Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Microenvironment in HBV-Related HCC
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Data Acquisition and HBV Infection-Associated FRGs Identification
2.2. Survival Analysis of the FRGs
2.3. Evaluation of the Prognostic Value of the FRGs
2.4. Computation of Immune Cellular Fraction and Prediction of Response to ICB
2.5. Analysis of the Correlations between Gene Expression and Signaling Pathways
2.6. Statistical Analysis
3. Results
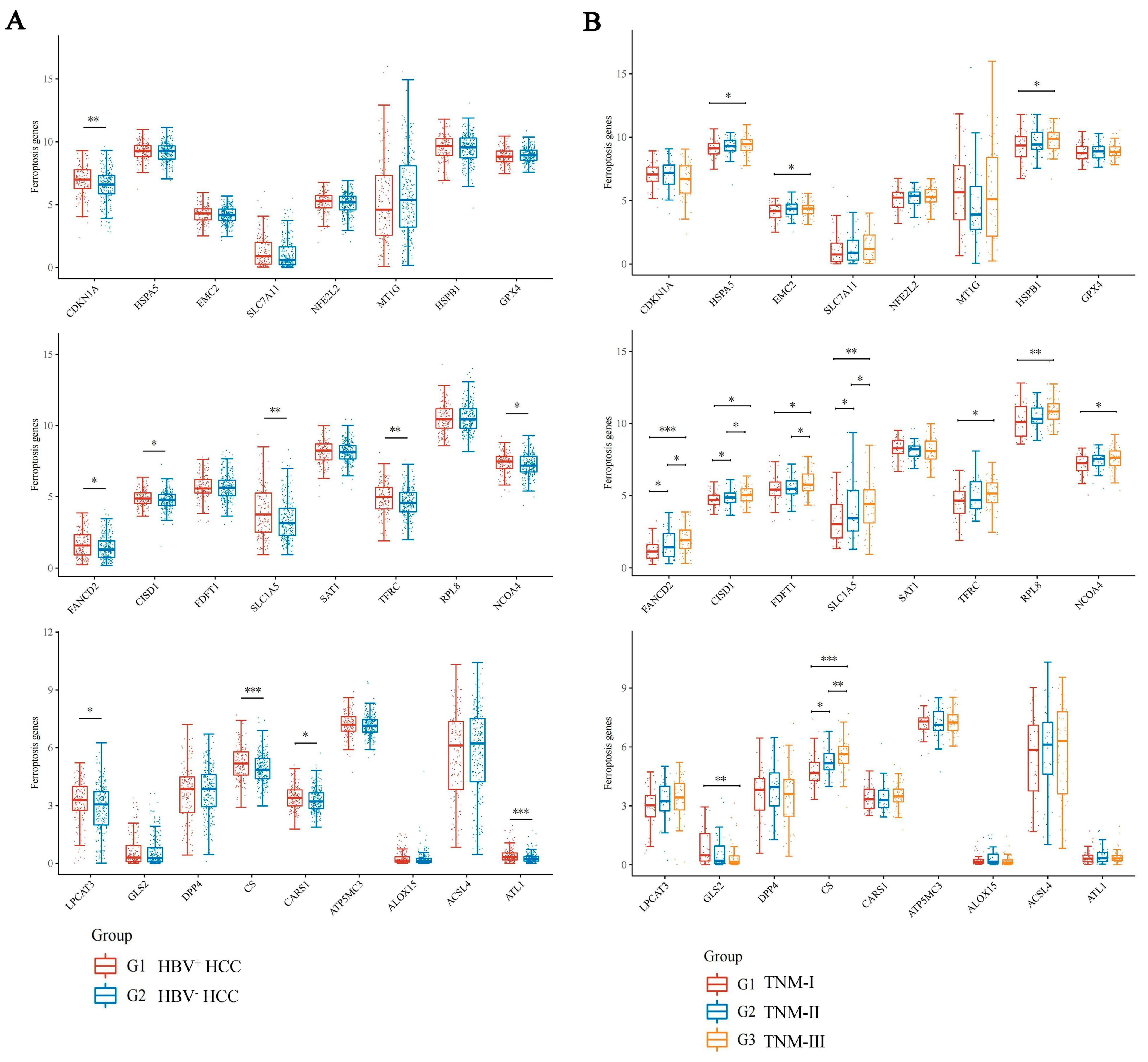
3.1. Screening of Ferroptosis Related Genes Associated with HBV Infection in HCC
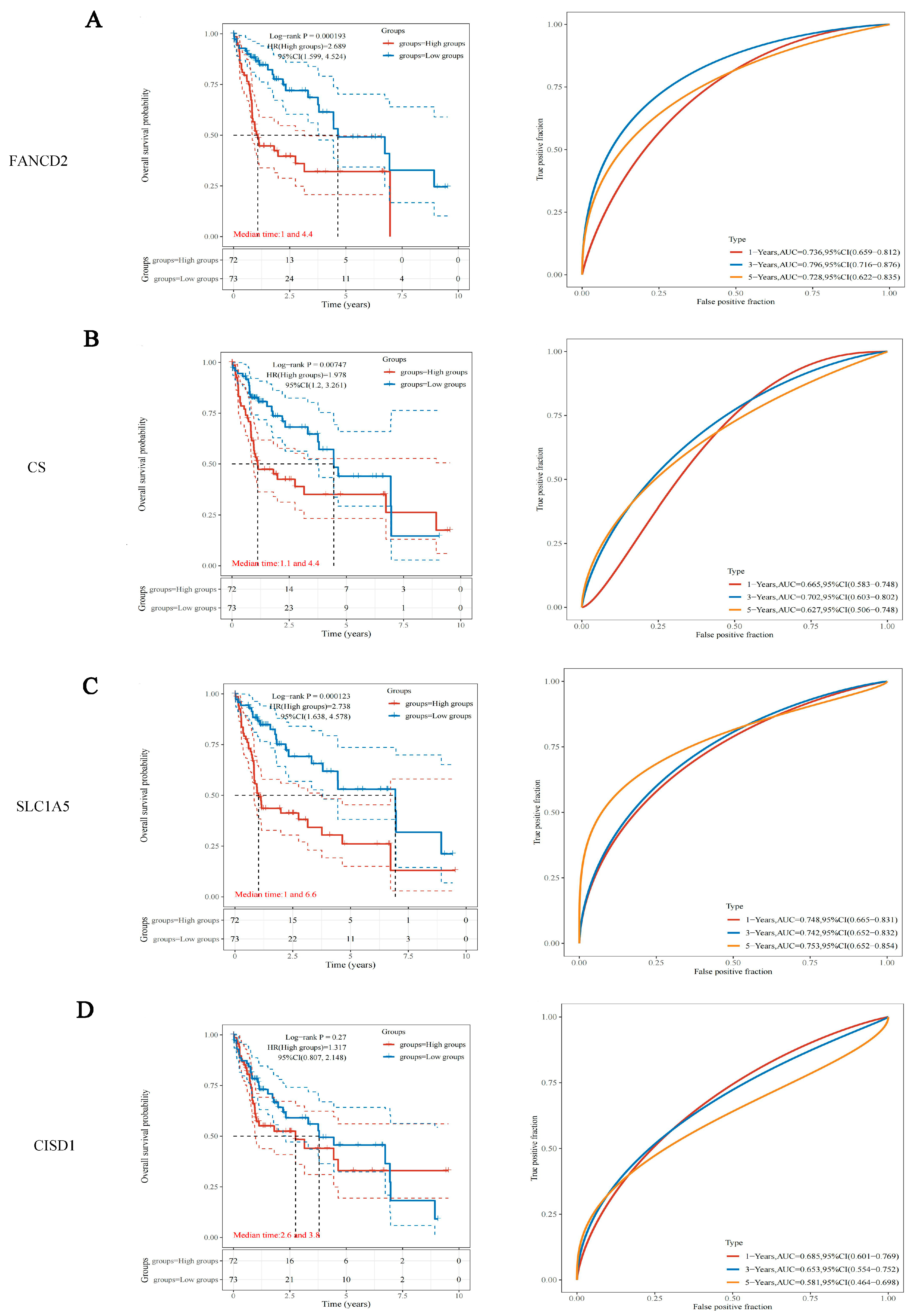
3.2. Survival Analysis of the 4 HBV Infection-Associated FRGs in HBV-Related HCC
3.3. SLC1A5 Combined with Clinicopathological Features of Nomogram Improves Prognosis and Survival Prediction of HBV-Related HCC
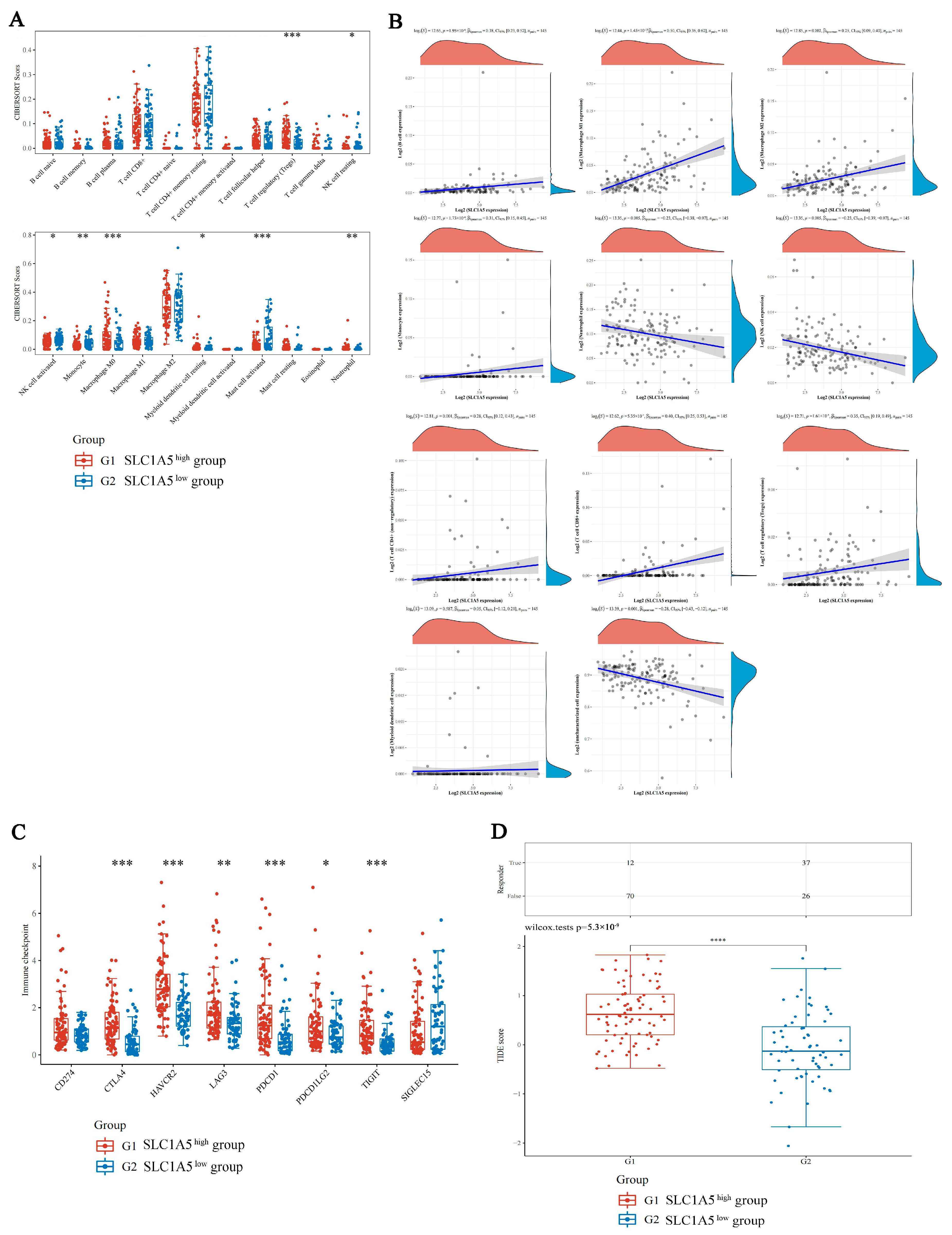
3.4. Comparison of Immune Characteristics and Potential ICB Response Using SLC1A5
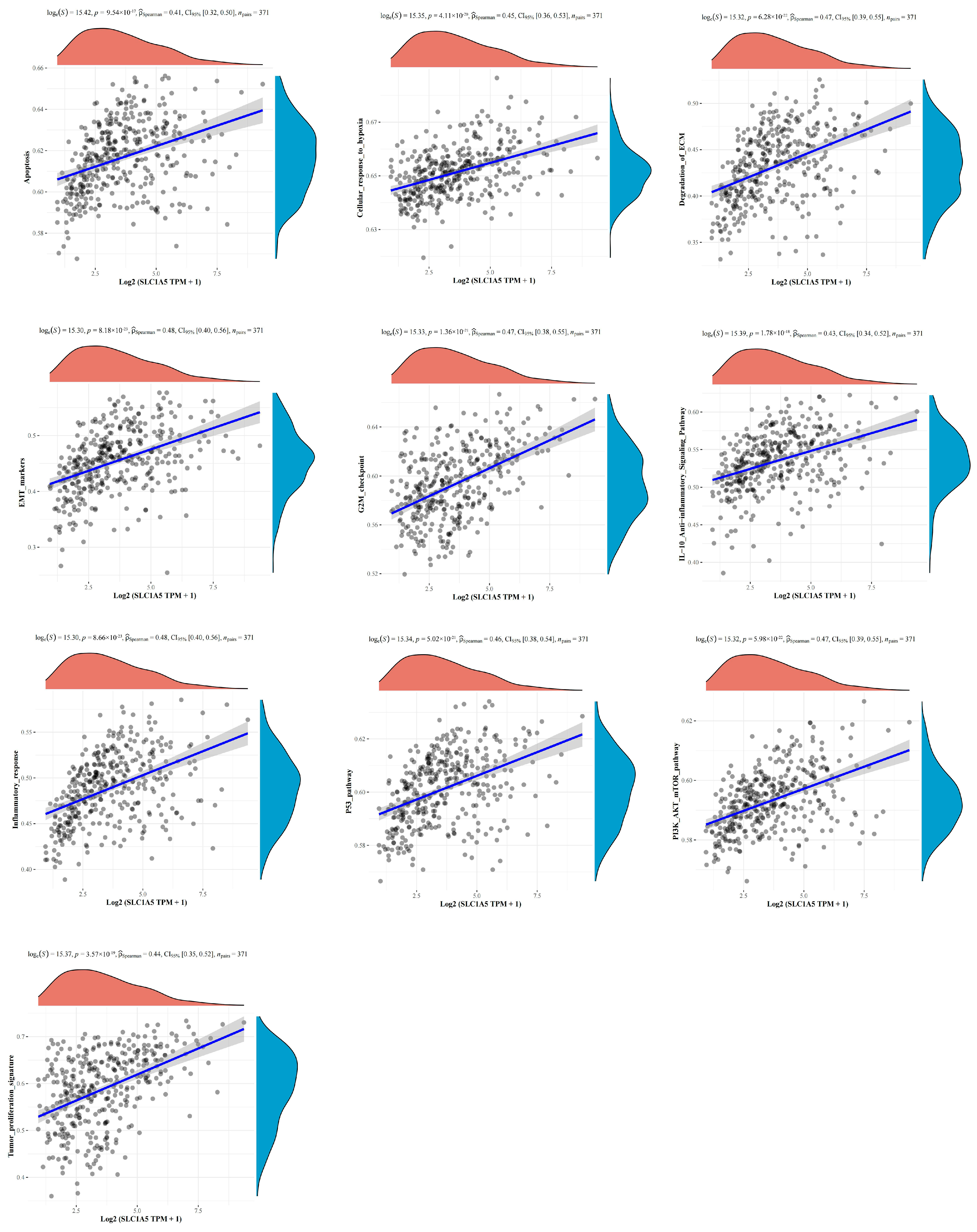
3.5. The Correlations between SLC1A5 and Signaling Pathways in HBV-Related HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular Carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Primer 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Wangensteen, K.J.; Chang, K.-M. Multiple Roles for Hepatitis B and C Viruses and the Host in the Development of Hepatocellular Carcinoma. Hepatol. Baltim. Md 2021, 73 (Suppl. 1), 27–37. