.017) | 0.539 (±0.049) | 0.864 (±0.017) | 0.514 (±0.023) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| RNN | 0.786 (±0.014) | 0.307 (±0.017) | 0.773 (±0.032) | 0.318 (±0.019) | 0.766 (±0.012) | 0.324 (±0.021) | 0.771 (±0.012) | 0.338 (±0.005) |
| GRU | 0.873 (±0.011) | 0.458 (±0.029) | 0.872 (±0.008) | 0.466 (±0.024) | 0.868 (±0.007) | 0.473 (±0.026) | 0.864 (±0.007) | 0.485 (±0.023) |
| LSTM | 0.826 (±0.032) | 0.375 (±0.047) | 0.832 (±0.026) | 0.398 (±0.025) | 0.838 (±0.020) | 0.414 (±0.036) | 0.811 (±0.022) | 0.399 (±0.036) |
| GRU-D | 0.858 (±0.015) | 0.426 (±0.046) | 0.853 (±0.023) | 0.424 (±0.044) | 0.856 (±0.017) | 0.442 (±0.035) | 0.845 (±0.018) | 0.438 (±0.029) |
| GRU-D++ | 0.898 (±0.018) | 0.523 (±0.051) | 0.901 (±0.016) | 0.542 (±0.045) | 0.895 (±0.019) | 0.543 (±0.050) | 0.897 (±0.008) | 0.575 (±0.027) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| LR | 0.707 (±0.002) | 0.234 (±0.001) | 0.709 (±0.002) | 0.244 (±0.001) | 0.711 (±0.002) | 0.254 (±0.002) | 0.716 (±0.002) | 0.274 (±0.002) |
| DT | 0.516 (±0.008) | 0.129 (±0.004) | 0.513 (±0.008) | 0.133 (±0.003) | 0.525 (±0.012) | 0.143 (±0.006) | 0.532 (±0.023) | 0.157 (±0.013) |
| RF | 0.683 (±0.004) | 0.211 (±0.006) | 0.680 (±0.011) | 0.217 (±0.007) | 0.694 (±0.008) | 0.230 (±0.009) | 0.693 (±0.004) | 0.251 (±0.003) |
| XGBoost | 0.676 (±0.010) | 0.220 (±0.009) | 0.675 (±0.008) | 0.227 (±0.005) | 0.683 (±0.013) | 0.239 (±0.005) | 0.692 (±0.008) | 0.264 (±0.005) |
| RNN | 0.761 (±0.014) | 0.277 (±0.010) | 0.777 (±0.015) | 0.290 (±0.009) | 0.775 (±0.022) | 0.304 (±0.015) | 0.772 (±0.006) | 0.323 (±0.006) |
| GRU | 0.856 (±0.010) | 0.413 (±0.021) | 0.843 (±0.008) | 0.406 (±0.015) | 0.848 (±0.016) | 0.426 (±0.026) | 0.837 (±0.027) | 0.434 (±0.058) |
| LSTM | 0.844 (±0.020) | 0.389 (±0.026) | 0.833 (±0.020) | 0.388 (±0.043) | 0.827 (±0.016) | 0.385 (±0.038) | 0.813 (±0.010) | 0.390 (±0.022) |
| GRU-D | 0.749 (±0.028) | 0.270 (±0.029) | 0.750 (±0.025) | 0.286 (±0.032) | 0.761 (±0.025) | 0.302 (±0.033) | 0.778 (±0.018) | 0.344 (±0.030) |
| GRU-D++ | 0.783 (±0.033) | 0.328 (±0.051) | 0.795 (±0.034) | 0.337 (±0.046) | 0.803 (±0.030) | 0.382 (±0.031) | 0.818 (±0.034) | 0.405 (±0.039) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| LR | 0.697 (±0.003) | 0.236 (±0.003) | 0.697 (±0.003) | 0.245 (±0.003) | 0.699 (±0.003) | 0.254 (±0.004) | 0.702 (±0.003) | 0.273 (±0.004) |
| DT | 0.515 (±0.046) | 0.132 (±0.019) | 0.546 (±0.030) | 0.148 (±0.016) | 0.537 (±0.032) | 0.148 (±0.013) | 0.453 (±0.097) | 0.138 (±0.016) |
| RF | 0.538 (±0.050) | 0.164 (±0.020) | 0.612 (±0.042) | 0.198 (±0.010) | 0.552 (±0.058) | 0.185 (±0.016) | 0.525 (±0.056) | 0.180 (±0.018) |
| XGBoost | 0.537 (±0.057) | 0.176 (±0.028) | 0.592 (±0.046) | 0.197 (±0.024) | 0.560 (±0.062) | 0.193 (±0.025) | 0.568 (±0.050) | 0.209 (±0.020) |
| RNN | 0.771 (±0.026) | 0.284 (±0.013) | 0.761 (±0.016) | 0.287 (±0.007) | 0.751 (±0.009) | 0.301 (±0.004) | 0.764 (±0.014) | 0.323 (±0.009) |
| GRU | 0.866 (±0.008) | 0.446 (±0.022) | 0.844 (±0.019) | 0.426 (±0.032) | 0.850 (±0.021) | 0.447 (±0.036) | 0.826 (±0.040) | 0.439 (±0.062) |
