Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review
Abstract
:1. Introduction
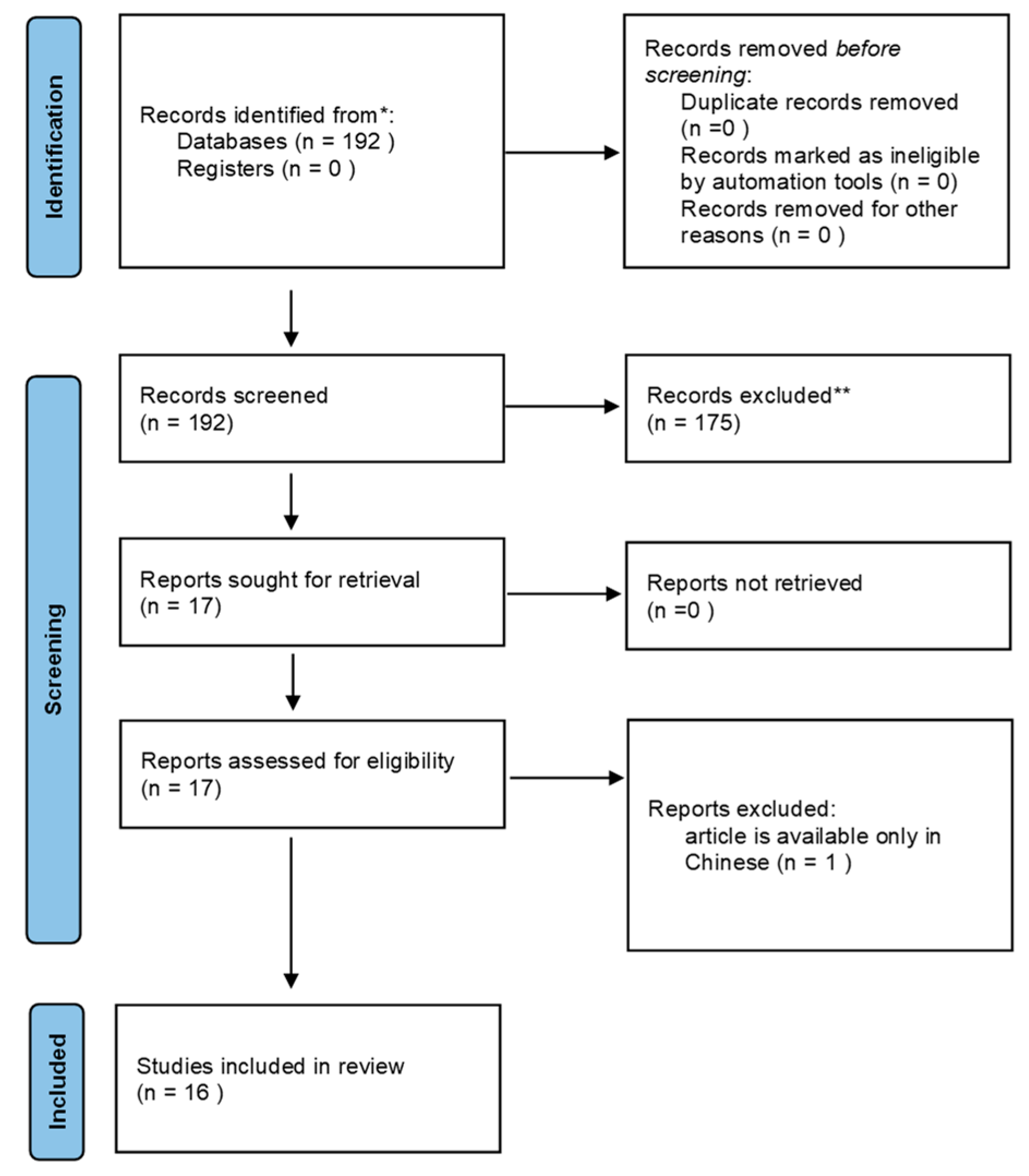
2. Methodology
3. Results
3.1. mpMRI
3.2. CT and CT/PET
| Title | Authors | Publication Year | Journal | Type | Number of Patients | Imaging Techniques | AIM | SEGM | AI | RQS | Best Results | Conclusions |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Preoperative prediction of pelvic lymph nodes metastasis in prostate cancer using an ADC-based radiomics model: comparison with clinical nomograms and PI-RADS assessment. | Liu X. et al. [17] | 2022 | Abdom Radiol (NY) | Retrospective | 474 (between 2017 and 2020). Another cohort of 128 Between 2020 and 2021. | 3T mpMRI | develop and test radiomics models based on manually corrected or automatically gained masks on ADC maps for PLNM prediction in Pca patients | 2d–3d U-Net automatic | AUC and DCA for comparison between the optimal radiomics model and MSKCC, Briganti 2017 nomograms, and PI-RADS assessment | 16 | The radiomics model based on the mask of automatically segmented prostate obtained the highest AUC among the four radiomics models (0.73 vs. 0.63 vs. 0.70 vs. 0.56). Briganti 2017, MSKCC nomograms, and PI-RADS assessment-yielded AUCs of 0.69, 0.71, and 0.70, respectively, and no significant differences were found compared with the optimal radiomics model (p = 0.605–0.955). | The radiomics model based on the mask of automatically segmented prostate offers a non-invasive method to predict PLNM for patients with PCa. It shows comparable accuracy to the current MKSCC and Briganti nomograms. |
| Utility of diffusion weighted imaging-based radiomics nomogram to predict pelvic lymph nodes metastasis in prostate cancer. | Liu X. et al. [18] | 2022 | BMC Med Imaging | Retrospective | 84 | 3T mpMRI | DWI-based radiomics for preoperative PLNM prediction in PCa patients | 3d U-Net of LN in DWI | Two preoperative PLNM prediction models with quantitative radiological LN features alone (Model 1) and combining radiological and radiomics features (Model 2) via multiple logistic regression. The visual assessments of junior (Model 3) and senior (Model 4) radiologists were compared. C-index of the nomogram analysis and DCA were used to evaluate performance and clinical usefulness. | 17 | No significant difference between AUCs of Models 1 and 2 (0.89 vs. 0.90; p = 0.573) in the held-out cohort. Model 2 showed the highest AUC (0.83, 95% CI 0.76, 0.89) for PLNM prediction in the LN subgroup with a short diameter ≤ 10 mm compared with Model 1 (0.78, 95% CI 0.70, 0.84), Model 3 (0.66, 95% CI 0.52, 0.77), and Model 4 (0.74, 95% CI 0.66, 0.88). The nomograms of Models 1 and 2 yielded C-index values of 0.804 and 0.910, respectively, in the held-out cohort. The C-index of the nomogram analysis (0.91) and decision curve analysis (DCA) curves confirmed the clinical usefulness and benefit of Model 2. | A DWI-based radiomics nomogram incorporating the LN radiomics signature with quantitative radiological features is promising for PLNM prediction in PCa patients, particularly for normal-sized LNM |
| Multiparametric MRI-based radiomics model to predict pelvic lymph node invasion for patients with prostate cancer | Zheng H. et al. [19] | 2022 | Eur Radiol. | Retrospective | 244 | 3T mpMRI | predicting lymph node invasion (LNI), via a radiomics-based machine learning approach. | manual segm of index lesion on T2w seq and ADC map | An integrative radiomics model (IRM) (SVM) performance was measured by AUC, sensitivity, specificity, NPV, and PPV. | 16 | Overall, 17 (10.6%) and 14 (16.7%) patients with LNI were included in training/validation set and testing set, respectively. Shape and first-order radiomics features showed usefulness in building the IRM (integrative radiomics model). The proposed IRM achieved an AUC of 0.915 (95% CI: 0.846–0.984) in the testing set, superior to pre-existing nomograms whose AUCs were from 0.698 to 0.724 (p < 0.05). | The proposed IRM could be potentially feasible to predict the risk of having LNI for patients with PCa. With the improved predictability, it could be utilized to assess which patients with PCa could safely avoid ePLND, thus reducing the number of unnecessary ePLND |