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global Epidemiology of NAFLD-Related HCC: Trends, Predictions, Risk Factors and Prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatol. Baltim. Md 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Kar, A.; Samanta, A.; Mukherjee, S.; Barik, S.; Biswas, A. The HBV Web: An Insight into Molecular Interactomes between the Hepatitis B Virus and Its Host En Route to Hepatocellular Carcinoma. J. Med. Virol. 2023, 95, e28436. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Ajoolabady, A.; Tang, D.; Kroemer, G.; Ren, J. Ferroptosis in Hepatocellular Carcinoma: Mechanisms and Targeted Therapy. Br. J. Cancer 2022, 128, 190–205. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Jiang, R.; Xue, R.; Yin, X.; Wu, M.; Meng, Q. Ferroptosis in Liver Disease: New Insights into Disease Mechanisms. Cell Death Discov. 2021, 7, 276. [Google Scholar] [CrossRef]
- Bekric, D.; Ocker, M.; Mayr, C.; Stintzing, S.; Ritter, M.; Kiesslich, T.; Neureiter, D. Ferroptosis in Hepatocellular Carcinoma: Mechanisms, Drug Targets and Approaches to Clinical Translation. Cancers 2022, 14, 1826. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Lin, B.; Zhou, M.; Wu, L.; Zheng, T. Role of Ferroptosis in Hepatocellular Carcinoma. J. Cancer Res. Clin. Oncol. 2018, 144, 2329–2337. [Google Scholar] [CrossRef]
- Liu, G.-Z.; Xu, X.-W.; Tao, S.-H.; Gao, M.-J.; Hou, Z.-H. HBx Facilitates Ferroptosis in Acute Liver Failure via EZH2 Mediated SLC7A11 Suppression. J. Biomed. Sci. 2021, 28, 67. [Google Scholar] [CrossRef] [PubMed]
- Hassanein, M.; Hoeksema, M.D.; Shiota, M.; Qian, J.; Harris, B.K.; Chen, H.; Clark, J.E.; Alborn, W.E.; Eisenberg, R.; Massion, P.P. SLC1A5 Mediates Glutamine Transport Required for Lung Cancer Cell Growth and Survival. