Abstract
Recent studies on the urine microbiome have highlighted the importance of the gut–vagina–bladder axis in recurrent urinary tract infection (rUTI). In particular, the role of Gardnerella as a covert pathogen that activates E. coli in animal experiments has been reported. Herein, we conducted a human bladder microbiome study to investigate the effect of Gardnerella on rUTI. Urine 16S ribosomal RNA gene sequencing via transurethral catheterization was conducted in the normal control group (NC) (n = 18) and rUTI group (n = 78). The positive detection rate of Gardnerella species did not differ between the NC and rUTI groups (22.2% vs. 18.0%, p = 0.677). In addition, the Gardnerella-positive NC and Gardnerella-positive rUTI groups showed similar levels of microbiome diversity. The Gardnerella-positive group was categorized into three subgroups: the Escherichia-dominant group, Gardnerella-dominant group, and Lactobacillus-dominant group. All of the Escherichia-dominant groups were associated with rUTI. The Gardnerella-dominant or Lactobacillus-dominant groups expressed rUTI with symptoms when risk factors such as the degree of Gardnerella proliferation or causative agents of bacterial vaginosis were present. The presence of Gardnerella in the urine is considered to be related to rUTI depending on other risk factors. New guideline recommendations regarding antibiotic selection based on a novel method to detect the cause of rUTI may be required to reduce antibiotic resistance.
1. Introduction
Urinary tract infections (UTI) are approximately eight times more common in women than in men [1], and 50–60% of adult women will experience at least one UTI in their lifetime [2,3]. Of these, 30% of UTI patients develop recurrent urinary tract infection (rUTI) within 6 months [2,3]. Clinically, UTI can develop into systemic diseases such as severe kidney disease (acute kidney injury, kidney atrophy, end-stage renal disease) and sepsis. Moreover, UTI is problematic as it can affect quality of life or cause depression [4,5,6]. UTI treatment is based on the assumption that urine is sterile, and antibiotics are the treatment of choice based on standard urine culture. However, in reality, the causative bacteria of many patients may not be detected by standard urine culture methods, and rUTI develops in approximately 25–30% of patients despite the use of antibiotics [7].
Traditionally, colonization of the vaginal introitus or periurethra by pathogenic gut bacteria is known to cause rUTI by retrograde infection [8,9]. Due to this gut–bladder axis etiology, the vaginal microbiome, particularly Gardnerella vaginalis, is frequently excluded as a causative agent of UTIs, although they can also cause the infection. The clinical significance of G. vaginalis has been underestimated to date, due to its low detection rate in traditional urine culture [10]. In general, uropathogenic Escherichia coli (UPEC) is known as the most common causative agent of rUTI, and thus clinical studies on polyinfection by Gram-positive bacteria or co-infective UTI by vaginal microbiota are few in number [11,12].
Recently, 16S ribosomal RNA gene amplification and sequencing and the enhanced quantitative urine culture (EQUC) technique have revealed that a complex bladder microbiome exists in addition to classical bacteria [13]. Through a previous pilot study, we demonstrated the difference in bacterial diversity and patterns between rUTI and acute uncomplicated cystitis [14]. Due to these new techniques, a new disease pathway through gut–vagina–bladder crosstalk has gained focus, apart from the traditional mechanism of UTI, which was mainly caused by gut–bladder crosstalk [8,9,15,16]. Such changes in study perspectives can provide good therapeutic clues, particularly in rUTI, which is difficult to treat with frequent recurrence.
Recent animal studies have shown that Gardnerella, a major pathogen of bacterial vaginosis, triggers UPEC and causes rUTI [15,17]. Based on these results, we conducted a 16S ribosomal RNA gene sequencing study to investigate the differences in the urinary microbiome of rUTI and the effect of Gardnerella, a major strain of bacterial vaginalis, in patients with rUTI.
