Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations
Abstract
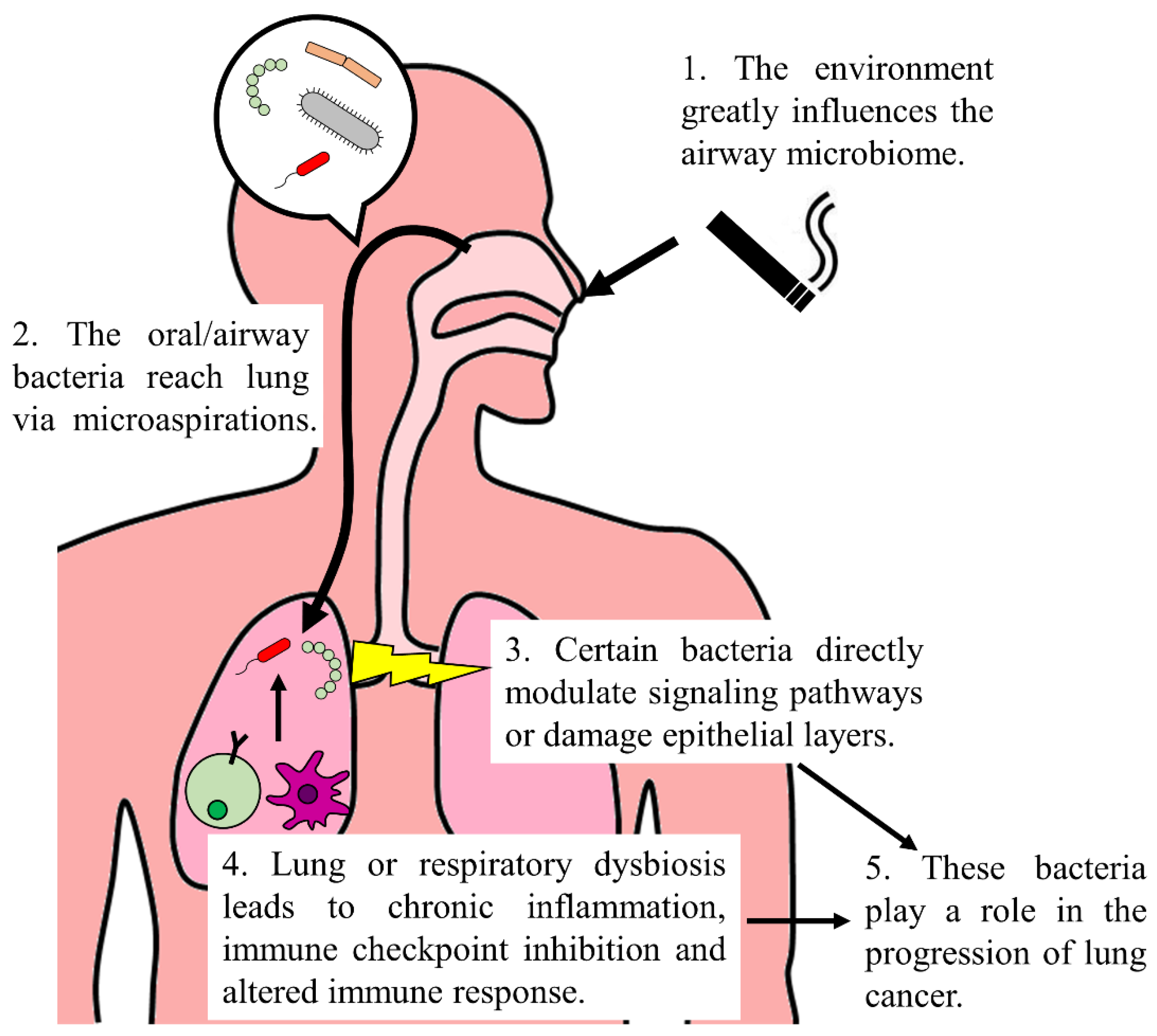
1. Introduction
2. Lung or Tumor-Resident Microbiome and Lung Cancers
2.1. Few Commonalities across Different Lung Microbiome Profilings
| Study | Sampling Site | Disease | Experiment Design | Sequencing Methods * | Diversity Variations in LC | Microbial Associations or Biomarkers |
|---|---|---|---|---|---|---|
| Yuan [24] | Tumor | LC | RM LC (n = 174) vs. non-RM LC (n = 134) | TCGA | RM LC has similar α-diversity, but reduced richness | Acidovorax, Clostridioides, Succinimonas, Shewanella, Leuconostoc and Dickeya are biomarkers for RM LC |
| Baranova [25] | Sputum | LUSC | Patients (n = 40) vs. healthy controls (n = 40); all male | 16S V3–V4 | Decreased β-diversity in LUSC; no changes in α-diversity | Streptococcus, Bacillus, Gemella and Haemophilus are enriched in LUSC patients |
| Wu [26] | BALF; tumor | LC as GGO | BALF from diseased lung and paired contralateral healthy lung (n = 11); lung GGO and paired adjacent normal tissues (n = 26) | 16S V4/16S V3/16S V3–V4/16S V4–V5 | No changes in α- and β-diversity | Significantly reduced Proteobacteria in LC tissues; In BALF of LC patients: reduced Rothia, and increased Lachnospiraceae, Bacteroides uniforms and Faecalibacterium prausnitzii |
| He [27] | Sputum | NSCLC and/or COPD | patients with NSCLC and COPD (CN, n = 67) vs. NSCLC (n = 9) vs. COPD (n = 14) | 16S V3–V4 | No significant differences in diversities | In CN patients: reduced Streptococcus, Veillonella, Moraxella and Actinomyces; and increased Neisseria and Acinetobacter |
| Vogtmann [28] | Oral wash | LC | Patients (n = 1306) | 16S V4 | Higher α-diversity is associated with lower LC risk | Increased Streptococcus and Peptoniphilus abundances are associated with increased LC risk, while Peptostreptococcus, Eubacterium yurii and Aggregatibacter are associated with reduced risk |
| Kim [29] | Tumor | NSCLC | Tumor tissue (n = 162) vs. adjacent normal tissue (n = 54) | 16S V4–V5 | Reduced diversity as LC progress | Increased Romboutsia, Novosphingobium, Acinetobacter and Prevotella in LC Increased Stenotrophomonas upon LC relapse |
| Qian [30] | BALF; airway protected brushing | sMPLC | Patients (n = 8) | 16S V3–V4 | Increased α-diversity in BALF | Clostridium, Actinobacteria, Fusobacterium and Rothia are enriched in the BALF of sMPLC lesions |
| Chen [19] | Tumor | LC | Tumor tissue (n = 34) vs. adjacent normal tissue (n = 29) | 16S | Lower α-diversity and higher β-diversity in LC tissues | In LC: increased Staphylococcus, Capnocytophaga, Lachnoanaerobaculum, Fusobacterium, Oligella, Rubellimicrobium, Marinococcus Sphingomonas and Sphingopyxis; and decreased Comamonas and Peptococcus |
| Masuhiro [31] | BALF | LC | Patients under PD-1 blockade treatment (n = 12) | 16S V3–V4 | Higher diversity in responders | Responders have higher Bacteroidetes and lower Proteobacteria |
| Zhang [32] | BALF; tumor | NSCLC | Patients (n = 6 for BALF; n = 37 for tumor tissues) | Pathogen-targeted sequencing (tumor and 4 BALF); 16S (2 BALF) | Higher diversity in BALF than in tumor | BALF and tumor tissues share Streptococcus pneumoniae, S. crista, S. constellatus, S. gordonii, Prevotella II, Haemophilus, H. haemolyticus, H. influenzae, Actinomyces Neesii, human herpes virus type 7 and Neisseria lactose |
| Marshall [33] | Epithelial brushing | Pre-cancer | A 10-year follow-up study of 393 patients: with incidence (n = 59), prevalence (n = 21), and no cancer (313) | 16S V4 | NA | The abundances of Bacilli, Lactobacillales, Streptococcus and Paenibacillus are associated with incident LC |
| Zeng [34] | BALF | NSCLC | LC (n = 46) vs. benign lung disease (n = 29) | 16S V3–V4 | Increased α-diversity during carcinogenesis and significant changes in β-diversity | Enrichment of phyla (Firmicutes and Bacteroidetes) and genera (Streptococcus, Prevotella and Veillonella) in NSCLC |
| Chu [35] | BALF | LC | Patients under PD-1 blockade treatment: responders (n = 19) vs. non-responders (n = 27) | 16S V3–V4 | Decreased diversity upon treatment | Increased abundance of Fusobacterium is associated with poor anti-PD-1 therapy response |
| Zhang [36] | Tumor | NSCLC | Patients (n = 53) | Pathogen-targeted sequencing | NA | At advanced stage: increased Serratia marcescens, Actinomyces neesii, Enterobacter cloacae and Haemophilus parainfluenzae; and decreased Staphylococcus haemolyticus and Streptococcus crista Survival prediction: Haemophilus parainfluenzae, Serratia marcescens, Acinetobacter jungii and Streptococcus constellation High PD-L1 expression: increased Acinetobacter jungii |
| Huang [37] | Sputum | NSCLC | Patients (n = 85) | 16S V3–V4 | Decreased α- and β-diversity at advanced stage | Early stage: Granulicatella and Actinobacillus are enriched Advanced stage: Actinomyces is enriched |
| Roy [38] | Saliva | LUAD | Patients (n = 5) and healthy control (n = 5) | 16S V3–V4 | No significant changes in α-diversity | Increased Rothia mucilaginosa, Veillonella dispar, Prevotella melaninogenica, Prevotella pallens, Prevotella copri, Haemophilus parainfluenzae, Neisseria bacilliformis and Aggregatibacter segnis in LUAD |