| Development of a Radiomic-Based Model Predicting Lymph Node Involvement in Prostate Cancer Patients | Bourbonne V. et al. [20] | 2021 | Cancers (Basel) | Retrospective | 280 | 3T e 1.5 T MRI | develop and internally validate a novel LNI risk prediction model based on radiomic features extracted from preoperative multimodal MRI | The index lesion was a semi-automatic 3D Slicer. | Radiomic features were extracted from the tumor volumes in ADC map and T2 seq (ComBat harmonization method for inter-site heterogeneity). A prediction model was trained using a neural network approach (Multilayer Perceptron Network) combining clinical, radiomic and all features. AUC and the C-Index for performance | 17 | The proposed model resulted in a C-Index of 0.89 in the testing set, which was higher than currently available models such as Partin, Roach, Yale, MSKCC, and Briganti 2012 and 2017. | The study suggests that radiomic features extracted from preoperative MRI scans combined with clinical features through a neural network could improve LNI risk prediction in PCa. |
| Radiomics-based prognosis classification for high-risk prostate cancer treated with radiotherapy | Franzese C. et al. [25] | 2022 | Strahlenther Onkol | Retrospective | 157 | Non-contrast CT | risk of metastatic progression in HR Pca patients after RT and ADT, considering MFS | Manual of gland only, prostate gland + seminal vesicles, seminal vesicles only. | 5 clinical features + 62 radiomics features were combined using R platform | 13 | Radiomic features were able to predict the risk of metastatic progression in high-risk prostate cancer. Combining the radiomic features and clinical characteristics can classify high-risk patients into favorable and unfavorable prognostic groups | |
| A CT-based radiomics model to detect prostate cancer lymph node metastases in PSMA radioguided surgery patients | Peeken JC. et al. [26] | 2020 | Eur J Nucl Med Mol Imaging | Retrospective | 80 | Contrast-CT | CT-based radiomic model to predict LNM status using a PSMA radioguided surgery (RGS) cohort with histological confirmation of all suspected lymph nodes (LNs). | LN segmentation was conducted manually using Eclipse 13.0 on the contrast-enhanced diagnostic CT. | 156 radiomic features analyzing texture, shape, intensity, and LBP were extracted. Least absolute shrinkage and selection operator (radiomic models) and logistic regression (conventional parameters) were used for modeling | 19 | A combined radiomic model achieved the best predictive performance with a testing AUC of 0.95 | The best radiomic model outperformed conventional measures for detection of LNM demonstrating an incremental value of radiomic features. |
| [18F]FMCH PET/CT biomarkers and similarity analysis to refine the definition of oligometastatic prostate cancer | Sollini M. et al. [32] | 2021 | EJNMMI Res. | Prospective | 92 | [18F]FMCH PET/CT | evaluate [18F]FMCH PET/CT radiomic analysis in patients with recurrent PCa after primary radical therapy, testing intra-patient lesions similarity in oligometastatic and plurimetastatic PCa, comparing the two most used definitions of oligometastatic disease | Lesions were semi-automatically segmented by the PET VCAR software (GE Healthcare, Waukesha, WI, USA) on a General Electric workstation | supervised and supervised clustering of patients for identifying the prevalence of metastasis | 15 | They found a comparable heterogeneity between patients with up to five lesions and plurimetastatic patients, while patients with up to three lesions were less heterogeneous than plurimetastatic patients, featuring different cell phenotypes in the two groups. Our results supported the use of a 3-lesion threshold to define oligometastatic PCa. | |
| Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate- and high-risk prostate cancer–a comparison study with histology reference | Zamboglou C. et al. [27] | 2019 | Theranostic | Prospective | 20 | PSMA PET | tumor discrimination and non-invasive characterization of Gleason score (GS) and pelvic lymph node status. | manually created segmentations of the intraprostatic tumor volume | Coregistered histopathological gross tumor volume. 133 radiomics feat from GTV-Histo of Pca and non-Pca. AUC evaluated the discrimination of Gleason and LN involvement | 23 | The texture feature QSZHGE was a statistically significant (p < 0.01) predictor for PCa patients with GS ≥ 8 tumors and pN1 status. | Radiomic features derived from PSMA PET discriminated between PCa and non-PCa tissue within the prostate. Additionally, the texture feature QSZHGE discriminated between GS 7 and GS ≥8 tumors and between patients with pN1 and pN0 disease. |
| Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer | Cysouw MCF et al. [28] | 2021 | Eur J Nucl Med Mol Imaging | Prospective | 76 | [18F]DCFPyL PET-CT | ability of ML-based analysis of quantitative [18F]DCFPyL PET metrics to predict metastatic disease or high-risk pathological tumor features | primary tumors were delineated using 50–70% peak isocontour thresholds on images with and without partial-volume correction | 408 extracted feat analyzed with RF; Gleason, LNI, mets, ECE. Models were trained with PET feat alone and radiomics ones | 24 | The radiomics-based machine learning models predicted LNI (AUC 0.86 ± 0.15, p < 0.01), nodal or distant metastasis (AUC 0.86 ± 0.14, p < 0.01), Gleason score (0.81 ± 0.16, p < 0.01), and ECE (0.76 ± 0.12, p < 0.01). The highest AUCs reached using standard PET metrics were lower than those of radiomics-based models. | Machine-learning-based analysis of quantitative [18F]DCFPyL PET metrics can predict LNI and high-risk pathological tumor features in primary PCa patients. These findings indicate that PSMA expression detected on PET is related to both primary tumor histopathology and metastatic tendency. |
| A machine learning-assisted decision-support model to better identify patients with prostate cancer requiring an extended pelvic lymph node dissection | Hou Y. et al. [23] | 2019 | BJU Int. | Retrospective | 248 | 3T mpMRI | develop an ML-assisted model for identifying the candidates for ePLND in PCa by integrating clinical, biopsy, and precisely defined MRI findings | No radiomic features | 18 integrated features using LR, SVM, and RF models were compared to an MSKCC nomogram. Performance evaluated with AUC and DCA | 7 | The predictive accuracy of ML-based models, with or without MRI-reported LNI, yielded similar AUCs, which were higher than the MSKCC nomogram (0.816; p-value < 0.001). | ML-based model below 15% cutoff, superior to MSKCC nomogram, allows up to 50% or more ePLNDs spared at the cost of missing < 3% LNIs. |
| Integration of clinicopathologic identification and deep transferrable image feature representation improves predictions of lymph node metastasis in prostate cancer | Hou Y et al. [24] | 2021 | EBioMedicine | Retrospective | 401 | 3TmpMRI | PLNM-Risk calculator to obtain a precisely informed decision about whether to perform extended pelvic lymph node dissection | PCa lesions manually segmented | set of radiologists’ interpretations, clinicopathological factors and newly refined imaging indicators from MR images with radiomics machine learning and deep transfer learning algorithms. Its clinical applicability was compared with Briganti and Memorial Sloan Kettering Cancer Center (MSKCC) nomograms. | 14 | PLNM-Risk achieved good diagnostic discrimination with areas under the receiver operating characteristic curve (AUCs) of 0.93 (95% CI, 0.90–0.96), 0.92 (95% CI, 0.84–0.97) and 0.76 (95% CI, 0.62–0.87) in the training/validation, internal test and external test cohorts, respectively. | PLNM-Risk calculator offers a noninvasive clinical biomarker to predict PLNM for patients with PCa. |