| LSTM | 0.840 (±0.008) | 0.380 (±0.010) | 0.821 (±0.033) | 0.381 (±0.029) | 0.798 (±0.039) | 0.368 (±0.043) | 0.791 (±0.023) | 0.376 (±0.021) |
| GRU-D | 0.811 (±0.020) | 0.408 (±0.036) | 0.853 (±0.013) | 0.469 (±0.016) | 0.845 (±0.035) | 0.468 (±0.048) | 0.810 (±0.059) | 0.431 (±0.115) |
| GRU-D++ | 0.868 (±0.037) | 0.492 (±0.078) | 0.888 (±0.012) | 0.530 (±0.034) | 0.889 (±0.014) | 0.546 (±0.040) | 0.874 (±0.032) | 0.543 (±0.062) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| RNN | 0.771 (±0.015) | 0.296 (±0.009) | 0.782 (±0.013) | 0.315 (±0.007) | 0.782 (±0.014) | 0.329 (±0.015) | 0.784 (±0.001) | 0.346 (±0.004) |
| GRU | 0.856 (±0.010) | 0.426 (±0.018) | 0.844 (±0.011) | 0.406 (±0.020) | 0.852 (±0.018) | 0.436 (±0.029) | 0.847 (±0.020) | 0.447 (±0.046) |
| LSTM | 0.853 (±0.009) | 0.406 (±0.022) | 0.851 (±0.012) | 0.408 (±0.028) | 0.837 (±0.025) | 0.405 (±0.025) | 0.835 (±0.001) | 0.418 (±0.009) |
| GRU-D | 0.852 (±0.010) | 0.424 (±0.021) | 0.85 (±0.012) | 0.439 (±0.031) | 0.857 (±0.016) | 0.449 (±0.037) | 0.845 (±0.006) | 0.459 (±0.024) |
| GRU-D++ | 0.881 (±0.025) | 0.500 (±0.073) | 0.867 (±0.020) | 0.475 (±0.059) | 0.878 (±0.016) | 0.504 (±0.048) | 0.875 (±0.011) | 0.518 (±0.039) |
References
- Araujo, S.M.; Sousa, P.; Dutra, I. Clinical decision support systems for pressure ulcer management: Systematic review. JMIR Med. Inform. 2020, 8, e21621. [Google Scholar] [CrossRef]
- PAN Pacific. Prevention and treatment of pressure ulcers: Quick reference guide. Natl. Press. Ulcer Advis. Panel 2014, 75, 1–72. [Google Scholar]
- Šín, P.; Hokynková, A.; Marie, N.; Andrea, P.; Krč, R.; Podroužek, J. Machine learning-based pressure ulcer prediction in modular critical care data. Diagnostics 2022, 12, 850. [Google Scholar] [CrossRef]
- Kim, E.; Ham, S.; Jung, B.K.; Park, J.-W.; Kim, J.; Lee, J.H. Effect of baicalin on wound healing in a mouse model of pressure ulcers. Int. J. Mol. Sci. 2023, 24, 329. [Google Scholar] [CrossRef]
- Park, S.-H.; Lee, Y.-S.; Kwon, Y.-M. Predictive validity of pressure ulcer risk assessment tools for elderly: A meta-analysis. West. J. Nurs. Res. 2016, 38, 459–483. [Google Scholar] [CrossRef]
- Weststrate, J.T.; Bruining, H.A. Pressure sores in an intensive care unit and related variables: A descriptive study. Intensive Crit. Care Nurs. 1996, 12, 280–284. [Google Scholar] [CrossRef]
- Cho, I.S.; Yoon, H.Y.; Im, P.S.; Sook, L.H. Availability of nursing data in an electronic nursing record system for a development of a risk assessment tool for pressure ulcers. J. Korean Soc. Med. Inform. 2008, 14, 161–168. [Google Scholar] [CrossRef]
- Cramer, E.M.; Seneviratne, M.G.; Sharifi, H.; Ozturk, A.; Hernandez-Boussard, T. Predicting the incidence of pressure ulcers in the intensive care unit using machine learning. eGEMs 2019, 7, 49. [Google Scholar] [CrossRef]
- Cicceri, G.; De Vita, F.; Bruneo, D.; Merlino, G.; Puliafito, A. A deep learning approach for pressure ulcer prevention using wearable computing. Hum. Centric Comput. Inf. Sci. 2020, 10, 5. [Google Scholar] [CrossRef]