| Dong [39] | Tumor | LC | Tumor tissues (n = 118) vs. adjacent normal tissue (n = 123) from 143 patients | 16S V3–V4 | No significant changes in α-diversity but significant differences in β-diversity | Massilia, Phenylobacterium and Pseudoxanthomonas are enriched in tumor tissue; Brevibacillus, Cupriavidus and Anaerococcus are enriched in normal tissues Massilia and Acidovorax are associated with TP53 mutation |
| Jang [40] | BALF | LC | Patients under PD-1 blockade treatment (n = 84) | 16S V3–V4 | No significant changes in α-diversity and β-diversity | High-PD-L1 group: dominated by Veillonella dispar; with reduced Neisseria Responders: dominated by Veillonella dispar Non-responders: dominated by Haemophilus influenzae and Neisseria perflava |
| Boesch [41] | Tumor | AdvancedNSCLC | Tumor tissues (n = 38) vs. adjacent normal tissue (n = 10) from patients with PD-1 blockade treatment | 16S V3–V4 | Increased diversity is associated with better survival | Gammaproteobacteria correlate with low PD-L1 expression and poor anti-PD-1 blockade treatment outcomes |
| Lu [42] | Sputum | NSCLC | Patients (n = 87) vs. healthy controls (n = 34) | 16S V3–V4 | Decreased α-diversity in NSCLC | NSCLC: increased Haemophilus parainfluenzae and Haemophilus influenzae Distant metastasis: decreased Capnocytophaga; and increased Pseudomonas, Coriobacteriaceae and Actinomyces |
| Chang [43] | Tumor | LC | Patients (n = 49) | 16S V4 | NA | Brevundimonas diminuta, Acinetobacter radioresistens Enterobacter cloacae, Mycobacterium chelonae, Mycobacterium franklinii, Staphylococcus sp., Bacillus megaterium, Pseudomonas aeruginosa and Rhodococcus erythropolis are enriched in LC and associated with poor prognosis |
| Shi [44] | Mouth rinse | LC | Patients (n = 156) vs. healthy control (n = 156) | 16S V4 | No significant changes in α-diversity and β-diversity | The abundances of families Lachnospiraceae, Peptostreptococcaceae, Erysipelotrichaceae and species Parvimonas micra are associated with decreased LC risk |
| Seixas [45] | BALF | LC, COPD and ILD | LC (n = 8) vs. COPD (n = 7) vs. ILD (n = 10) | 16S V4 | No significant changes in diversity between cancer and non-cancer | Streptococcus and Prevotella are associated with LC Haemophilus is associated with COPD |
| Zheng [22] | BALF | NSCLC | Patients (n = 32) vs. non-cancer controls (n = 15) | 16S | Decreased diversity in NSCLC | LC: increased Lactobacillus rossiae, Burkholderia mallei and Bacteroides pyogenes; decreased Paenibacillus odorifer, Pseudomonas entomophila and Magnetospirillum gryphiswaldense |
| Zhang [46] | Sputum; stool | Metastatic NSCLC | Patients (n = 75) at baseline and during immune checkpoint inhibitors treatment | 16S | α-diversity between the gut and respiratory microbiota is not related Only increased α-diversity in the gut is associated with better treatment outcomes | Streptococcus in sputum as a biomarker for good treatment response |
| Dumont-Leblond [47] | Tumor | NSCLC | Tumor tissues vs. adjacent normal tissue from 29 patients | 16S V3–V4 | Higher β diversity differences among different patients than tissues from the same patient. Higher α-diversity in tumor tissues | LC has an increased abundance of pathogenic and pro-inflammatory bacteria: Escherichia-Shigella, Faecalibacterium, Pseudomonas, unclassified Enterobacteriaceae, Alloprevotella and Brevundimonas High Phascolarctobacterium in LUSC |
| Ma [48] | Tumor | LUAD as SSN or SN | Tumor tissues vs. adjacent normal tissue (n = 10 pairs for SSN; n = 25 pairs for SN) | 16S V3–V4 | SSN has higher microbiome richness and diversity Tumor and normal tissues have similar diversity and richness | Increased Actinobacteria, Proteobacteria, Parvibaculales, Parvibaculaceae, Parvibaculum, Renibacterium and Ancylobacter; and decreased Firmicutes, Bacteroidetes and Lactobacillus in LUAD |
| Leng [49] | Tumor; sputum | LC | Tumor tissues vs. adjacent normal tissue (n = 31 pairs); sputum from NSCLC patients (n = 17) vs. cancer-free smoker controls (n = 10) | Droplet digital PCR for 25 NSCLC-associated bacterial genera | NA | Enrichment of Acidovorax, Streptococus and Veillonella in sputum of LUSC Enrichment of Capnocytophaga in sputum of LUAD |
| Druzhinin [50] | Sputum | LC | Patients (n = 66) vs. healthy controls (n = 62); all male | 16S V3–V4 | Decreased β diversity in LC patients | Increased Streptococcus, Bacillus, Gemella and Haemophilus in LC patients Chromosomal aberration frequency is positively associated with increased Bacteroides, Lachnoanaerobaculum, Porphyromonas, Mycoplasma and Fusobacterium; and decreased Granulicatella. Micronuclei frequency is negatively associated with increased Megasphaera and Selenomonas bovis |
| Hosgood [51] | Oral rinse | LC | Patients (n = 114) vs. healthy controls (n = 114) | Metagenomic shotgun sequencing | Individuals with lower α-diversity had an increased risk of lung cancer No significant changes in β-diversity | Decreased risk of LC: a higher abundance of Spirochaetia and Bacteroidetes Increased risk of LC: Bacilli class and Lactobacillales order |
| Tsay [52] | Lower airway brushing; buccal brushing | LC | Patients (n = 83) | 16S V4 | α-diversity is similar across different stages of NSCLC. Higher α-diversity in lower airways than in upper airways | Veillonella parvula is associated with LC progression, IL-17 expression and the activation of the immune checkpoint Increased Moraxella, Fusobacterium, Pseudomonas and Haemophilus; and decreased Actinomycetales in advanced LC Streptococcus, Prevotella and Veillonella enrichment is related to poor prognosis |
