Landscape and Predictive Significance of the Structural Classification of EGFR Mutations in Chinese NSCLCs: A Real-World Study
Abstract
1. Introduction
2. Materials and Methods
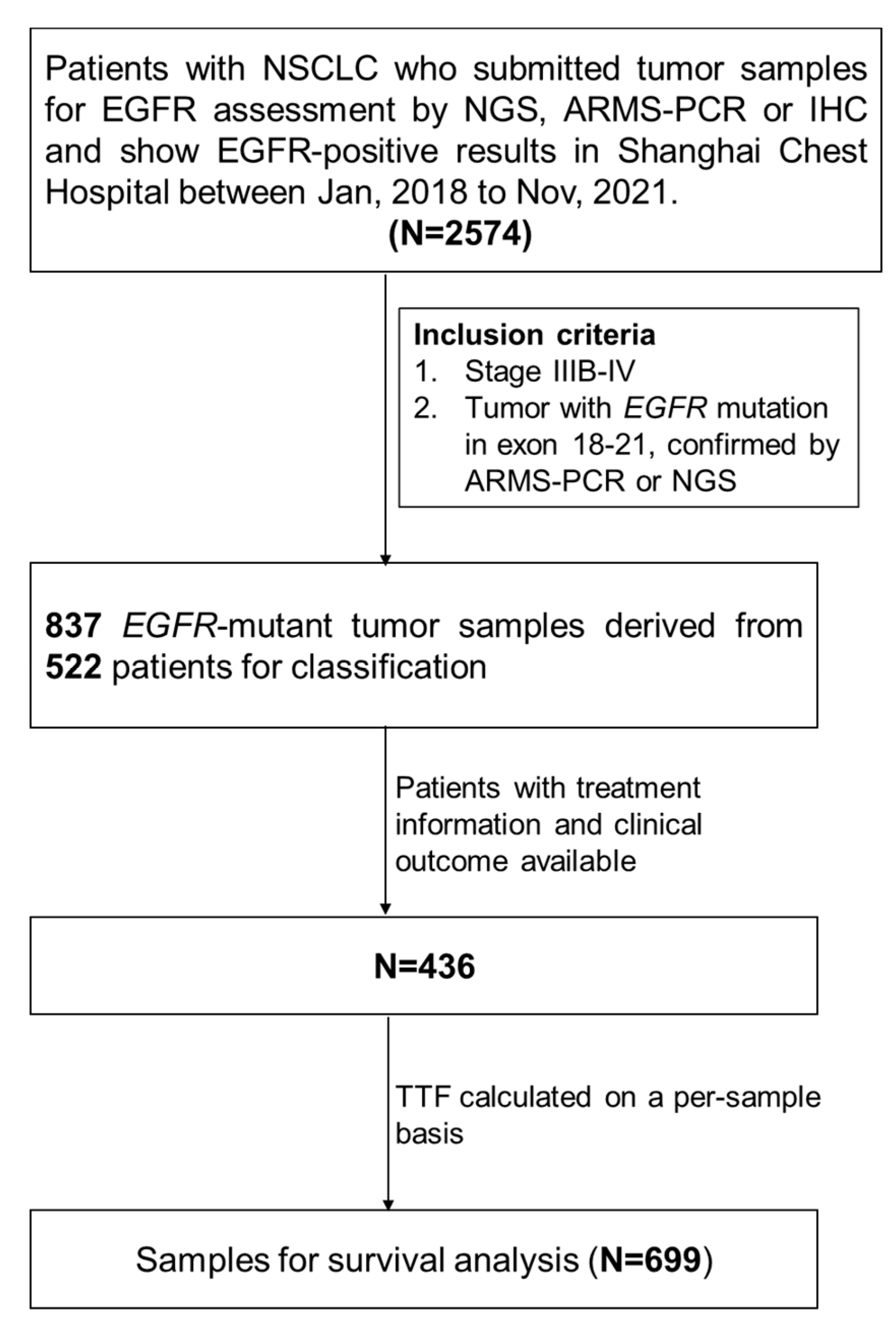
2.1. Patients and Study Design
2.2. ARMS-PCR
2.3. NGS
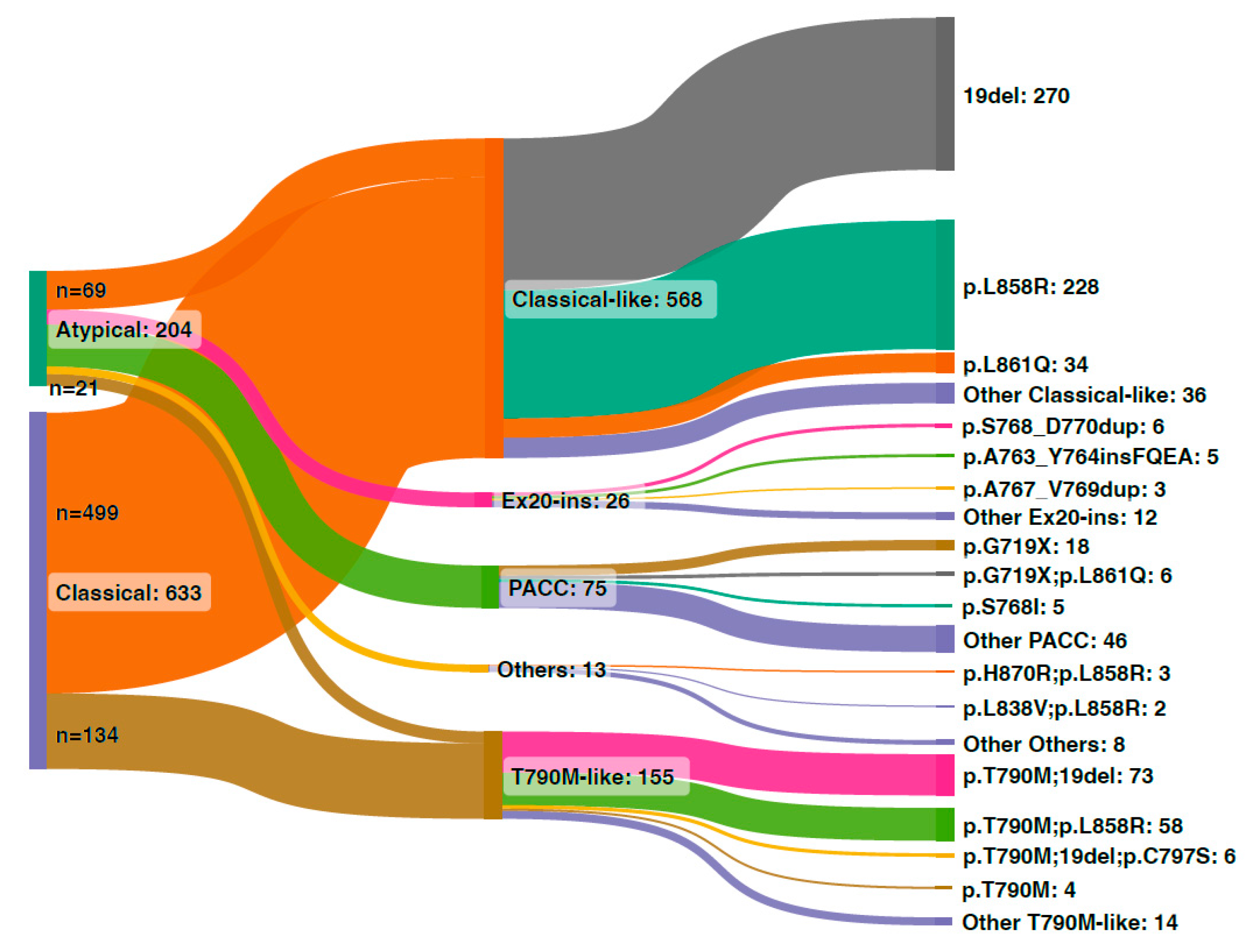
2.4. Classification of EGFR Mutations
2.5. Statistical Analyses
3. Results
3.1. Patients’ Characteristics
3.2. Classical and Atypical EGFR Mutations in the Chinese Cohort
3.3. Structural Classification of EGFR Mutations in the Chinese Cohort
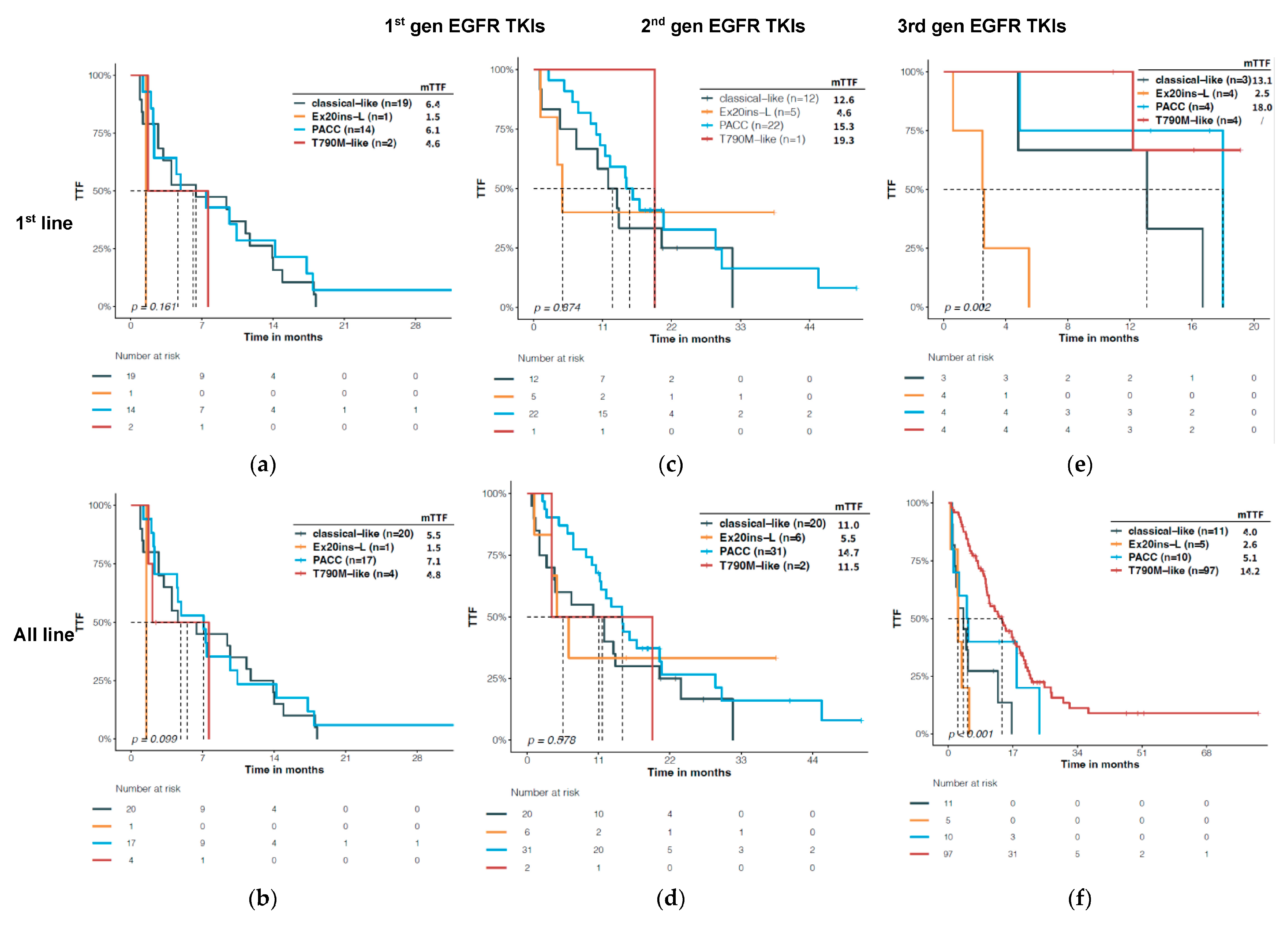
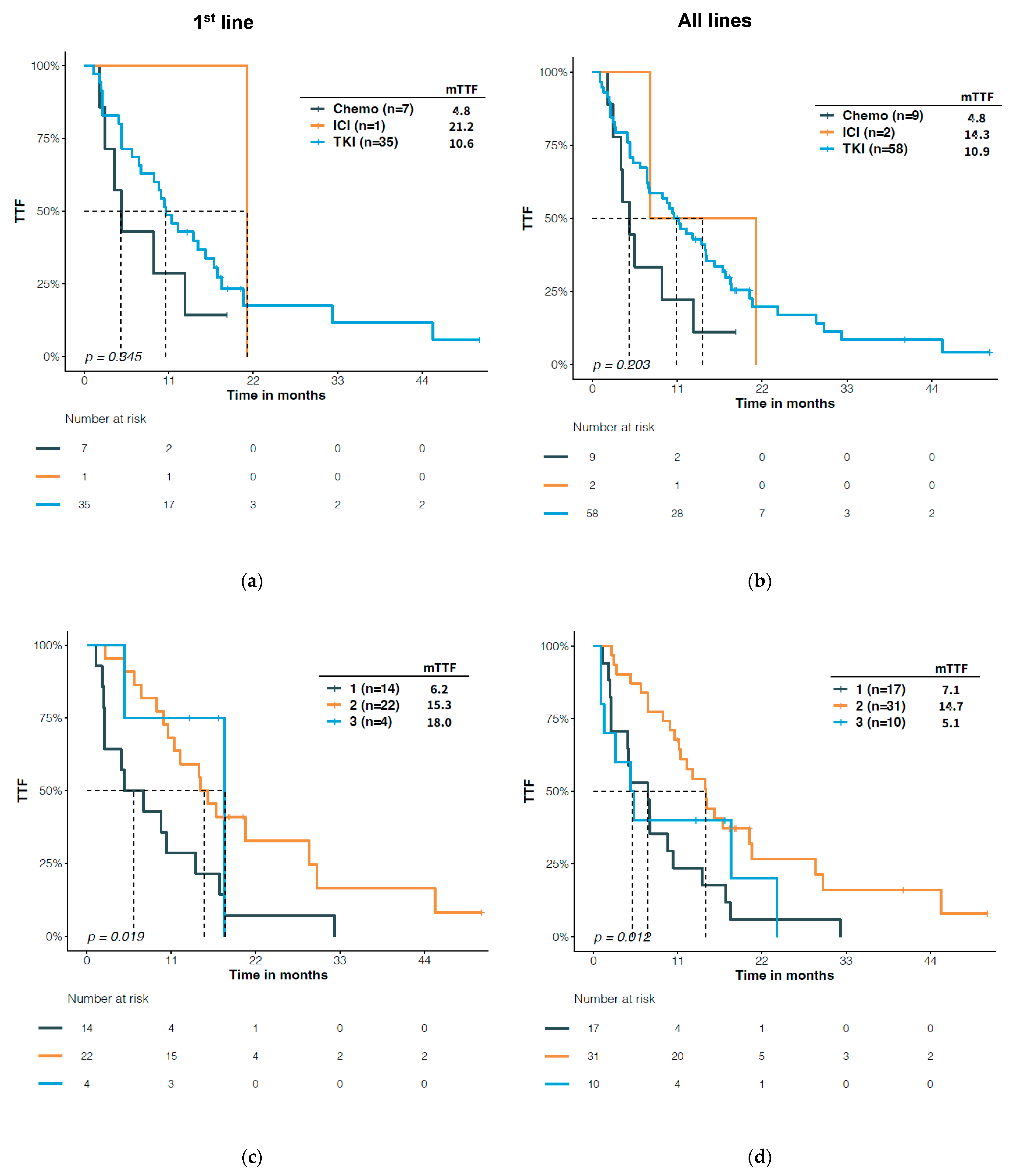
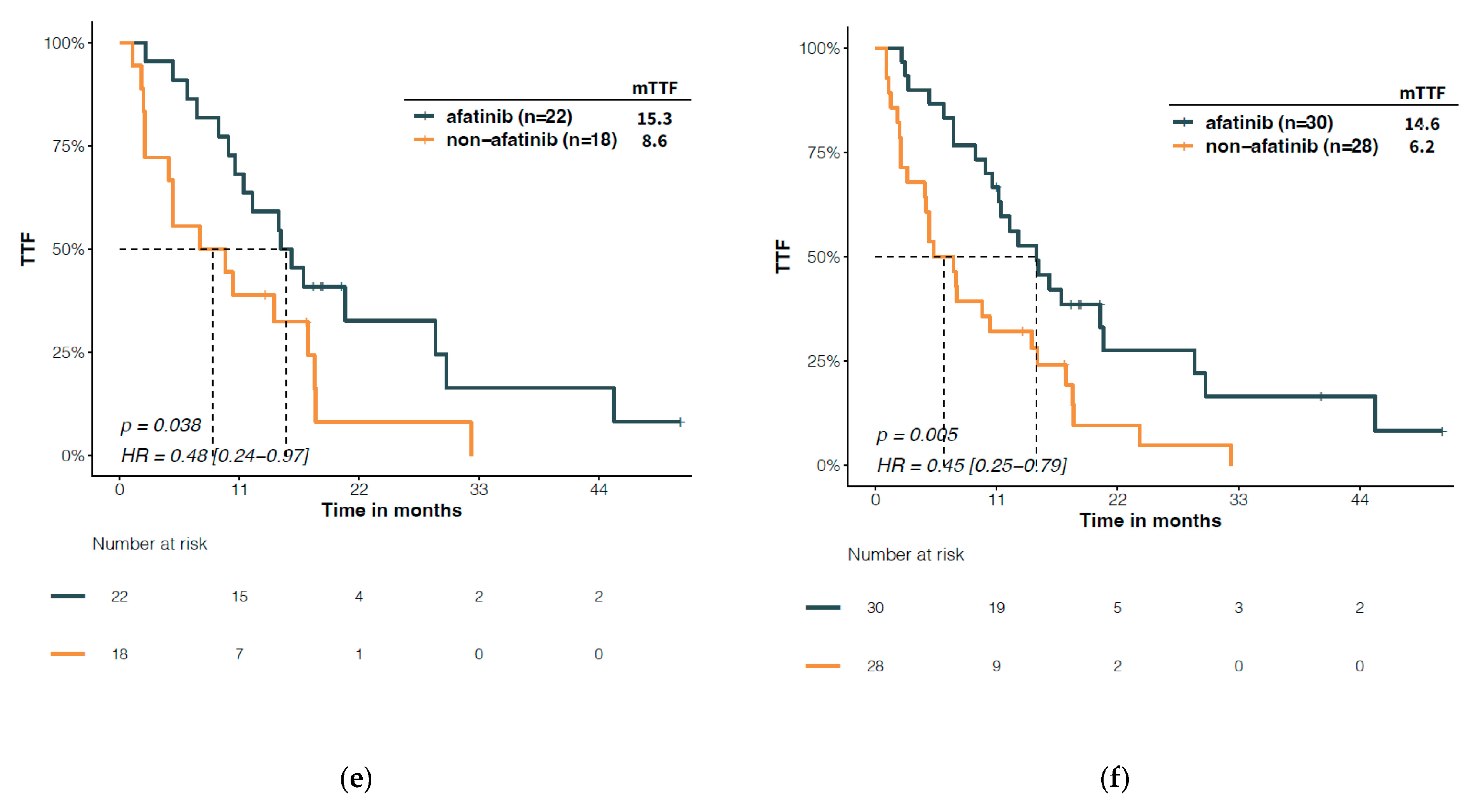
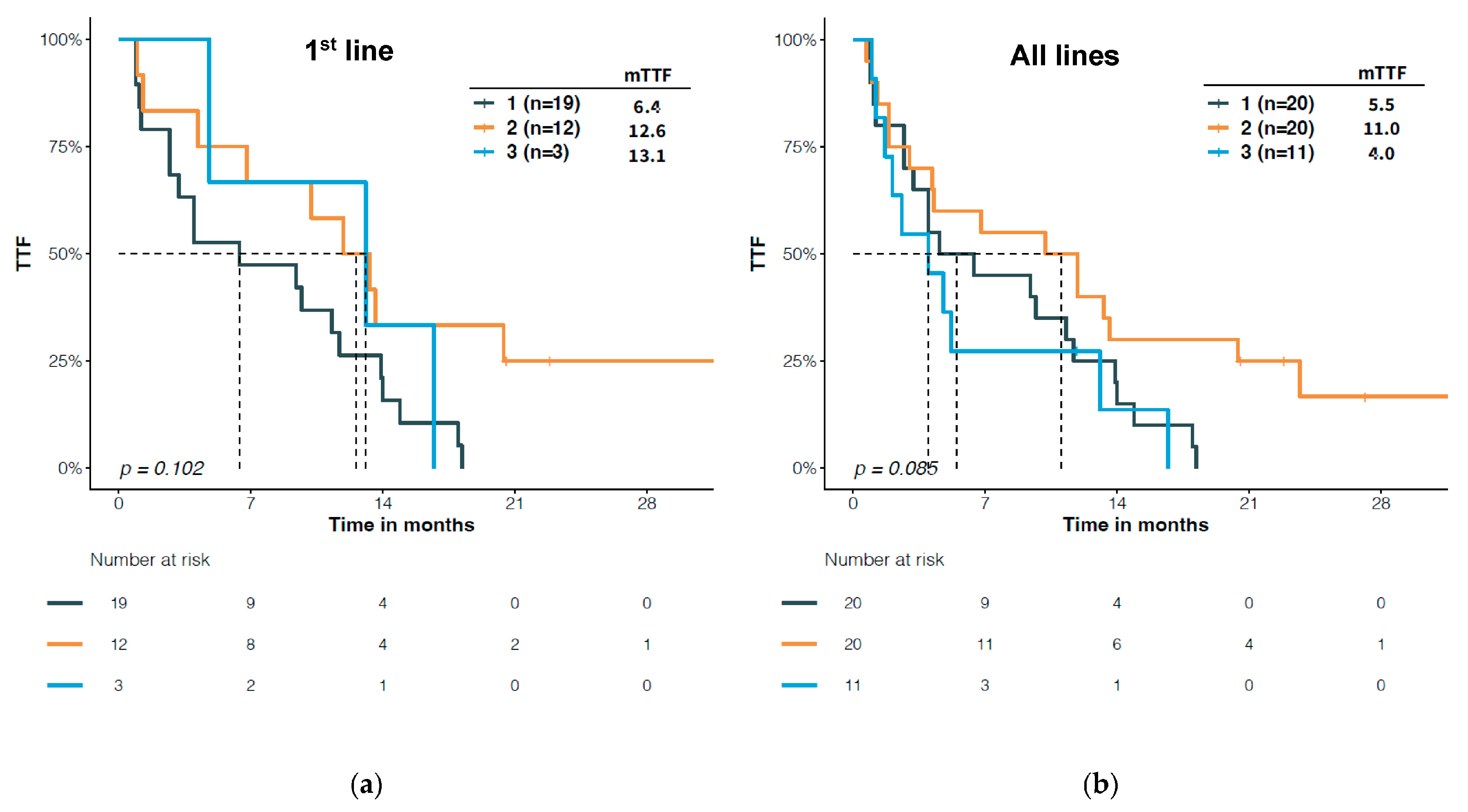