| Freely Available, Fully Automated AI-Based Analysis of Primary Tumour and Metastases of Prostate Cancer in Whole-Body [18F]-PSMA-1007 PET-CT. | Trägårdh E et al. [29] | 2022 | Diagnostics (Basel) | Retrospective | 660 | [18F]PSMA PET-CT | develop and validate a fully automated AI- based method for detection and quantification of suspected prostate tumor/local recurrence, LN, and bone mets from [18F]PSMA-1007 PET-CT | manual | evaluation of total lesion volume and uptake by CNN vs. nuclear medicine physician | 7 | sensitivity of the AI method: 79% for detecting prostate tumor/recurrence, 79% for lymph node metastases, and 62% for bone metastases. On average, nuclear medicine physicians’ corresponding sensitivities were 78%, 78%, and 59%, respectively. | The development of an AI-based method for prostate cancer detection with sensitivity on par with nuclear medicine physicians was possible. |
| Semiautomated pelvic lymph node treatment response evaluation for patients with advanced prostate cancer: based on MET-RADS-P guidelines | Liu X et al. [20] | 2023 | Cancer Imaging. | Retrospective | 162 | 3T mpMRI | Deep-learning-based algorithm for semiautomated treatment response assessment of pelvic lymph nodes. | Automated segmentation of pelvic lymph nodes. | The performance of the deep learning algorithm was evaluated using the Dice similarity coefficient (DSC) and volumetric similarity (VS). Kappa statistics were used to assess the accuracy and consistency of the treatment response assessment by the deep learning model and two radiologists | 8 | The accuracies of automated segmentation-based assessment were 0.92 (95% CI: 0.85–0.96), 0.91 (95% CI: 0.86–0.95), and 75% (95% CI: 0.46–0.92) for target lesions, nontarget lesions, and nonpathological lesions, respectively | The deep-learning-based semiautomated algorithm showed high accuracy for the treatment response assessment of pelvic lymph nodes and demonstrated comparable performance with radiologists. |
| Freely available artificial intelligence for pelvic lymph node metastases in PSMA PET-CT that performs on par with nuclear medicine physicians | Trägårdh E et al. [30] | 2022 | Eur J Nucl Med Mol Imaging | Retrospective | 211 | [18F]PSMA-1007 PET-CT | develop and validate a CNN for detection of PLNM in [18F]PSMA-1007 PET-CT from patients with high-risk PCa. | manual by 3 readers | Suspected pelvic lymph node metastases were marked by three independent readers vs. CNN | 9 | Sensitivity of the AI method for detecting pelvic lymph node metastases was 82%, and the corresponding sensitivity for the human readers was 77% on average. | This study shows that AI can obtain a sensitivity on par with that of physicians with a reasonable number of false positives. |
| Development and validation of the 3D U-Net algorithm for segmentation of pelvic lymph nodes on diffusion-weighted images | Liu X. et al. [21] | 2021 | BMC Med Imaging | Retrospective | 393 | 3T mpMRI | evaluate the feasibility of the 3D U-Net algorithm for automated detection and segmentation of LNs on DWI | manual | Segmentation performance was assessed using the Dice score, positive predictive value (PPV), true positive rate (TPR), volumetric similarity (VS), Hausdorff distance (HD), the Average distance (AVD), and the Mahalanobis distance (MHD) with manual annotation of pelvic LNs as the reference. The accuracy with which the suspicious metastatic LNs (short diameter > 0.8 cm) were detected was evaluated using the area under the curve (AUC) at the patient level, and the precision, recall, and F1-score were determined at the lesion level. | 11 | The precision, recall, and F1-score for the detection of suspicious LNs were 0.97, 0.98, and 0.97, respectively. In the temporal validation dataset, the AUC of the model for identifying PCa patients with suspicious LNs was 0.963 (95% CI: 0.892–0.993). | The 3D U-Net algorithm can accurately detect and segment pelvic LNs based on DWI images. |