2. Materials and Methods
2.1. Patients and Study Protocol
Between April 2020 and June 2021, we collected information on patients who underwent the urine next-generation sequencing (NGS) test, 16S ribosomal RNA gene amplification. Patients who fulfilled the following inclusion criteria were eligible for this study: (a) patients 20 years of age or older, (b) who underwent urinalysis, urine culture, and urine NGS. Patients with single acute uncomplicated cystitis, anatomical or structural abnormalities such as a prolonged indwelling catheter, pregnancy, or urinary stone were excluded from the study. In addition, patients with intrauterine contraceptive devices, vaginitis, and vaginal discharge were excluded. To prevent the study results from being affected by changes in the microbiological environment in the bladder due to antibiotics, patients who were prescribed antibiotics for recurrent UTI or who had taken them within the last 4 weeks were excluded from this study. As a result, 96 patients who met the criteria were included in the study.
The patients were divided into two groups according to the following definitions. The normal group consisted of patients displaying no cystitis symptoms for at least the past year, and those that underwent a urine NSG test for the purpose of examination at a health promotion center, with no abnormal findings in abdominal image or urinalysis. The rUTI group was defined as consecutive patients who visited the outpatient clinic with symptomatic rUTI between April 2020 and June 2021. rUTI was defined as positive repetitive urine cultures (twice in six months, or three times in a year) with typical cystitis symptoms (low abdominal discomfort or pain, dysuria, frequency, urgency, hematuria, postvoid sense of residual urine). The previous urine culture results of rUTI patients were verified as medical records, and the urine culture positive rate was 52%. The most common causative bacteria were E. coli 64%, Klebsiella 10%, Enterococcus 8%, S. aureus 4%, Streptococcus 3%, and others, such as Proteus. However, in 48% of rUTI cases, the causative organism was not identified in urine culture even though the patient complained of urinary symptoms. Therefore, we found that it is difficult to diagnose the cause of rUTI patients using traditional culture methods only. This suggests that strains other than the traditionally known strains may affect rUTI. Therefore, in a total of 78 patients (30 patients with 2 or more occurrences in 6 months and 48 patients with 3 or more occurrences within 1 year), NGS sequencing and urine culture were performed simultaneously at the time of the next UTI recurrence.
The study protocol was approved by the Institutional Review Board of Soonchunhyang University Bucheon Hospital (IRB number SCHBC 2021-10-011-01). The study protocol conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki. Informed consent was waived from the IRB of Soonchunhyang University Bucheon Hospital due to the retrospective design of this study.
2.2. DNA Extraction and 16S rDNA Sequencing
The overall process is similar to the previous research protocol [14]. All urine samples were collected by transurethral catheters. Upon collection, samples were immediately sent to the lab to be stored and refrigerated with boric acid, and transported to the genetic lab within one day. The urine specimens (25 mL) were centrifuged at 3300× g rpm for 30 min, and the urinary pellets were harvested and processed for DNA extraction using the MagMAX™ Microbiome Ultra Nucleic Acid Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. To check the contamination of the DNA extraction process, we performed a negative DNA extraction consisting of only reagents without urine specimens during the test setup.
Prepared DNA was used for 16S library construction using the NEXTflex 16S V4 Amplicon-Seq (Bioo Scientific, Austin, TX, USA), according to the manufacturer’s protocol. In detail, the amplification cycle was 8 cycles for PCR I amplification and 22 cycles for PCR II amplification, and each PCR clean-up used AMPure XP beads to purify the 16S V4 amplicon assay from free primers and primer dimer species. Final library products were quantified using Agilent D1000 screentape (Agilent Technologies, Santa Clara, CA, USA) and diluted to 4nM. Diluted library were pooled together and sequenced with the Miseq system (Illumina) using a paired-end 500-cycle kit.
2.3. Bioinformatics Analysis and Data Processing
We used QIIME 2 to analyze the 16S sequence data. Demultiplexed and primer-trimmed data were quality-filtered and denoised using DADA2 [18,19]. Amplicon sequence variants (ASVs) with fewer than 10 reads or present in only a single sample were removed, and taxonomy was assigned to each ASV using the naive Bayes machine learning taxonomy classifiers in the q2 feature classifier against the NCBI RefSeq database with taxonomic weight assembly using q2 clawback [20,21].
Contamination was removed separately for each pair of urine sample and negative control with the R package microDecon [22], which uses the proportions of contaminant operational taxonomic units (OTUs) or amplicon sequence variants (ASVs) in blank samples to systematically identify and remove contaminant reads from metabarcoding data sets. Although we applied decontamination bioinformatic pipelines, it was considered that the applying a relative abundance cut-off 1% was appropriate to more strictly identify the core microbiome as referring to other studies targeting low-biomass samples [23,24].