| Zhuo [53] | BALF | LC | From cancerous lung and the contralateral non-cancerous lung (n = 50) | 16S V3–V4 | No significant changes in α- and β-diversity | Increased risk of LC: genera Weissella and Spiroplasma Decreased risk of LC: phylum Bacteroidetes (class Bacteroidia and order Bacteroidales) |
| Kovaleva [54] | Tumor | NSCLC | Tumor tissues vs. adjacent normal tissue (n = 89) | 16S V3–V4 | Tumor tissues have similar α-diversity, but reduce overall bacterial load | High bacterial load with increased iNOS expression is a favorable prognostic factor; High bacterial load with increased FOXP3+ cells is associated with poor prognosis Increased Propionibacterium is associated with lower iNOS expression |
| Cheng [55] | BALF | LC | Patients (n = 32) vs. benign pulmonary diseases (n = 22) | 16S V3–V4 | Similar richness and evenness in LC | TM7-3, Gemmiger, Capnocytophaga, Sediminibacterium, Blautia and Oscillospira are enriched in LC |
| Mao [56] | Tumor | LC | Tumor tissues vs. adjacent normal tissue (n = 55) | 16S V3–V4 | Reduced α-diversity in LC; but no significant changes in β-diversity | Propionibacterium is significantly reduced in tumor tissues Other reduced genera include: unclassified Comamonadaceae, unclassified Enterobacteriaceae, Rhodobacter, Psychrobacter, Phormidium, Propionibacterium, Microbacterium and Finegoldia |
| Bello [57] | Bronchial biopsy; saliva | Central LC | Patients (n = 25): saliva and biopsies of affected and contralateral bronchi vs. healthy controls (n = 16): saliva and single bronchi biopsy | 16S V3–V4 | The diversity of salivary sample is comparable in patients and controls | Streptococcus has dominated in both affected and contralateral bronchi of patients Pseudomonas is dominated in control Increased abundance of Streptococcus, Rothia, Gemella and Lactobacillus in patients’ saliva |
| Druzhinin [58] | Sputum | LC | Patients (n = 17) vs. healthy control (n = 17) | 16S V3–V4 | No significant differences in α-diversity | Increased genera Haemophilus and Bergeyella; and decreased genera Atopobium, Stomatobaculum, Treponema and Porphyromonas in LC patients Chromosomal aberration frequency is negatively associated with the genus Atopobium and positively associated with the species Alloprevotella |
| Wong [59] | Tumor | LC | Tumor tissues vs. adjacent normal tissue (n = 497 for LUAD and 433 for LUSC) | TCGA | NA | The LC-associated microbiome is age and gender-specific Escherichia coli str. K-12 substrain W3110 is associated with the survival of aged LUAD patients |
| Reinhold [21] | Tumor; PO swab; BALF | LC | Patients undergoing surgery (n = 13) | 16S V3–V4 | Decreased α-diversity in the upper airways | High Prevotella, Veillonella and Streptococcus in the upper airways and BALF High Pseudomonas, Propionibacteria, Proteobacteria and Actinobacteria in lung cancer tissues |
| Bingula [60] | Saliva; BAL (from excised lobe); tumor | NSCLC | saliva, BAL, peritumoral tissues, tumor tissues and adjacent normal tissue from 18 patients | 16S V3–V4 | Unique β-diversity of BAL Diversity varies depending on lobe location | Tissue samples: dominated by Phylum Proteobacteria BAL: dominated by class Clostridia Saliva: dominated by class Bacilli |
| Patnaik [61] | Saliva; BALF; tumor | Early recurrentNSCLC | Pre-surgery saliva and BALF; tumor tissues and adjacent normal tissue from 48 patients undergoing surgery | 16S | Higher diversity in saliva and BALF; Tumor tissues and adjacent normal tissue have similar diversity | Recurrence is associated with increased genus Delftia and decreased Bifidobacterium in saliva; as well as increased Staphylococcus and decreased Bacillus and Anaerobacillus in tumor tissues |
| Ekanayake [62] | BALF; PO swab | LC and BRS | Patients (n = 20 for LC and n = 20 for BRS) vs. healthy controls (n = 20) | 16S V3–V4 | Increased diversity in patients | Enterococcus faecalis, Corynebacterium tuberculostearicum and Keratinibaculum paraultunense are LC-specific |
| Huang [63] | Bronchial washing fluid; sputum | LC | Bronchial washing fluid (n = 40) and sputum (n = 52) from LC patients | 16S V3–V4 | No significant difference in α- and β- diversity between LUAD and LUSC | All from Bronchial washing fluid samples: Genera Veillonell, Megasphaera, Actinomyces and Arthrobacter are enriched in LUAD without metastasis Genera Capnocytophaga and Rothia are enriched in LUSC with metastasis Streptococcus is decreased in LUAD upon metastasis Veillonella and Rothia are increased in LUSC upon metastasis |
| Jin [64] | BALF | LC | Patients (n = 91) vs. nonmalignant pulmonary diseases (n = 29) vs. healthy controls (n = 30); a validation cohort of 85 patients | Metagenomics | Diversity and richness are reduced in LC patients β-diversity is different between LC patients and healthy controls | Haemophilus influenzae shows the greatest difference between LC patients and healthy controls |
| Gomes [65] | BALF | LC | Patients (n = 49) vs. healthy controls (n = 54) | 16S V3-V6 | LUSC has a higher diversity than LUAD | Biomarkers for LUAD: Acinetobacter, Propionibacterium, Phenylobacterium, Brevundimonas and Staphylococcus Biomarkers for LUSC: Enterobacter, Serratia, Klebsiella, Kluyvera, Morganella, Achromobacter and Capnocytophaga |