3.4. Predictive Significance of Structural Classification of EGFR Mutations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Zhou, C.; Hu, C.P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y.; et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Kitadai, R.; Okuma, Y. Treatment Strategies for Non-Small Cell Lung Cancer Harboring Common and Uncommon EGFR Mutations: Drug Sensitivity Based on Exon Classification, and Structure-Function Analysis. Cancers 2022, 14, 2519. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Cho, E.N.; Park, H.S.; Hong, J.Y.; Lim, S.; Youn, J.P.; Hwang, S.Y.; Chang, Y.S. Compound EGFR mutation is frequently detected with co-mutations of actionable genes and associated with poor clinical outcome in lung adenocarcinoma. Cancer Biol. Ther. 2016, 17, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Sequist, L.V.; Geater, S.L.; Tsai, C.M.; Mok, T.S.; Schuler, M.; Yamamoto, N.; Yu, C.J.; Ou, S.H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Minegishi, Y.; Yoshizawa, H.; Maemondo, M.; Inoue, A.; Sugawara, S.; Isobe, H.; Harada, M.; Ishii, Y.; Gemma, A.; et al. Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J. Thorac. Oncol. 2014, 9, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Lim, S.H.; An, H.J.; Kim, K.H.; Park, K.U.; Kang, E.J.; Choi, Y.H.; Ahn, M.S.; Lee, M.H.; Sun, J.M.; et al. Osimertinib for Patients With Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09). J. Clin. Oncol. 2020, 38, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, X.; Dai, Y.; Wu, D.; Liu, F.; Yang, Z.; Song, B.; Xie, L.; Yang, L.; Zhao, W.; et al. Concordance Study of a 520-Gene Next-Generation Sequencing-Based Genomic Profiling Assay of Tissue and Plasma Samples. Mol. Diagn. Ther. 