2.4. Statistical Analysis
The proportions of Gardnerella (+) urinary microbiota samples of the NC group and rUTI group were compared using the Chi-squared test. Next, we compared the characteristics of the microbial community between the Gardnerella (+) NC group and Gardnerella (+) rUTI group. Differences in alpha diversity between the Gardnerella (+) NC group and Gardnerella (+) rUTI group were analyzed based on Shannon’s diversity index using the Wilcoxon test. Principal coordinates analysis based on weighted Unifrac distances was used to construct a visualization of the data. Permutational multivariate analysis of variance (PERMANOVA) [23], implemented in the adonis function of the R/vegan package (v2.5–2, R version 4.1.0; The R Foundation for Statistical Computing, Vienna, Austria), was performed to identify the microbial community dissimilarity of the Gardnerella (+) NC group and Gardnerella (+) rUTI group. To discriminate Gardnerella (+) urine samples into subgroups according to microbial community similarity regardless of the disease group, we used the k-medoids clustering algorithm, which clusters samples with the smallest total pairwise distance [25]. The number of clusters was assessed with the silhouette method [26]. In addition, hierarchical clustering using complete linkage was performed to visualize relationships among the samples based on the similarity of microbial composition. The procedure was also operated on a phylogenetically informed distance matrix, which was computed using the weighted UniFrac metric. Through hierarchical clustering, a heatmap of the relative abundance of the compositional genera based on Spearman´s correlation coefficient was represented and each subgroup was named on the basis of the dominant member of the respective subgroup.
3. Results
3.1. Baseline Characteristics