| Ren [66] | Tumor | LUAD as GGN | Tumor tissues (n = 10) vs. adjacent normal tissue (n = 5) | Whole genome sequencing | High β diversity variation among patients | No significant differences in microbiome compositions between GGNs and normal tissues (except LUAD) |
| Zhang [67] | Saliva | NSCLC | Patients (n = 39) vs. healthy controls (n = 20) | 16S V1-V2 | A higher richness and lower diversity in NSCLC patients | In NSCLC: increased Veillonella, Streptococcus, Lautropia, Leptotrichia, Rothia and Aggregatibacter; and decreased Prevotella_7, Fusobacterium, Porphyromonas, Alloprevotella, Prevotella, Bacteroides and Faecalibacterium Veillonella is positively associated with the Neutrophil-lymphocyte ratio Streptococcus is negatively associated with the lymphocyte-monocyte ratio |
| Wang [68] | Saliva; BALF | PBC | Patients (n = 51) vs. healthy controls (n = 15) | 16S V4 | Patients have lower diversity in both saliva and BALF samples | Treponema (in saliva) and Filifactor (in both saliva and BALF) are potential biomarkers for LC |
| Hosgood [69] | Sputum | LC | Patients (n = 45) vs. healthy controls (n = 45) | 16S V1-V2 | Lower α-diversity is associated with an increased risk of LC | Decreased relative abundance of Fusobacteria is a risk factor for LC |
| Peters [70] | Tumor | NSCLC | Tumor tissues vs. remote normal tissue (n = 19 pairs) | 16S V4 | Tumor tissues have reduced richness and diversity | Increased Koribacteraceae; and decreased Bacteroidaceae, Lachnospiraceae and Ruminococcaceae in normal tissues are associated with a better survival outcome |
| Yang [71] | Saliva | LC | Patients (n = 75) vs. healthy controls (n = 172); all female | 16S V1-V2 | Tumor tissues have reduced richness and diversity | Increased Sphingomonas and Blastomonas in LC patients |
| Liu [72] | Tumor | LC | LC-only (n = 11) vs. emphysema-only (n = 10) vs. both LC and emphysema (n = 19); all heavy smokers | 16S V4 | The emphysema-only group has a lower diversity | LC vs. emphysema-only: decreased Proteobacteria (primarily the genera Acinetobacter and Acidovorax); and increased Firmicutes (Streptococcus) and Bacteroidetes (Prevotella) |
| Greathouse [73] | Tumor | LC | Patients (n = 143) vs. healthy controls (n = 33) TCGA was used as a validation cohort | 16S V3-V5 | Control tissues have lower α-diversity | Acidovorax, Klebsiella, Rhodoferax and Anaerococcus are enriched in LUSC only |
| Tsay [74] | Lower airway brushing; buccal brushing | LC | Patients (n = 39) vs. non-cancer patients (n = 36) vs. healthy controls (n = 10) | 16S V4 | No differences (α- and β-diversity) in buccal samples Significant changes in β-diversity in Lower airway samples between LC and non-cancer/healthy controls | Streptococcus and Veillonella are highly enriched in the lower airways of LC patients and are associated with ERK and PI3K signaling pathway activation |
| Liu [75] | Bronchial specimen brushing | LC | Diseased lung and paired contralateral healthy lung (n = 24 pairs) vs. healthy controls (n = 8) | 16S V3–V4 | α-diversity reduces from healthy site to noncancerous to cancerous site | Genera Streptococcus and Neisseria are significantly more abundant in LC Genera Staphylococcus and Dialister are significantly more abundant in healthy controls |
| Cameron [76] | Sputum | LC | Patients (n = 4) vs. non-cancer controls (n = 6) | 16S | No significant changes in α-diversity | Streptococcus viridans and Granulicatella adiacens are significantly increased in LC patients |
| Lee [77] | BALF | LC | Patients (n = 20) vs. benign diseases (n = 8) | 16S V1-V3 | Increased diversity in LC | Phyla Firmicutes and TM7 are significantly increased in LC patients |
| Yu [23] | Tumor | LC | Tumor tissues (n = 31) vs. remote normal tissue (n = 165) | 16S V3–V4 | α-diversity is increased with environmental exposures, residence population, smoking and disease history LC has reduced diversity | Biomarkers for advanced LC: Genus Thermus Biomarkers for LC metastasis: Genus Legionella |
| Yan [78] | Saliva | LC | Patients (n = 10 for LUAD and n = 10 for LUSC) vs. healthy controls (n = 10) | 16S V3 and V6 | NA | Capnocytophaga and Veillonella are promising biomarkers for LUSC The abundance of Capnocytophaga, Selenomonas, Veillonella and Neisseria in saliva is significantly changed in LC patients |
| Hosgood [79] | Sputum; oral rinse | LC | Patients (n = 8) vs. healthy controls (n = 8) | 16S V1-V2 | The diversity between LC and control is similar in buccal samples, but significantly different in sputum | Granulicatella, Abiotrophia and Streptococcus are enriched in the sputum of LC patients |
2.2. Potential Reasons for the Few Commonalities across Studies
3. Airway or Respiratory Tract Microbiome and Lung Cancers
4. Frequently Altered Bacterial Genera in Lung Cancer Patients
4.1. Veillonella
4.2. Prevotella
4.3. Streptococcus
4.4. Acidovorax
4.5. Haemophilus
4.6. Capnocytophaga
4.7. Other Commonly Identified Lung Cancer-Associated Microbes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Peek, R.M., Jr.; Wilson, K.T. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e1326. [Google Scholar] [CrossRef] [PubMed]
- Kalaora, S.; Nagler, A.; Nejman, D.; Alon, M.; Barbolin, C.; Barnea, E.; Ketelaars, S.L.C.; Cheng, K.; Vervier, K.; Shental, N.; et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021, 592, 138–143. [Google Scholar] [CrossRef] [PubMed]
- WHO. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 23 October 2022).