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Han, M.; Zhang, X.; He, Y.; Yuan, C.; Xiong, Y.; Li, X.; Zeng, C.; Lu, K.; Zhu, H.; et al. Discoidin Domain Receptor 1 Promotes Hepatocellular Carcinoma Progression through Modulation of SLC1A5 and the MTORC1 Signaling Pathway. Cell. Oncol. Dordr. 2022, 45, 163–178. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, D.; Hong, M.; Liu, J.; Li, Y.; Hao, J.; Lu, L.; Yin, Z.; Wu, Y. Apoptosis, Pyroptosis, and Ferroptosis Conspiringly Induce Immunosuppressive Hepatocellular Carcinoma Microenvironment and Γδ T-Cell Imbalance. Front. Immunol. 2022, 13, 845974. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. Ferroptosis Turns 10: Emerging Mechanisms, Physiological Functions, and Therapeutic Applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef]
- Zhou, N.; Bao, J. FerrDb: A Manually Curated Resource for Regulators and Markers of Ferroptosis and Ferroptosis-Disease Associations. Database J. Biol. Databases Curation 2020, 2020, baaa021. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, Q.; Zuo, Z.-X.; Yuan, S.-Q.; Yu, K.; Zhang, Q.; Zhang, X.; Sheng, H.; Ju, H.-Q.; Cheng, H.; et al. Systematic Analysis of the Aberrances and Functional Implications of Ferroptosis in Cancer. iScience 2020, 23, 101302. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Cao, J.Y.; Dixon, S.J. Mechanisms of Ferroptosis. Cell. Mol. Life Sci. CMLS 2016, 73, 2195–2209. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yang, T.; Peng, Z.; Xiao, H.; Jiang, N.; Zhang, L.; Ca, D.; Wu, P.; Pan, Q. SLC1A5 Silencing Inhibits Esophageal Cancer Growth via Cell Cycle Arrest and Apoptosis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 48, 397. [Google Scholar] [CrossRef]