The clinical characteristics and the urinalysis results of the patients are presented in Table 1. The patients were all female and the mean age was 54.5 ± 14.9 years. The NC group consisted of 18 patients, and the rUTI group was of 78 patients. The mean age of the rUTI group was slightly higher than that of the control group, but it was not statistically significant (56.2 vs. 47.1 years, p = 0.291). There were no differences between the groups in terms of menopause or diabetes.

Table 1.
Baseline characteristics of patients.
3.2. Gardnerella Positive Detection Rate in the NC and rUTI Groups
Next, we compared the positive detection rate of Gardnerella in the NC and rUTI groups (Table 2). Gardnerella was detected in 18 of 96 patients (18.8%). Regarding the positive detection rate, there was no significant difference between the two groups, with 22.2% in the NC group and 18.0% in the rUTI group (p = 0.677).

Table 2.
Positive detection rate of Gardnerella species.
3.3. Microbiome Diversity of the NC and rUTI Groups
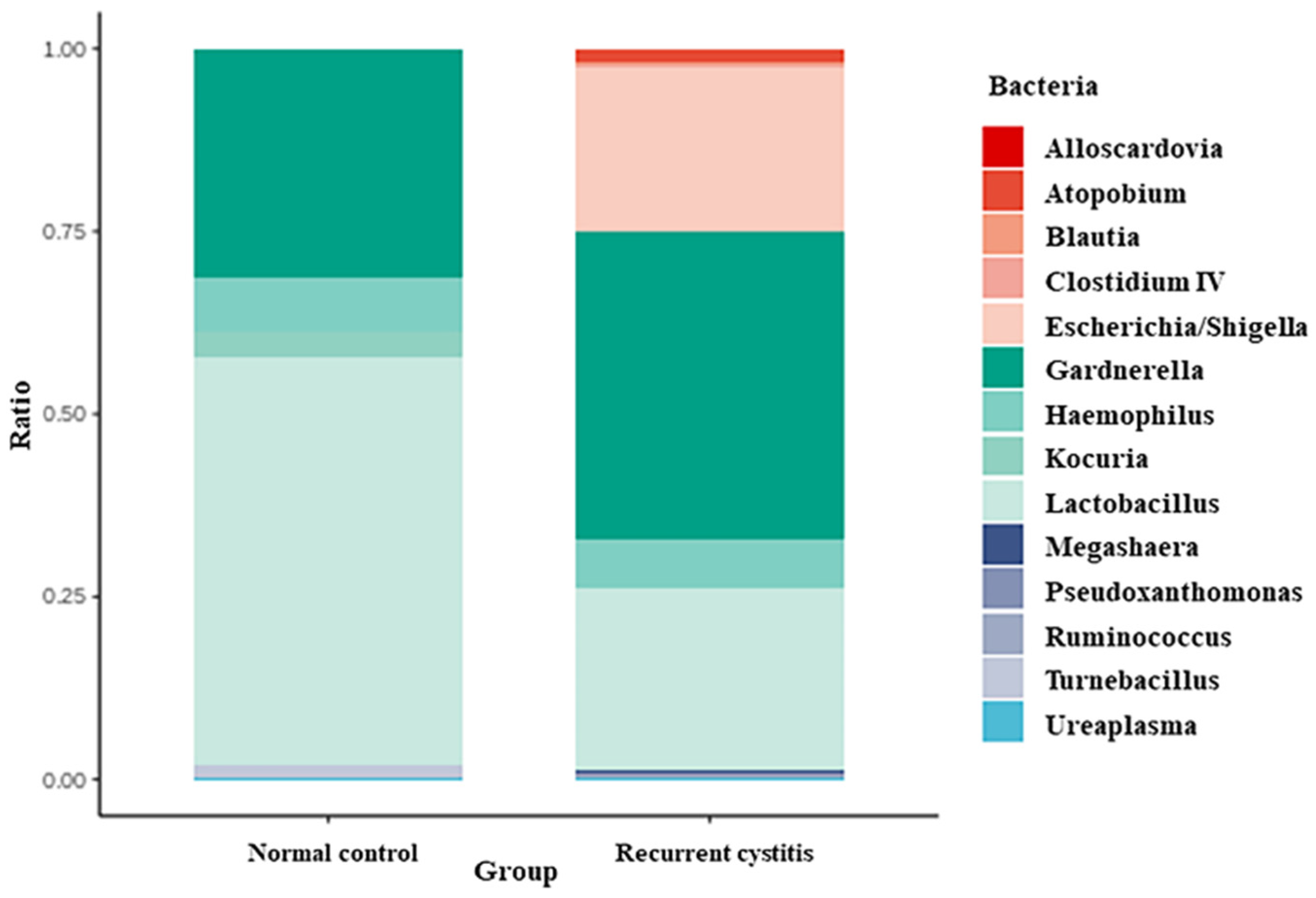
Bacteria frequently detected in Gardnerella-positive patients were classified according to the NC group and rUTI group, respectively (Table 3, Figure 1). In the Gardnerella-positive NC group, Lactobacillus (55.67%), Gardnerella (30.94%), Haemophilus (7.58%), and Kocuria (3.6%) groups were frequently detected. On the other hand, in the Gardnerella-positive rUTI group, Gardnerella (42.18%), Lactobacillus (24.87%), Escherichia (22.57%), and Haemophilus (6.52%) groups were frequently detected. In particular, Atopobium, Megasphaera, and Ureaplasma, known to be associated with bacterial vaginosis, were detected only in the rUTI group.

Table 3.
Contribution of most abundant urinary bacterial genera to Gardnerella positive group.
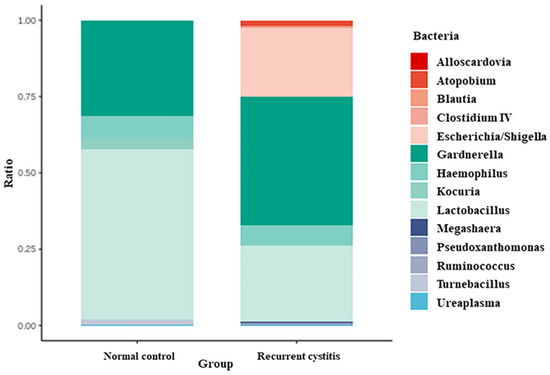
Figure 1.
Relative abundance of urinary microbiota in Gardnerella (+) normal control group and Gardnerella (+) recurrent UTI group.