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhang, R.; Rao, J.; Xiao, Y.; Zhang, Z.; Yang, B.; Cao, D.; Zhong, H.; Ning, P.; Shang, Y.; et al. Transcriptionally Active Lung Microbiome and Its Association with Bacterial Biomass and Host Inflammatory Status. mSystems 2018, 3, e00199-18. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, Y.; Ding, X.; Yang, Z.; He, L.; Ning, M.; Yang, Z.; He, D.; Yang, L.; Liu, Z.; et al. A Multi-Omics Study of Familial Lung Cancer: Microbiome and Host Gene Expression Patterns. Front. Immunol. 2022, 13, 827953. [Google Scholar] [CrossRef]
- Wong-Rolle, A.; Dong, Q.; Zhu, Y.; Divakar, P.; Hor, J.L.; Kedei, N.; Wong, M.; Tillo, D.; Conner, E.A.; Rajan, A.; et al. Spatial meta-transcriptomics reveal associations of intratumor bacteria burden with lung cancer cells showing a distinct oncogenic signature. J. Immunother. Cancer 2022, 10, e004698. [Google Scholar] [CrossRef]
- Reinhold, L.; Mollering, A.; Wallis, S.; Palade, E.; Schafer, K.; Dromann, D.; Rupp, J.; Graspeuntner, S.; Dalhoff, K. Dissimilarity of Airway and Lung Tissue Microbiota in Smokers undergoing Surgery for Lung Cancer. Microorganisms 2020, 8, 794. [Google Scholar] [CrossRef]
- Zheng, L.; Sun, R.; Zhu, Y.; Li, Z.; She, X.; Jian, X.; Yu, F.; Deng, X.; Sai, B.; Wang, L.; et al. Lung microbiome alterations in NSCLC patients. Sci. Rep. 2021, 11, 11736. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016, 17, 163. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, Z.; Li, C.; Lv, K.; Tian, G.; Tang, M.; Ji, L.; Yang, J. Bacterial biomarkers capable of identifying recurrence or metastasis carry disease severity information for lung cancer. Front. Microbiol. 2022, 13, 1007831. [Google Scholar] [CrossRef] [PubMed]
- Baranova, E.; Druzhinin, V.; Matskova, L.; Demenkov, P.; Volobaev, V.; Minina, V.; Larionov, A.; Titov, V. Sputum Microbiome Composition in Patients with Squamous Cell Lung Carcinoma. Life 2022, 12, 1365. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Tang, J.; Zhuang, R.; Meng, D.; Zhang, L.; Gu, C.; Teng, X.; Zhu, Z.; Liu, J.; Pang, J.; et al. The microbiome of lower respiratory tract and tumor tissue in lung cancer manifested as radiological ground-glass opacity. Front. Bioeng. Biotechnol. 2022, 10, 892613. [Google Scholar] [CrossRef] [PubMed]
- He, J.Q.; Chen, Q.; Wu, S.J.; Wang, D.Q.; Zhang, S.Y.; Zhang, S.Z.; Chen, R.L.; Wang, J.F.; Wang, Z.; Yu, C.H. Potential Implications of the Lung Microbiota in Patients with Chronic Obstruction Pulmonary Disease and Non-Small Cell Lung Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 937864. [Google Scholar] [CrossRef]
- Vogtmann, E.; Hua, X.; Yu, G.; Purandare, V.; Hullings, A.G.; Shao, D.; Wan, Y.; Li, S.; Dagnall, C.L.; Jones, K.; et al. The oral microbiome and lung cancer risk: An analysis of 3 prospective cohort studies. J. Natl. Cancer Inst. 2022, 114, 1501–1510. [Google Scholar] [CrossRef]
- Kim, O.H.; Choi, B.Y.; Kim, D.K.; Kim, N.H.; Rho, J.K.; Sul, W.J.; Lee, S.W. The microbiome of lung cancer tissue and its association with pathological and clinical parameters. Am. J. Cancer Res. 2022, 12, 2350–2362. [Google Scholar]
- Qian, K.; Deng, Y.; Krimsky, W.S.; Feng, Y.G.; Peng, J.; Tai, Y.H.; Peng, H.; Jiang, L.H. Airway Microbiota in Patients With Synchronous Multiple Primary Lung Cancer: The Bacterial Topography of the Respiratory Tract. Front. Oncol. 2022, 12, 811279. [Google Scholar] [CrossRef]
- Masuhiro, K.; Tamiya, M.; Fujimoto, K.; Koyama, S.; Naito, Y.; Osa, A.; Hirai, T.; Suzuki, H.; Okamoto, N.; Shiroyama, T.; et al. Bronchoalveolar lavage fluid reveals factors contributing to the efficacy of PD-1 blockade in lung cancer. JCI Insight 2022, 7, e157915. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Han, Y.; Zhao, X.; Sun, Y. Lung microbiota features of stage III and IV non-small cell lung cancer patients without lung infection. Transl. Cancer Res. 2022, 11, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E.A.; Filho, F.S.L.; Sin, D.D.; Lam, S.; Leung, J.M.; Lam, W.L. Distinct bronchial microbiome precedes clinical diagnosis of lung cancer. Mol. Cancer 2022, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Zhao, C.; Yu, M.; Chen, H.; Pan, Y.; Wang, Y.; Bao, H.; Ma, H.; Ma, S. Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered 2022, 13, 6665–6677. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Cheng, Z.; Yin, Z.; Xu, J.; Wu, F.; Jin, Y.; Yang, G. Airway Fusobacterium is Associated with Poor Response to Immunotherapy in Lung Cancer. Onco Targets Ther. 2022, 15, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Sun, Y.; Wang, S.; Liang, H.; Han, Y. Intratumoral Microbiota Impacts the First-Line Treatment Efficacy and Survival in Non-Small Cell Lung Cancer Patients Free of Lung Infection. J. Healthc. Eng. 2022, 2022, 5466853. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.H.; He, J.; Su, X.F.; Wen, Y.N.; Zhang, S.J.; Liu, L.Y.; Zhao, H.; Ye, C.P.; Wu, J.H.; Cai, S.; et al. The airway microbiota of non-small cell lung cancer patients and its relationship to tumor stage and EGFR gene mutation. Thorac. Cancer 2022, 13, 858–869. [Google Scholar] [CrossRef]