| Artificial intelligence-based detection of lymph node metastases by PET/CT predicts prostate cancer-specific survival. | Borrelli P. et al. [31] | 2021 | Clin Physiol Funct Imaging | Retrospective | 399 | PET/CT | developing and validating an AI-based tool for detection of lymph node lesions on PET/CT | automatically by Organ CNN | The tool consisted of CNN using complete PET/CT scans as inputs. In the test set, the AI-based LN detections were compared to those of two independent readers. The association with PCa-specific survival was investigated. | 7 | The AI-based tool detected more lymph node lesions than Reader B (98 vs. 87/117; p = 0.045) using Reader A as reference. AI-based tool and Reader A showed similar performance (90 vs. 87/111; p = 0.63) using Reader B as reference. | This study shows the feasibility of using an AI-based tool for automated and objective interpretation of PET/CT images that can provide assessments of lymph node lesions comparable with that of experienced readers, and prognostic information in PCa patients. |
| Title | Authors | Journal | Machine Learning Algorithms and Methods |
|---|---|---|---|
| Preoperative prediction of pelvic lymph nodes metastasis in prostate cancer using an ADC-based radiomics model: comparison with clinical nomograms and PI-RADS assessment. | Liu X. et al. [17] | Abdom Radiol (NY) |
|
| Utility of diffusion-weighted imaging-based radiomics nomogram to predict pelvic lymph node metastasis in prostate cancer. | Liu X. et al. [18] | BMC Med Imaging |
|
| Multiparametric MRI-based radiomics model to predict pelvic lymph node invasion for patients with prostate cancer | Zheng H. et al. [19] | Eur Radiol. |
|
| Development of a Radiomic-Based Model Predicting Lymph Node Involvement in Prostate Cancer Patients | Bourbonne V. et al. [20] | Cancers (Basel) |
|
| Radiomics-based prognosis classification for high-risk prostate cancer treated with radiotherapy | Franzese C. et al. [25] | Strahlenther Onkol |
A homemade radiomics model (type not specified) based on features extracted using the software package LifeX |
| A CT-based radiomics model to detect prostate cancer lymph node metastases in PSMA radioguided surgery patients | Peeken JC. et al. [26] | Eur J Nucl Med Mol Imaging | Two different modeling approaches:
|
| [18F]FMCH PET/CT biomarkers and similarity analysis to refine the definition of oligometastatic prostate cancer | Sollini M. et al. [32] | EJNMMI Res. | Did not provide explicit details about specific machine learning algorithms used for predictive modeling. It primarily discusses radiomics feature extraction, univariate analysis, similarity analysis, and unsupervised clustering. |
| Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate- and high-risk prostate cancer–a comparison study with histology reference | Zamboglou C. et al. [27] | Theranostic | Extraction of radiomic features from PET/CT images. The machine learning algorithms used are not specified |
| Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer | Cysouw MCF et al. [28] | Eur J Nucl Med Mol Imaging |
|
| A machine-learning-assisted decision-support model to better identify patients with prostate cancer requiring an extended pelvic lymph node dissection | Hou Y. et al. [23] | BJU Int. |
|
| Integration of clinicopathologic identification and deep transferrable image feature representation improves predictions of lymph node metastasis in prostate cancer | Hou Y et al. [24] | EBioMedicine |
|
| Freely Available, Fully Automated AI-Based Analysis of Primary Tumour and Metastases of Prostate Cancer in Whole-Body [18F]-PSMA-1007 PET-CT. | Trägårdh E et al. [29] | Diagnostics (Basel) |
|