2022, 26, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cao, J.; Song, Q.; Wang, L.; Xiong, Y.; Chen, R. Structure-based classification of EGFR mutations in operable pre-invasive and invasive non-small cell lung cancer: A cross-sectional study. J. Thorac. Dis. 2022, 14, 3508–3516. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Marten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Banno, E.; Togashi, Y.; Nakamura, Y.; Chiba, M.; Kobayashi, Y.; Hayashi, H.; Terashima, M.; de Velasco, M.A.; Sakai, K.; Fujita, Y.; et al. Sensitivities to various epidermal growth factor receptor-tyrosine kinase inhibitors of uncommon epidermal growth factor receptor mutations L861Q and S768I: What is the optimal epidermal growth factor receptor-tyrosine kinase inhibitor? Cancer Sci. 2016, 107, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Mok, T.; Peters, S.; Popat, S.; Ahn, M.J.; de Marinis, F. Recent Advances on the Role of EGFR Tyrosine Kinase Inhibitors in the Management of NSCLC With Uncommon, Non Exon 20 Insertions, EGFR Mutations. J. Thorac. Oncol. 2021, 16, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Janning, M.; Suptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ramalingam, S.S.; Kim, T.M.; Kim, S.W.; Yang, J.C.; Riely, G.J.; Mekhail, T.; Nguyen, D.; Garcia Campelo, M.R.; Felip, E.; et al. Treatment Outcomes and Safety of Mobocertinib in Platinum-Pretreated Patients with EGFR Exon 20 Insertion-Positive Metastatic Non-Small Cell Lung Cancer: A Phase 1/2 Open-label Nonrandomized Clinical Trial. JAMA Oncol. 2021, 7, e214761. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Haura, E.B.; Leighl, N.B.; Mitchell, P.; Shu, C.A.; Girard, N.; Viteri, S.; Han, J.Y.; Kim, S.W.; Lee, C.K.; et al. Amivantamab in EGFR Exon 20 Insertion-Mutated Non-Small-Cell Lung Cancer Progressing on Platinum Chemotherapy: Initial Results from the CHRYSALIS Phase I Study. J. Clin. Oncol. 2021, 39, 3391–3402. [Google Scholar] [CrossRef] [PubMed]


| Overall (n = 436) | |
|---|---|
| Sex | |
| Female | 234 (53.7%) |
| Male | 202 (46.3%) |
| Smoking history | |
| No | 343 (78.7%) |
| Yes | 93 (21.3%) |
| Histology | |
| ADC | 414 (94.9%) |
| ADSQ | 4 (0.9%) |
| NOS | 9 (2.1%) |
| SCC | 9 (2.1%) |
| Stage | |
| IIIB | 6 (1.4%) |
| IIIC | 9 (2.0%) |
| IVA | 108 (24.8%) |
| IVB | 32 (7.3%) |
| IVC | 281 (64.4%) |
| Age | |
| Median [IQR] | 61.0 [53.8, 67.0] |
| Therapy | |
| 1st gen EGFR TKI | 302 |
| 1st-line a | 273 (90.4%) |