- Luo, M.; Wu, L.; Zhang, K.; Wang, H.; Zhang, T.; Gutierrez, L.; O’Connell, D.; Zhang, P.; Li, Y.; Gao, T.; et al. MiR-137 Regulates Ferroptosis by Targeting Glutamine Transporter SLC1A5 in Melanoma. Cell Death Differ. 2018, 25, 1457–1472. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; Santos-Jiménez, J.D.L.; Márquez, J. Dysregulation of Glutaminase and Glutamine Synthetase in Cancer. Cancer Lett. 2019, 467, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, Z.; Tu, M.; Meng, W.; Gao, H.; Li, M.D.; Li, L. Correlation Between Prognostic Biomarker SLC1A5 and Immune Infiltrates in Various Types of Cancers Including Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 608641. [Google Scholar] [CrossRef]
- van Geldermalsen, M.; Wang, Q.; Nagarajah, R.; Marshall, A.D.; Thoeng, A.; Gao, D.; Ritchie, W.; Feng, Y.; Bailey, C.G.; Deng, N.; et al. ASCT2/SLC1A5 Controls Glutamine Uptake and Tumour Growth in Triple-Negative Basal-like Breast Cancer. Oncogene 2016, 35, 3201–3208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, R.; Shuai, Y.; Huang, Y.; Jin, R.; Wang, X.; Luo, J. ASCT2 (SLC1A5)-Dependent Glutamine Uptake Is Involved in the Progression of Head and Neck Squamous Cell Carcinoma. Br. J. Cancer 2020, 122, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhou, J.; Li, L.; Wu, X.; Shi, Y.; Cui, W.; Zhang, S.; Hu, Q.; Wang, J.; Bai, H.; et al. SLC1A5 Enhances Malignant Phenotypes through Modulating Ferroptosis Status and Immune Microenvironment in Glioma. Cell Death Dis. 2022, 13, 1071. [Google Scholar] [CrossRef]