| Semiautomated pelvic lymph node treatment response evaluation for patients with advanced prostate cancer: based on MET-RADS-P guidelines | Liu X et al. [20] | Cancer Imaging. |
|
| Freely available artificial intelligence for pelvic lymph node metastases in PSMA PET-CT that performs on par with nuclear medicine physicians | Trägårdh E et al. [30] | Eur J Nucl Med Mol Imaging |
|
| Development and validation of the 3D U-Net algorithm for segmentation of pelvic lymph nodes on diffusion-weighted images | Liu X. et al. [21] | BMC Med Imaging |
|
| Artificial-intelligence-based detection of lymph node metastases by PET/CT predicts prostate cancer-specific survival. | Borrelli P. et al. [31] | Clin Physiol Funct Imaging |
|
4. Discussion
- -
- Limited number of articles and retrospective design: the low number of articles analyzing LNI in PCa, coupled with the majority being retrospective in design, poses limitations in terms of sample size and potential biases in data collection.
- -
- Variability and lack of standardization: There is a lack of standardized approaches in various aspects, including the software used for analysis, as well as the type and number of features analyzed. This variability hinders the comparability and reproducibility of results. However, this extreme variability also indicates how many aspects are susceptible to evaluation and how much room there is for growth.
- -
- Manual segmentation and reproducibility: the reliance on manual segmentation for feature extraction introduces subjectivity and variability, leading to reduced reproducibility of results. The lack of standardized segmentation methods hampers the comparability of findings across different studies.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Fleischmann, A.; Boormans, J.L.; Fassan, M.; Nottegar, A.; Lucato, P.; Stubbs, B.; Solmi, M.; Porcaro, A.; Veronese, N.; et al. Extranodal extension of lymph node metastasis influences recurrence in prostate cancer: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 2374. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA. Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Hricak, H.; Choyke, P.L.; Eberhardt, S.C.; Leibel, S.A.; Scardino, P.T. Imaging prostate cancer: A multidisciplinary perspective. Radiology 2007, 243, 28–53. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Ploussard, G.; Valerio, M.; Mattei, A.; Fiori, C.; Fossati, N.; Stabile, A.; Beauval, J.-B.; Malavaud, B.; Roumiguié, M.; et al. A Novel Nomogram to Identify Candidates for Extended Pelvic Lymph Node Dissection Among Patients with Clinically Localized Prostate Cancer Diagnosed with Magnetic Resonance Imaging-targeted and Systematic Biopsies. Eur. Urol. 2019, 75, 506–514. [Google Scholar] [CrossRef]
- Mottet, N.; Bellmunt, J.; Bolla, M.; Briers, E.; Cumberbatch, M.G.; De Santis, M.; Fossati, N.; Gross, T.; Henry, A.M.; Joniau, S.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017, 71, 618–629. [Google Scholar] [CrossRef]
- Fossati, N.; Willemse, P.-P.M.; Van den Broeck, T.; van den Bergh, R.C.N.; Yuan, C.Y.; Briers, E.; Bellmunt, J.; Bolla, M.; Cornford, P.; De Santis, M.; et al. The Benefits and Harms of Different Extents of Lymph Node Dissection During Radical Prostatectomy for Prostate Cancer: A Systematic Review. Eur. Urol. 2017, 72, 84–109. [Google Scholar] [CrossRef]
- Haiquel, L.; Cathelineau, X.; Sanchez-Salas, R.; Macek, P.; Secin, F. Pelvic lymph node dissection in high-risk prostate cancer. Int. Braz J Urol Off. J. Braz. Soc. Urol. 2022, 48, 54–66. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Smith, C.P.; Czarniecki, M.; Mehralivand, S.; Stoyanova, R.; Choyke, P.L.; Harmon, S.; Turkbey, B. Radiomics and radiogenomics of prostate cancer. Abdom. Radiol. N. Y. 2019, 44, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Artificial Intelligence in Breast Cancer MRI: Applications for TNM Staging. Available online: https://www.iris.unicampus.it/handle/20.500.12610/71823 (accessed on 16 October 2023).