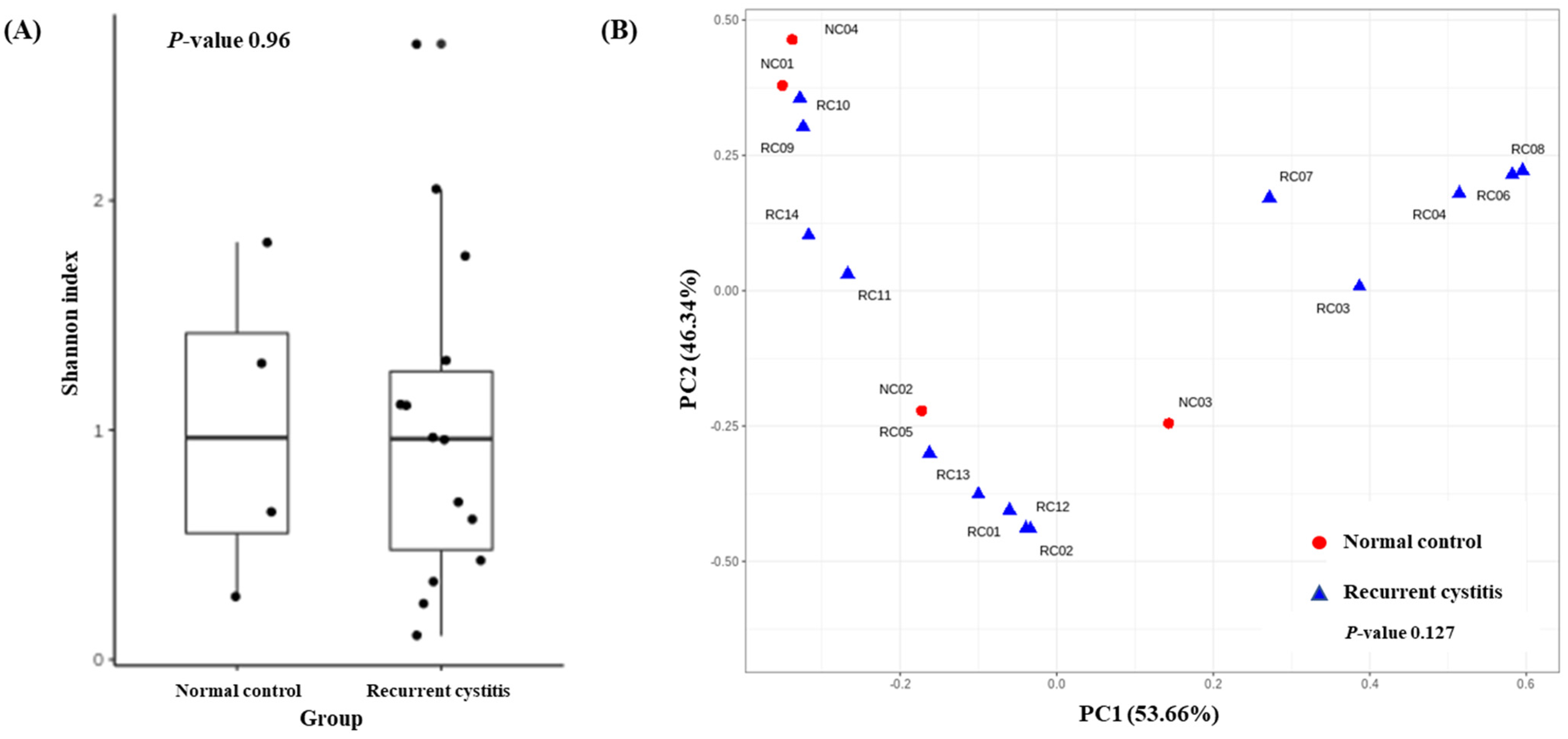
To determine the distribution of various microorganisms present in one sample (alpha diversity), we calculated the Shannon index (Figure 2A). There was no significant difference regarding alpha diversity between the rUTI group and NC group (p = 0.96). To determine whether the microbial community was different between the two cystitis groups (beta diversity), we evaluated the weighted UniFrac distances (Figure 2B). The composition of the microbiome did not differ between the two groups (p = 0.127).