- Xu, F.; Wang, H.; Pei, H.; Zhang, Z.; Liu, L.; Tang, L.; Wang, S.; Ren, B.-C. SLC1A5 Prefers to Play as an Accomplice Rather Than an Opponent in Pancreatic Adenocarcinoma. Front. Cell Dev. Biol. 2022, 10, 800925. [Google Scholar] [CrossRef]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.-X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.-L. Metabolism. Differential Regulation of MTORC1 by Leucine and Glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; You, M.; Fu, J.; Tian, M.; Zhong, X.; Du, C.; Hong, Z.; Zhu, Z.; Liu, J.; Markowitz, G.J.; et al. Intratumoral Stem-like CCR4+ Regulatory T Cells Orchestrate the Immunosuppressive Microenvironment in HCC Associated with Hepatitis B. J. Hepatol. 2022, 76, 148–159. [Google Scholar] [CrossRef] [PubMed]
- van Buuren, N.; Ramirez, R.; Turner, S.; Chen, D.; Suri, V.; Aggarwal, A.; Moon, C.; Kim, S.; Kornyeyev, D.; Bui, N.; et al. Characterization of the Liver Immune Microenvironment in Liver Biopsies from Patients with Chronic HBV Infection. JHEP Rep. Innov. Hepatol. 2022, 4, 100388. [Google Scholar] [CrossRef] [PubMed]
- Friedmann Angeli, J.P.; Krysko, D.V.; Conrad, M. Ferroptosis at the Crossroads of Cancer-Acquired Drug Resistance and Immune Evasion. Nat. Rev. Cancer 2019, 19, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Sun, S.; Johnson, T.; Qi, R.; Zhang, S.; Zhang, J.; Yang, K. The Glutathione Peroxidase Gpx4 Prevents Lipid Peroxidation and Ferroptosis to Sustain Treg Cell Activation and Suppression of Antitumor Immunity. Cell Rep. 2021, 35, 109235. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Sena, L.A.; Chandel, N.S. Mitochondria in the Regulation of Innate and Adaptive Immunity. Immunity 2015, 42, 406–417. [Google Scholar] [CrossRef]