- Santucci, D.; Faiella, E.; Cordelli, E.; Calabrese, A.; Landi, R.; de Felice, C.; Beomonte Zobel, B.; Grasso, R.F.; Iannello, G.; Soda, P. The Impact of Tumor Edema on T2-Weighted 3T-MRI Invasive Breast Cancer Histological Characterization: A Pilot Radiomics Study. Cancers 2021, 13, 4635. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; de Cobelli, O.; Musi, G.; Del Giudice, F.; Carrieri, G.; Busetto, G.M.; Falagario, U.G.; Sciarra, A.; Maggi, M.; Crocetto, F.; et al. Radiomics in prostate cancer: An up-to-date review. Ther. Adv. Urol. 2022, 14, 17562872221109020. [Google Scholar] [CrossRef] [PubMed]
- Santucci, D.; Faiella, E.; Cordelli, E.; Sicilia, R.; de Felice, C.; Zobel, B.B.; Iannello, G.; Soda, P. 3T MRI-Radiomic Approach to Predict for Lymph Node Status in Breast Cancer Patients. Cancers 2021, 13, 2228. [Google Scholar] [CrossRef] [PubMed]
- Cordelli, E.; Sicilia, R.; Santucci, D.; de Felice, C.; Quattrocchi, C.C.; Zobel, B.B.; Iannello, G.; Soda, P. Radiomics-Based Non-Invasive Lymph Node Metastases Prediction in Breast Cancer. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020; pp. 486–491. [Google Scholar]
- Liu, X.; Wang, X.; Zhang, Y.; Sun, Z.; Zhang, X.; Wang, X. Preoperative prediction of pelvic lymph nodes metastasis in prostate cancer using an ADC-based radiomics model: Comparison with clinical nomograms and PI-RADS assessment. Abdom. Radiol. N. Y. 2022, 47, 3327–3337. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, J.; Wu, J.; Zhang, Y.; Wang, X.; Zhang, X.; Wang, X. Utility of diffusion weighted imaging-based radiomics nomogram to predict pelvic lymph nodes metastasis in prostate cancer. BMC Med. Imaging 2022, 22, 190. [Google Scholar] [CrossRef]
- Zheng, H.; Miao, Q.; Liu, Y.; Mirak, S.A.; Hosseiny, M.; Scalzo, F.; Raman, S.S.; Sung, K. Multiparametric MRI-based radiomics model to predict pelvic lymph node invasion for patients with prostate cancer. Eur. Radiol. 2022, 32, 5688–5699. [Google Scholar] [CrossRef]
- Bourbonne, V.; Jaouen, V.; Nguyen, T.A.; Tissot, V.; Doucet, L.; Hatt, M.; Visvikis, D.; Pradier, O.; Valéri, A.; Fournier, G.; et al. Development of a Radiomic-Based Model Predicting Lymph Node Involvement in Prostate Cancer Patients. Cancers 2021, 13, 5672. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Han, C.; Cui, Y.; Huang, J.; Wang, X.; Zhang, X.; Wang, X. Development and validation of the 3D U-Net algorithm for segmentation of pelvic lymph nodes on diffusion-weighted images. BMC Med. Imaging 2021, 21, 170. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, Z.; Wang, K.; Zhang, Y.; Li, J.; Wang, X.; Zhang, X.; Wang, X. Semiautomated pelvic lymph node treatment response evaluation for patients with advanced prostate cancer: Based on MET-RADS-P guidelines. Cancer Imaging Off. Publ. Int. Cancer Imaging Soc. 2023, 23, 7. [Google Scholar] [CrossRef]
- Hou, Y.; Bao, M.-L.; Wu, C.-J.; Zhang, J.; Zhang, Y.-D.; Shi, H.-B. A machine learning-assisted decision-support model to better identify patients with prostate cancer requiring an extended pelvic lymph node dissection. BJU Int. 2019, 124, 972–983. [Google Scholar] [CrossRef]
- Hou, Y.; Bao, J.; Song, Y.; Bao, M.-L.; Jiang, K.-W.; Zhang, J.; Yang, G.; Hu, C.-H.; Shi, H.-B.; Wang, X.-M.; et al. Integration of clinicopathologic identification and deep transferrable image feature representation improves predictions of lymph node metastasis in prostate cancer. EBioMedicine 2021, 68, 103395. [Google Scholar] [CrossRef]