- Roy, P.; Sarma, A.; Kataki, A.C.; Rai, A.K.; Chattopadhyay, I. Salivary microbial dysbiosis may predict lung adenocarcinoma: A pilot study. Indian J. Pathol. Microbiol. 2022, 65, 123–128. [Google Scholar] [CrossRef]
- Dong, H.; Tan, Q.; Xu, Y.; Zhu, Y.; Yao, Y.; Wang, Y.; Li, C.; Li, H.; Zhang, G.; Xiong, Y.; et al. Convergent alteration of lung tissue microbiota and tumor cells in lung cancer. iScience 2022, 25, 103638. [Google Scholar] [CrossRef]
- Jang, H.J.; Choi, J.Y.; Kim, K.; Yong, S.H.; Kim, Y.W.; Kim, S.Y.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; Park, M.S.; et al. Relationship of the lung microbiome with PD-L1 expression and immunotherapy response in lung cancer. Respir. Res. 2021, 22, 322. [Google Scholar] [CrossRef]
- Boesch, M.; Baty, F.; Albrich, W.C.; Flatz, L.; Rodriguez, R.; Rothschild, S.I.; Joerger, M.; Fruh, M.; Brutsche, M.H. Local tumor microbial signatures and response to checkpoint blockade in non-small cell lung cancer. Oncoimmunology 2021, 10, 1988403. [Google Scholar] [CrossRef]
- Lu, H.; Gao, N.L.; Tong, F.; Wang, J.; Li, H.; Zhang, R.; Ma, H.; Yang, N.; Zhang, Y.; Wang, Y.; et al. Alterations of the Human Lung and Gut Microbiomes in Non-Small Cell Lung Carcinomas and Distant Metastasis. Microbiol. Spectr. 2021, 9, e0080221. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Hsu, M.H.; Tu, S.J.; Yen, J.C.; Lee, Y.T.; Fang, H.Y.; Chang, J.G. Metatranscriptomic Analysis of Human Lung Metagenomes from Patients with Lung Cancer. Genes 2021, 12, 1458. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, Y.; Xie, H.; Wang, X.; Wu, J.; Long, J.; Courtney, R.; Shu, X.O.; Zheng, W.; Blot, W.J.; et al. Association of oral microbiota with lung cancer risk in a low-income population in the Southeastern USA. Cancer Causes Control 2021, 32, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Seixas, S.; Kolbe, A.R.; Gomes, S.; Sucena, M.; Sousa, C.; Vaz Rodrigues, L.; Teixeira, G.; Pinto, P.; Tavares de Abreu, T.; Barbara, C.; et al. Comparative analysis of the bronchoalveolar microbiome in Portuguese patients with different chronic lung disorders. Sci. Rep. 2021, 11, 15042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Sun, Z.; Cao, Y.; Mu, Z.; Ji, X. Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci. 2021, 112, 3005–3017. [Google Scholar] [CrossRef] [PubMed]
- Dumont-Leblond, N.; Veillette, M.; Racine, C.; Joubert, P.; Duchaine, C. Non-small cell lung cancer microbiota characterization: Prevalence of enteric and potentially pathogenic bacteria in cancer tissues. PLoS ONE 2021, 16, e0249832. [Google Scholar] [CrossRef]
- Ma, Y.; Qiu, M.; Wang, S.; Meng, S.; Yang, F.; Jiang, G. Distinct tumor bacterial microbiome in lung adenocarcinomas manifested as radiological subsolid nodules. Transl. Oncol. 2021, 14, 101050. [Google Scholar] [CrossRef]
- Leng, Q.; Holden, V.K.; Deepak, J.; Todd, N.W.; Jiang, F. Microbiota Biomarkers for Lung Cancer. Diagnostics 2021, 11, 407. [Google Scholar] [CrossRef]
- Druzhinin, V.G.; Matskova, L.V.; Demenkov, P.S.; Baranova, E.D.; Volobaev, V.P.; Minina, V.I.; Larionov, A.V.; Titov, V.A.; Fucic, A. Genetic damage in lymphocytes of lung cancer patients is correlated to the composition of the respiratory tract microbiome. Mutagenesis 2021, 36, 143–153. [Google Scholar] [CrossRef]
- Hosgood, H.D.; Cai, Q.; Hua, X.; Long, J.; Shi, J.; Wan, Y.; Yang, Y.; Abnet, C.; Bassig, B.A.; Hu, W.; et al. Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax 2021, 76, 256–263. [Google Scholar] [CrossRef]
- Tsay, J.J.; Wu, B.G.; Sulaiman, I.; Gershner, K.; Schluger, R.; Li, Y.; Yie, T.A.; Meyn, P.; Olsen, E.; Perez, L.; et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov. 2021, 11, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.; An, T.; Zhang, C.; Wang, Z. Characterization of Microbiota in Cancerous Lung and the Contralateral Non-Cancerous Lung Within Lung Cancer Patients. Front. Oncol. 2020, 10, 1584. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, O.; Podlesnaya, P.; Rashidova, M.; Samoilova, D.; Petrenko, A.; Zborovskaya, I.; Mochalnikova, V.; Kataev, V.; Khlopko, Y.; Plotnikov, A.; et al. Lung Microbiome Differentially Impacts Survival of Patients with Non-Small Cell Lung Cancer Depending on Tumor Stroma Phenotype. Biomedicines 2020, 8, 349. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Z.; Wang, J.; Ding, C.; Sun, C.; Liu, P.; Xu, X.; Liu, Y.; Chen, B.; Gu, B. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 2020, 9, 693–704. [Google Scholar] [CrossRef]
- Mao, Q.; Ma, W.; Wang, Z.; Liang, Y.; Zhang, T.; Yang, Y.; Xia, W.; Jiang, F.; Hu, J.; Xu, L. Differential flora in the microenvironment of lung tumor and paired adjacent normal tissues. Carcinogenesis 2020, 41, 1094–1103. [Google Scholar] [CrossRef]
- Bello, S.; Vengoechea, J.J.; Ponce-Alonso, M.; Figueredo, A.L.; Minchole, E.; Rezusta, A.; Gambo, P.; Pastor, J.M.; Javier, G.; Del Campo, R. Core Microbiota in Central Lung Cancer With Streptococcal Enrichment as a Possible Diagnostic Marker. Arch. Bronconeumol. 2021, 57, 681–689. [Google Scholar] [CrossRef]