| Later-line | 29 (9.6%) |
| 2nd gen EGFR TKI | 124 |
| 1st-line a | 54 (43.5%) |
| Later-line | 70 (56.5%) |
| 3rd gen EGFR TKI | 241 |
| 1st-line a | 56 (23.2%) |
| Later-line | 185 (76.8%) |
| Chemotherapy | 93 |
| 1st-line b | 31 (33.3%) |
| Later-line | 62 (66.7%) |
| ICI | 36 |
| 1st-line b | 8 (22.2%) |
| Later-line | 28 (77.8%) |
| Other | 81 |
| 1st-line b | 14 (17.3%) |
| Later-line | 67 (82.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, L.; Huang, H.; Xu, Z.; Niu, X.; Li, Z.; Xia, L.; Yu, Y.; Lu, S. Landscape and Predictive Significance of the Structural Classification of EGFR Mutations in Chinese NSCLCs: A Real-World Study. J. Clin. Med. 2023, 12, 236. https://doi.org/10.3390/jcm12010236
Gu L, Huang H, Xu Z, Niu X, Li Z, Xia L, Yu Y, Lu S. Landscape and Predictive Significance of the Structural Classification of EGFR Mutations in Chinese NSCLCs: A Real-World Study. Journal of Clinical Medicine. 2023; 12(1):236. https://doi.org/10.3390/jcm12010236
Chicago/Turabian StyleGu, Linping, Huayan Huang, Zhangwendi Xu, Xiaomin Niu, Ziming Li, Liliang Xia, Yongfeng Yu, and Shun Lu. 2023. "Landscape and Predictive Significance of the Structural Classification of EGFR Mutations in Chinese NSCLCs: A Real-World Study" Journal of Clinical Medicine 12, no. 1: 236. https://doi.org/10.3390/jcm12010236
APA StyleGu, L., Huang, H., Xu, Z., Niu, X., Li, Z., Xia, L., Yu, Y., & Lu, S. (2023). Landscape and Predictive Significance of the Structural Classification of EGFR Mutations in Chinese NSCLCs: A Real-World Study. Journal of Clinical Medicine, 12(1), 236. https://doi.org/10.3390/jcm12010236