- Walther, F.; Heinrich, L.; Schmitt, J.; Eberlein-Gonska, M.; Roessler, M. Prediction of inpatient pressure ulcers based on routine healthcare data using machine learning methodology. Sci. Rep. 2022, 12, 5044. [Google Scholar] [CrossRef]
- Choi, J.E.; Hwang, S.-K. Predictive validity of pressure ulcer risk assessment scales among patients in a trauma intensive care unit. J. Korean Crit. Care Nurs. 2019, 12, 26–38. [Google Scholar] [CrossRef]
- Qaseem, A.; Mir, T.P.; Starkey, M.; Denberg, T.D.; Clinical Guidelines Committee of the American College of Physicians. Risk assessment and prevention of pressure ulcers: A clinical practice guideline from the american college of physicians. Ann. Intern. Med. 2015, 162, 359–369. [Google Scholar] [CrossRef]
- Dweekat, O.Y.; Lam, S.S.; McGrath, L. An integrated system of multifaceted machine learning models to predict if and when hospital-acquired pressure injuries (bedsores) occur. Int. J. Environ. Res. Public Health 2023, 20, 828. [Google Scholar] [CrossRef]
- Silva, A.; Metrôlho, J.; Ribeiro, F.; Fidalgo, F.; Santos, O.; Dionisio, R. A review of intelligent sensor-based systems for pressure ulcer prevention. Computers 2022, 11, 6. [Google Scholar] [CrossRef]
- Cox, J.; Schallom, M.; Jung, C. Identifying risk factors for pressure injury in adult critical care patients. Am. J. Crit. Care 2020, 29, 204–213. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Khan, I.H. Current status and applications of artificial intelligence (ai) in medical field: An overview. Curr. Med. Res. Pract. 2019, 9, 231–237. [Google Scholar] [CrossRef]
- Ribeiro, F.; Fidalgo, F.; Silva, A.; Metrôlho, J.; Santos, O.; Dionisio, R. Literature review of machine-learning algorithms for pressure ulcer prevention: Challenges and opportunities. Informatics 2021, 8, 76. [Google Scholar] [CrossRef]
- Song, J.; Gao, Y.; Yin, P.; Li, Y.; Li, Y.; Zhang, J.; Su, Q.; Fu, X.; Pi, H. The random forest model has the best accuracy among the four pressure ulcer prediction models using machine learning algorithms. Risk Manag. Healthc. Policy 2021, 14, 1175–1187. [Google Scholar] [CrossRef]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Che, Z.; Purushotham, S.; Cho, K.; Sontag, D.; Liu, Y. Recurrent Neural Networks for Multivariate Time Series with Missing Values. Sci. Rep. 2018, 8, 6085. [Google Scholar] [CrossRef]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Horng, S.; Celi, L.A.; Mark, R. “MIMIC-IV” (Version 2.0). PhysioNet 2022. [Google Scholar] [CrossRef]
- Cihan, P.; Ozger, Z.B. A New Heuristic Approach for Treating Missing Value: ABCimp. Elektron. Ir Elektrotechnika 2019, 25, 48–54. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Y.; Cai, X.; Yuan, X. E2GAN: End-to-End Generative Adversarial Network for Multivariate Time Series Imputation. In Proceedings of the 28th International Joint Conference on Artificial Intelligence, Palo Alto, CA, USA, 10–16 August 2019. [Google Scholar]
- Luo, Y.; Cai, X.; Zhang, Y.; Xu, J. Multivariate Time Series Imputation with Generative Adversarial Networks. Adv. Neural Inf. Process. Syst. 2018, 31, 1–12. [Google Scholar]