- Druzhinin, V.G.; Matskova, L.V.; Demenkov, P.S.; Baranova, E.D.; Volobaev, V.P.; Minina, V.I.; Apalko, S.V.; Churina, M.A.; Romanyuk, S.A.; Shcherbak, S.G.; et al. Taxonomic diversity of sputum microbiome in lung cancer patients and its relationship with chromosomal aberrations in blood lymphocytes. Sci. Rep. 2020, 10, 9681. [Google Scholar] [CrossRef]
- Wong, L.M.; Shende, N.; Li, W.T.; Castaneda, G.; Apostol, L.; Chang, E.Y.; Ongkeko, W.M. Comparative Analysis of Age- and Gender-Associated Microbiome in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Cancers 2020, 12, 1447. [Google Scholar] [CrossRef]
- Bingula, R.; Filaire, E.; Molnar, I.; Delmas, E.; Berthon, J.Y.; Vasson, M.P.; Bernalier-Donadille, A.; Filaire, M. Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: A cross-sectional clinical trial. Respir. Res. 2020, 21, 129. [Google Scholar] [CrossRef]
- Patnaik, S.K.; Cortes, E.G.; Kannisto, E.D.; Punnanitinont, A.; Dhillon, S.S.; Liu, S.; Yendamuri, S. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2021, 161, 419–429.e416. [Google Scholar] [CrossRef]
- Ekanayake, A.; Madegedara, D.; Chandrasekharan, V.; Magana-Arachchi, D. Respiratory Bacterial Microbiota and Individual Bacterial Variability in Lung Cancer and Bronchiectasis Patients. Indian J. Microbiol. 2020, 60, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Su, X.; Yuan, M.; Zhang, S.; He, J.; Deng, Q.; Qiu, W.; Dong, H.; Cai, S. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am. J. Cancer Res. 2019, 9, 2047–2063. [Google Scholar] [PubMed]
- Jin, J.; Gan, Y.; Liu, H.; Wang, Z.; Yuan, J.; Deng, T.; Zhou, Y.; Zhu, Y.; Zhu, H.; Yang, S.; et al. Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer patients: A multiple comparative study design with independent validation. Lung Cancer 2019, 136, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.; Cavadas, B.; Ferreira, J.C.; Marques, P.I.; Monteiro, C.; Sucena, M.; Sousa, C.; Vaz Rodrigues, L.; Teixeira, G.; Pinto, P.; et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019, 9, 12838. [Google Scholar] [CrossRef]
- Ren, Y.; Su, H.; She, Y.; Dai, C.; Xie, D.; Narrandes, S.; Huang, S.; Chen, C.; Xu, W. Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules. Transl. Lung Cancer Res. 2019, 8, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Luo, J.; Dong, X.; Zhao, S.; Hao, Y.; Peng, C.; Shi, H.; Zhou, Y.; Shan, L.; Sun, Q.; et al. Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J. Cancer 2019, 10, 1651–1662. [Google Scholar] [CrossRef]
- Wang, K.; Huang, Y.; Zhang, Z.; Liao, J.; Ding, Y.; Fang, X.; Liu, L.; Luo, J.; Kong, J. A Preliminary Study of Microbiota Diversity in Saliva and Bronchoalveolar Lavage Fluid from Patients with Primary Bronchogenic Carcinoma. Med. Sci. Monit. 2019, 25, 2819–2834. [Google Scholar] [CrossRef]
- Hosgood, H.D., 3rd; Mongodin, E.F.; Wan, Y.; Hua, X.; Rothman, N.; Hu, W.; Vermeulen, R.; Seow, W.J.; Rohan, T.; Xu, J.; et al. The respiratory tract microbiome and its relationship to lung cancer and environmental exposures found in rural china. Environ. Mol. Mutagen. 2019, 60, 617–623. [Google Scholar] [CrossRef]
- Peters, B.A.; Hayes, R.B.; Goparaju, C.; Reid, C.; Pass, H.I.; Ahn, J. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 731–740. [Google Scholar] [CrossRef]
- Yang, J.; Mu, X.; Wang, Y.; Zhu, D.; Zhang, J.; Liang, C.; Chen, B.; Wang, J.; Zhao, C.; Zuo, Z.; et al. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front. Oncol. 2018, 8, 520. [Google Scholar] [CrossRef]
- Liu, Y.; O’Brien, J.L.; Ajami, N.J.; Scheurer, M.E.; Amirian, E.S.; Armstrong, G.; Tsavachidis, S.; Thrift, A.P.; Jiao, L.; Wong, M.C.; et al. Lung tissue microbial profile in lung cancer is distinct from emphysema. Am. J. Cancer Res. 2018, 8, 1775–1787. [Google Scholar] [PubMed]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I.; et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Tsay, J.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Tao, L.L.; Zhang, J.; Zhu, Y.G.; Zheng, Y.; Liu, D.; Zhou, M.; Ke, H.; Shi, M.M.; Qu, J.M. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer 2018, 142, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.J.S.; Lewis, K.E.; Huws, S.A.; Hegarty, M.J.; Lewis, P.D.; Pachebat, J.A.; Mur, L.A.J. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS ONE 2017, 12, e0177062. [Google Scholar] [CrossRef]
- Lee, S.H.; Sung, J.Y.; Yong, D.; Chun, J.; Kim, S.Y.; Song, J.H.; Chung, K.S.; Kim, E.Y.; Jung, J.Y.; Kang, Y.A.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef]
- Yan, X.; Yang, M.; Liu, J.; Gao, R.; Hu, J.; Li, J.; Zhang, L.; Shi, Y.; Guo, H.; Cheng, J.; et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 2015, 5, 3111–3122. [Google Scholar]
- Hosgood, H.D., 3rd; Sapkota, A.R.; Rothman, N.; Rohan, T.; Hu, W.; Xu, J.; Vermeulen, R.; He, X.; White, J.R.; Wu, G.; et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 2014, 55, 643–651. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Han, Y.; Zhao, X.; Sun, Y. Characteristics of pathogenic microbes in lung microenvironment of lung cancer patients without respiratory infection. J. BUON 2021, 26, 1862–1870. [Google Scholar]