- Ma, X.; Xiao, L.; Liu, L.; Ye, L.; Su, P.; Bi, E.; Wang, Q.; Yang, M.; Qian, J.; Yi, Q. CD36-Mediated Ferroptosis Dampens Intratumoral CD8+ T Cell Effector Function and Impairs Their Antitumor Ability. Cell Metab. 2021, 33, 1001–1012.e5. [Google Scholar] [CrossRef]
- Xu, S.; Chaudhary, O.; Rodríguez-Morales, P.; Sun, X.; Chen, D.; Zappasodi, R.; Xu, Z.; Pinto, A.F.M.; Williams, A.; Schulze, I.; et al. Uptake of Oxidized Lipids by the Scavenger Receptor CD36 Promotes Lipid Peroxidation and Dysfunction in CD8+ T Cells in Tumors. Immunity 2021, 54, 1561–1577.e7. [Google Scholar] [CrossRef]
- Rotte, A. Combination of CTLA-4 and PD-1 Blockers for Treatment of Cancer. J. Exp. Clin. Cancer Res. CR 2019, 38, 255. [Google Scholar] [CrossRef]
- Zhu, D.; Wu, S.; Li, Y.; Zhang, Y.; Chen, J.; Ma, J.; Cao, L.; Lyu, Z.; Hou, T. Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Infiltrates in Stomach Adenocarcinoma. Cancer Cell Int. 2022, 22, 124. [Google Scholar] [CrossRef]
- Xue, R.; Zhang, Q.; Cao, Q.; Kong, R.; Xiang, X.; Liu, H.; Feng, M.; Wang, F.; Cheng, J.; Li, Z.; et al. Liver Tumour Immune Microenvironment Subtypes and Neutrophil Heterogeneity. Nature 2022, 612, 141–147. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-Associated Macrophages in Tumor Metastasis: Biological Roles and Clinical Therapeutic Applications. J. Hematol. Oncol. J. Hematol Oncol 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Franco, F.; Ho, P.-C. Metabolic Regulation of Tregs in Cancer: Opportunities for Immunotherapy. Trends Cancer 2017, 3, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Somasundaram, A.; Bruno, T.C.; Workman, C.J.; Vignali, D.A.A. Therapeutic Targeting of Regulatory T Cells in Cancer. Trends Cancer 2022, 8, 944–961. [Google Scholar] [CrossRef]