- Zhang, Y.; Zhou, B.; Cai, X.; Guo, W.; Ding, X.; Yuan, X. Missing value imputation in multivariate time series with end-to-end generative adversarial networks. Inf. Sci. 2021, 551, 67–82. [Google Scholar] [CrossRef]
- Fortuin, V.; Baranchuk, D.; Rätsch, G.; Mandt, S. GP-VAE: Deep Probabilistic Time Series Imputation. In Proceedings of the International Conference on Artificial Intelligence and Statistics, Online, 26–28 August 2020; PMLR: London, UK, 2020. [Google Scholar]

| Variables | MIMIC-IV (Internal) | KNUH (External) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Development Cohort | Validation Cohort | Validation Cohort | |||||||
| Control (n = 51,600) | Pressure Ulcer (n = 2140) | p-Value | Control (n = 12,876) | Pressure Ulcer (n = 559) | p-Value | Control (n = 6317) | Pressure Ulcer (n = 559) | p-Value | |
| Demographics | |||||||||
| Male, n (%) | 28,600 (55.43%) | 1265 (59.11%) | <0.001 | 7125 (55.34%) | 352 (62.97%) | <0.001 | 1575 (44.82%) | 164 (65.86%) | <0.001 |
| Age * | 63.74 (±17.10) | 65.44 (±15.22) | <0.001 | 63.76 (±16.89) | 64.98 (±16.25) | 0.096 | 61.32 (±20.90) | 72.60 (±13.08) | <0.001 |
| Height (cm) * | 168.56 (±9.08) | 169.14 (±10.34) | 0.004 | 168.49 (±9.07) | 169.52 (±10.52) | 0.009 | 161.75 (±8.57) | 162.32 (±9.39) | 0.314 |
| Body Weight (kg) * | 81.92 (±35.39) | 85.49 (±28.56) | <0.001 | 81.98 (±27.48) | 83.94 (±24.21) | 0.097 | 61.89 (±14.38) | 56.78 (±14.33) | <0.001 |
| BMI * | 28.79 (±13.01) | 29.79 (±9.35) | <0.001 | 28.84 (±9.44) | 29.12 (±7.59) | 0.481 | 23.61 (±4.88) | 21.46 (±4.77) | <0.001 |
| Underlying Disease | |||||||||
| DM, n (%) | 15,243 (29.54%) | 728 (34.02%) | <0.001 | 3661 (28.44%) | 192 (34.35%) | 0.003 | 221 (3.50%) | 38 (6.80%) | <0.001 |
| CVD, n (%) | 8236 (15.96%) | 465 (21.730%) | <0.001 | 2019 (15.68%) | 118 (21.11%) | <0.001 | 793 (12.55%) | 111 (19.86%) | <0.001 |
| Hypertension, n (%) | 32,541 (63.06%) | 1385 (64.72%) | 0.125 | 8081 (62.76%) | 374 (66.91%) | 0.052 | 141 (2.23%) | 10 (1.79%) | 0.593 |
| Arterial Disease, n (%) | 5958 (11.55%) | 385 (17.99%) | <0.001 | 1496 (11.62%) | 100 (17.89%) | <0.001 | 71 (1.12%) | 10 (1.79%) | 0.233 |
| Skin Disease, n (%) | 4852 (9.40%) | 510 (23.83%) | <0.001 | 1235 (9.59%) | 136 (24.33%) | <0.001 | 101 (1.60%) | 55 (9.84%) | <0.001 |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| LR | 0.818 (±0.000) | 0.433 (±0.001) | 0.814 (±0.000) | 0.438 (±0.001) | 0.810 (±0.000) | 0.442 (±0.001) | 0.804 (±0.000) | 0.450 (±0.001) |
| DT | 0.569 (±0.007) | 0.145 (±0.006) | 0.567 (±0.007) | 0.151 (±0.005) | 0.566 (±0.005) | 0.156 (±0.004) | 0.569 (±0.004) | 0.172 (±0.003) |
| RF | 0.818 (±0.000) | 0.407 (±0.005) | 0.813 (±0.001) | 0.412 (±0.004) | 0.808 (±0.001) | 0.414 (±0.004) | 0.801 (±0.001) | 0.424 (±0.004) |
| XGBoost | 0.814 (±0.002) | 0.404 (±0.006) | 0.807 (±0.003) | 0.404 (±0.006) | 0.805 (±0.006) | 0.410 (±0.012) | 0.797 (±0.003) | 0.424 (±0.011) |
| Simple RNN | 0.860 (±0.021) | 0.489 (±0.037) | 0.864 (±0.003) | 0.506 (±0.015) | 0.859 (±0.010) | 0.511 (±0.022) | 0.837 (±0.003) | 0.507 (±0.015) |
| GRU | 0.918 (±0.003) | 0.657 (±0.011) | 0.913 (±0.002) | 0.657 (±0.009) | 0.905 (±0.001) | 0.649 (±0.008) | 0.885 (±0.003) | 0.630 (±0.007) |