- Franzese, C.; Cozzi, L.; Badalamenti, M.; Baldaccini, D.; D’Agostino, G.; Fogliata, A.; Navarria, P.; Franceschini, D.; Comito, T.; Clerici, E.; et al. Radiomics-based prognosis classification for high-risk prostate cancer treated with radiotherapy. Strahlenther. Onkol. 2022, 198, 710–718. [Google Scholar] [CrossRef]
- Peeken, J.C.; Shouman, M.A.; Kroenke, M.; Rauscher, I.; Maurer, T.; Gschwend, J.E.; Eiber, M.; Combs, S.E. A CT-based radiomics model to detect prostate cancer lymph node metastases in PSMA radioguided surgery patients. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Zamboglou, C.; Carles, M.; Fechter, T.; Kiefer, S.; Reichel, K.; Fassbender, T.F.; Bronsert, P.; Koeber, G.; Schilling, O.; Ruf, J.; et al. Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate- and high-risk prostate cancer—A comparison study with histology reference. Theranostics 2019, 9, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Cysouw, M.C.F.; Jansen, B.H.E.; van de Brug, T.; Oprea-Lager, D.E.; Pfaehler, E.; de Vries, B.M.; van Moorselaar, R.J.A.; Hoekstra, O.S.; Vis, A.N.; Boellaard, R. Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Trägårdh, E.; Enqvist, O.; Ulén, J.; Jögi, J.; Bitzén, U.; Hedeer, F.; Valind, K.; Garpered, S.; Hvittfeldt, E.; Borrelli, P.; et al. Freely Available, Fully Automated AI-Based Analysis of Primary Tumour and Metastases of Prostate Cancer in Whole-Body [18F]-PSMA-1007 PET-CT. Diagnostics 2022, 12, 2101. [Google Scholar] [CrossRef]
- Trägårdh, E.; Enqvist, O.; Ulén, J.; Hvittfeldt, E.; Garpered, S.; Belal, S.L.; Bjartell, A.; Edenbrandt, L. Freely available artificial intelligence for pelvic lymph node metastases in PSMA PET-CT that performs on par with nuclear medicine physicians. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3412–3418. [Google Scholar] [CrossRef]
- Borrelli, P.; Larsson, M.; Ulén, J.; Enqvist, O.; Trägårdh, E.; Poulsen, M.H.; Mortensen, M.A.; Kjölhede, H.; Høilund-Carlsen, P.F.; Edenbrandt, L. Artificial intelligence-based detection of lymph node metastases by PET/CT predicts prostate cancer-specific survival. Clin. Physiol. Funct. Imaging 2021, 41, 62–67. [Google Scholar] [CrossRef]
- Sollini, M.; Bartoli, F.; Cavinato, L.; Ieva, F.; Ragni, A.; Marciano, A.; Zanca, R.; Galli, L.; Paiar, F.; Pasqualetti, F.; et al. [18F]FMCH PET/CT biomarkers and similarity analysis to refine the definition of oligometastatic prostate cancer. EJNMMI Res. 2021, 11, 119. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faiella, E.; Vaccarino, F.; Ragone, R.; D’Amone, G.; Cirimele, V.; Piccolo, C.L.; Vertulli, D.; Grasso, R.F.; Zobel, B.B.; Santucci, D. Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review. J. Clin. Med. 2023, 12, 7032. https://doi.org/10.3390/jcm12227032
Faiella E, Vaccarino F, Ragone R, D’Amone G, Cirimele V, Piccolo CL, Vertulli D, Grasso RF, Zobel BB, Santucci D. Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review. Journal of Clinical Medicine. 2023; 12(22):7032. https://doi.org/10.3390/jcm12227032
Chicago/Turabian StyleFaiella, Eliodoro, Federica Vaccarino, Raffaele Ragone, Giulia D’Amone, Vincenzo Cirimele, Claudia Lucia Piccolo, Daniele Vertulli, Rosario Francesco Grasso, Bruno Beomonte Zobel, and Domiziana Santucci. 2023. "Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review" Journal of Clinical Medicine 12, no. 22: 7032. https://doi.org/10.3390/jcm12227032
APA StyleFaiella, E., Vaccarino, F., Ragone, R., D’Amone, G., Cirimele, V., Piccolo, C. L., Vertulli, D., Grasso, R. F., Zobel, B. B., & Santucci, D. (2023). Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review. Journal of Clinical Medicine, 12(22), 7032. https://doi.org/10.3390/jcm12227032