- Yan, Y.; Huang, L.; Liu, Y.; Yi, M.; Chu, Q.; Jiao, D.; Wu, K. Metabolic Profiles of Regulatory T Cells and Their Adaptations to the Tumor Microenvironment: Implications for Antitumor Immunity. J. Hematol. Oncol. J. Hematol Oncol 2022, 15, 104. [Google Scholar] [CrossRef]
- Giannini, E.G.; Aglitti, A.; Borzio, M.; Gambato, M.; Guarino, M.; Iavarone, M.; Lai, Q.; Levi Sandri, G.B.; Melandro, F.; Morisco, F.; et al. Overview of Immune Checkpoint Inhibitors Therapy for Hepatocellular Carcinoma, and The ITA.LI.CA Cohort Derived Estimate of Amenability Rate to Immune Checkpoint Inhibitors in Clinical Practice. Cancers 2019, 11, 1689. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.; Liu, Y.; Huang, J. Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Microenvironment in HBV-Related HCC. J. Clin. Med. 2023, 12, 1715. https://doi.org/10.3390/jcm12051715
Su H, Liu Y, Huang J. Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Microenvironment in HBV-Related HCC. Journal of Clinical Medicine. 2023; 12(5):1715. https://doi.org/10.3390/jcm12051715
Chicago/Turabian StyleSu, Hanwen, Youyi Liu, and Jingtao Huang. 2023. "Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Microenvironment in HBV-Related HCC" Journal of Clinical Medicine 12, no. 5: 1715. https://doi.org/10.3390/jcm12051715
APA StyleSu, H., Liu, Y., & Huang, J. (2023). Ferroptosis-Related Gene SLC1A5 Is a Novel Prognostic Biomarker and Correlates with Immune Microenvironment in HBV-Related HCC. Journal of Clinical Medicine, 12(5), 1715. https://doi.org/10.3390/jcm12051715