| LSTM | 0.909 (±0.005) | 0.625 (±0.020) | 0.903 (±0.004) | 0.619 (±0.019) | 0.900 (±0.007) | 0.620 (±0.020) | 0.880 (±0.005) | 0.605 (±0.013) |
| GRU-D | 0.944 (±0.003) | 0.737 (±0.008) | 0.938 (±0.002) | 0.723 (±0.009) | 0.930 (±0.003) | 0.712 (±0.005) | 0.909 (±0.002) | 0.686 (±0.007) |
| GRU-D++ | 0.945 (±0.003) | 0.742 (±0.005) | 0.940 (±0.002) | 0.730 (±0.004) | 0.933 (±0.001) | 0.722 (±0.005) | 0.912 (±0.003) | 0.699 (±0.007) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| Braden scale | 0.730 | 0.201 | 0.730 | 0.212 | 0.731 | 0.223 | 0.732 | 0.245 |
| GRU-D++ | 0.945 (±0.003) | 0.742 (±0.005) | 0.940 (±0.002) | 0.730 (±0.004) | 0.933 (±0.001) | 0.722 (±0.005) | 0.912 (±0.003) | 0.699 (±0.007) |
| GRU-D++10 | 0.923 (±0.004) | 0.670 (±0.012) | 0.918 (±0.003) | 0.665 (±0.004) | 0.911 (±0.005) | 0.658 (±0.003) | 0.888 (±0.007) | 0.637 (±0.008) |
| GRU-D++ (w/o Braden) | 0.934 (±0.002) | 0.671 (±0.009) | 0.928 (±0.002) | 0.660 (±0.010) | 0.920 (±0.001) | 0.648 (±0.010) | 0.901 (±0.002) | 0.631 (±0.011) |
| On Time | 12 h In Advance | 24 h In Advance | 48 h In Advance | |||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| GRU-D++ | 0.807 (±0.015) | 0.350 (±0.026) | 0.827 (±0.018) | 0.380 (±0.031) | 0.826 (±0.035) | 0.398 (±0.054) | 0.848 (±0.016) | 0.445 (±0.030) |
| GRU-D++ (rescale) | 0.895 (±0.032) | 0.534 (±0.078) | 0.892 (±0.022) | 0.538 (±0.044) | 0.889 (±0.017) | 0.539 (±0.049) | 0.864 (±0.017) | 0.514 (±0.023) |
| GRU-D++ (fine-tune) | 0.898 (±0.018) | 0.523 (±0.051) | 0.901 (±0.016) | 0.542 (±0.045) | 0.895 (±0.019) | 0.543 (±0.050) | 0.897 (±0.008) | 0.575 (±0.027) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Kim, T.-H.; Kim, D.; Lee, D.; Kim, D.; Heo, J.; Kang, S.; Ha, T.; Kim, J.; Moon, D.H.; et al. In-Advance Prediction of Pressure Ulcers via Deep-Learning-Based Robust Missing Value Imputation on Real-Time Intensive Care Variables. J. Clin. Med. 2024, 13, 36. https://doi.org/10.3390/jcm13010036
Kim M, Kim T-H, Kim D, Lee D, Kim D, Heo J, Kang S, Ha T, Kim J, Moon DH, et al. In-Advance Prediction of Pressure Ulcers via Deep-Learning-Based Robust Missing Value Imputation on Real-Time Intensive Care Variables. Journal of Clinical Medicine. 2024; 13(1):36. https://doi.org/10.3390/jcm13010036
Chicago/Turabian StyleKim, Minkyu, Tae-Hoon Kim, Dowon Kim, Donghoon Lee, Dohyun Kim, Jeongwon Heo, Seonguk Kang, Taejun Ha, Jinju Kim, Da Hye Moon, and et al. 2024. "In-Advance Prediction of Pressure Ulcers via Deep-Learning-Based Robust Missing Value Imputation on Real-Time Intensive Care Variables" Journal of Clinical Medicine 13, no. 1: 36. https://doi.org/10.3390/jcm13010036
APA StyleKim, M., Kim, T.-H., Kim, D., Lee, D., Kim, D., Heo, J., Kang, S., Ha, T., Kim, J., Moon, D. H., Heo, Y., Kim, W. J., Lee, S.-J., Kim, Y., Park, S. W., Han, S.-S., & Choi, H.-S. (2024). In-Advance Prediction of Pressure Ulcers via Deep-Learning-Based Robust Missing Value Imputation on Real-Time Intensive Care Variables. Journal of Clinical Medicine, 13(1), 36. https://doi.org/10.3390/jcm13010036