- Rodriguez, R.M.; Hernandez, B.Y.; Menor, M.; Deng, Y.; Khadka, V.S. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput. Struct. Biotechnol. J. 2020, 18, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Pragman, A.A.; Wendt, C. Biomarkers and the microbiome in the detection and treatment of early-stage non-small cell lung cancer. Semin. Oncol. 2022, 49, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Ying, K.L.; Brasky, T.M.; Freudenheim, J.L.; McElroy, J.P.; Nickerson, Q.A.; Song, M.A.; Weng, D.Y.; Wewers, M.D.; Whiteman, N.B.; Mathe, E.A.; et al. Saliva and Lung Microbiome Associations with Electronic Cigarette Use and Smoking. Cancer Prev. Res. 2022, 15, 435–446. [Google Scholar] [CrossRef]
- Berger, G.; Wunderink, R.G. Lung microbiota: Genuine or artifact? Isr. Med. Assoc. J. 2013, 15, 731–733. [Google Scholar]
- Ran, Z.; Liu, J.; Wang, F.; Xin, C.; Shen, X.; Zeng, S.; Song, Z.; Xiong, B. Analysis of Pulmonary Microbial Diversity in Patients with Advanced Lung Cancer Based on High-throughput Sequencing Technology. Zhongguo Fei Ai Za Zhi 2020, 23, 1031–1038. [Google Scholar] [CrossRef]
- Scrima, M.; De Marco, C.; Fabiani, F.; Franco, R.; Pirozzi, G.; Rocco, G.; Ravo, M.; Weisz, A.; Zoppoli, P.; Ceccarelli, M.; et al. Signaling networks associated with AKT activation in non-small cell lung cancer (NSCLC): New insights on the role of phosphatydil-inositol-3 kinase. PLoS ONE 2012, 7, e30427. [Google Scholar] [CrossRef]
- Gustafson, A.M.; Soldi, R.; Anderlind, C.; Scholand, M.B.; Qian, J.; Zhang, X.; Cooper, K.; Walker, D.; McWilliams, A.; Liu, G.; et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010, 2, 26ra25. [Google Scholar] [CrossRef]
- Pustelny, C.; Komor, U.; Pawar, V.; Lorenz, A.; Bielecka, A.; Moter, A.; Gocht, B.; Eckweiler, D.; Musken, M.; Grothe, C.; et al. Contribution of Veillonella parvula to Pseudomonas aeruginosa-mediated pathogenicity in a murine tumor model system. Infect. Immun. 2015, 83, 417–429. [Google Scholar] [CrossRef]
- Kononen, E.; Gursoy, U.K. Oral Prevotella Species and Their Connection to Events of Clinical Relevance in Gastrointestinal and Respiratory Tracts. Front. Microbiol. 2021, 12, 798763. [Google Scholar] [CrossRef]
- Shimizu, M.; Miyanaga, A.; Seike, M.; Matsuda, K.; Matsumoto, M.; Noro, R.; Fujita, K.; Mano, Y.; Furuya, N.; Kubota, K.; et al. The respiratory microbiome associated with chronic obstructive pulmonary disease comorbidity in non-small cell lung cancer. Thorac. Cancer 2022, 13, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Adar, S.D.; Huffnagle, G.B.; Curtis, J.L. The respiratory microbiome: An underappreciated player in the human response to inhaled pollutants? Ann. Epidemiol. 2016, 26, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.I.; Cooks, T.; Vega-Valle, E.; Vetizou, M.; Rose, U.; Miyanaga, A.; Trehan, A.; Oike, T.; Ryan, B.M.; Sen, S.J.; et al. Abstract PR07: Role of the microbiota in inflammation and lung cancer. Clin. Cancer Res. 2018, 24, PR07. [Google Scholar] [CrossRef]
- Zhang, C.; von Muhlinen, N.; Romina, A.; Stone, J.; Goldszmid, R.; Harris, C. Acidovorax temperans modulates the host immune microenvironment to promote lung cancer development. Cancer Res. 2021, 81, 1782. [Google Scholar] [CrossRef]
- Kosikowska, U.; Biernasiuk, A.; Rybojad, P.; Los, R.; Malm, A. Haemophilus parainfluenzae as a marker of the upper respiratory tract microbiota changes under the influence of preoperative prophylaxis with or without postoperative treatment in patients with lung cancer. BMC Microbiol. 2016, 16, 62. [Google Scholar] [CrossRef]
- Middleton, A.M.; Dowling, R.B.; Mitchell, J.L.; Watanabe, S.; Rutman, A.; Pritchard, K.; Tillotson, G.; Hill, S.L.; Wilson, R. Haemophilus parainfluenzae infection of respiratory mucosa. Respir. Med. 2003, 97, 375–381. [Google Scholar] [CrossRef]
- Thirumala, R.; Rappo, U.; Babady, N.E.; Kamboj, M.; Chawla, M. Capnocytophaga lung abscess in a patient with metastatic neuroendocrine tumor. J. Clin. Microbiol. 2012, 50, 204–207. [Google Scholar] [CrossRef][Green Version]
- Dohlman, A.B.; Klug, J.; Mesko, M.; Gao, I.H.; Lipkin, S.M.; Shen, X.; Iliev, I.D. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell 2022, 185, 3807–3822.e12. [Google Scholar] [CrossRef]
- Cai, H.Z.; Zhang, H.; Yang, J.; Zeng, J.; Wang, H. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J. Med. Virol. 2021, 93, 5126–5133. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Huang, J. Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations. J. Clin. Med. 2022, 11, 7298. https://doi.org/10.3390/jcm11247298
Huang J, Huang J. Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations. Journal of Clinical Medicine. 2022; 11(24):7298. https://doi.org/10.3390/jcm11247298
Chicago/Turabian StyleHuang, Jiawen, and Juan Huang. 2022. "Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations" Journal of Clinical Medicine 11, no. 24: 7298. https://doi.org/10.3390/jcm11247298
APA StyleHuang, J., & Huang, J. (2022). Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations. Journal of Clinical Medicine, 11(24), 7298. https://doi.org/10.3390/jcm11247298