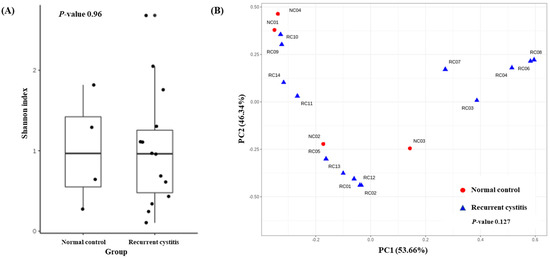
Figure 2.
(A) Alpha diversity and (B) principal coordinate analysis based on weighted UniFrac distances in Gardnerella (+) normal control group and Gardnerella (+) recurrent UTI group.
3.4. Three Urotypes of Bladder Microbiome Associated with Gardnerella
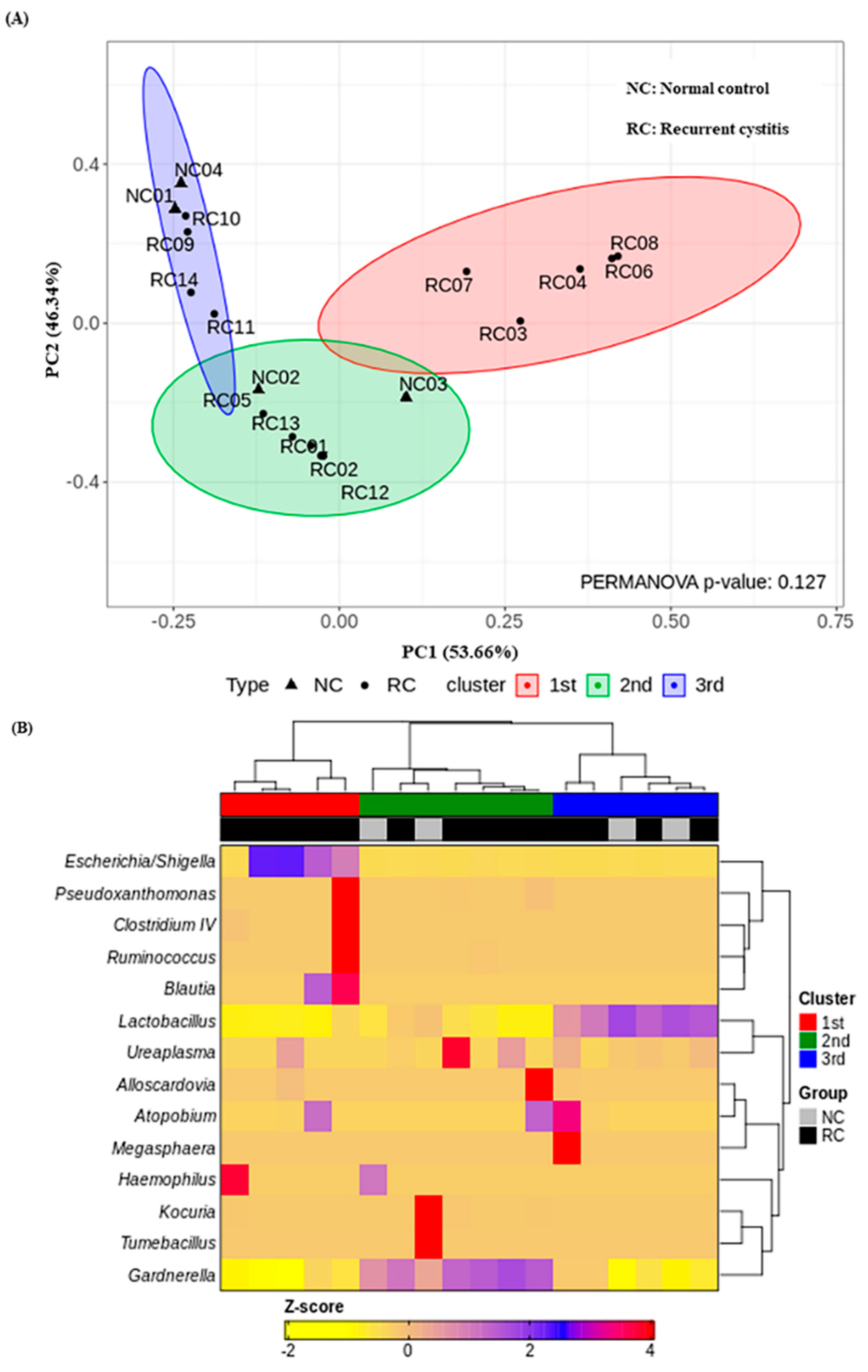
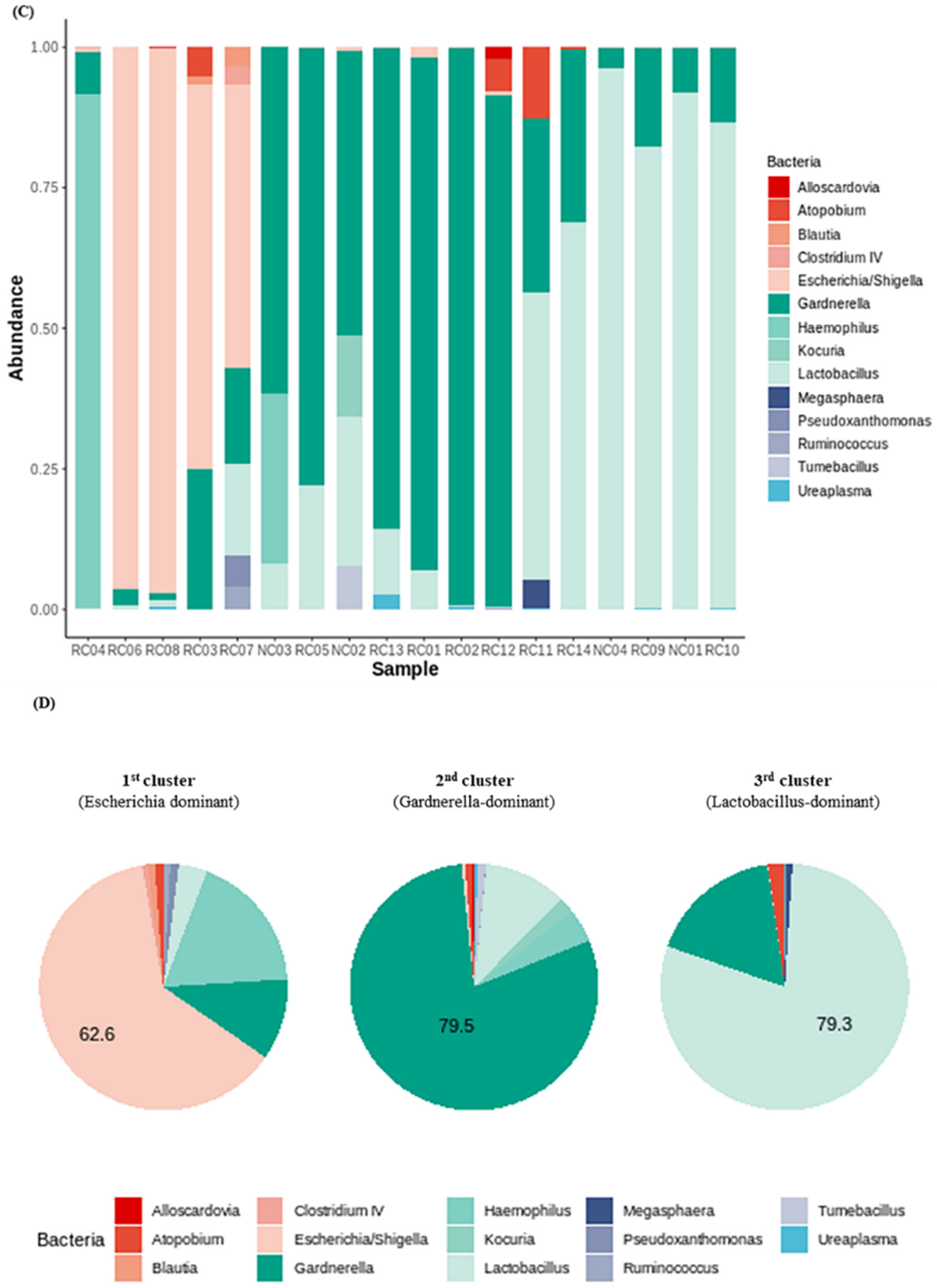
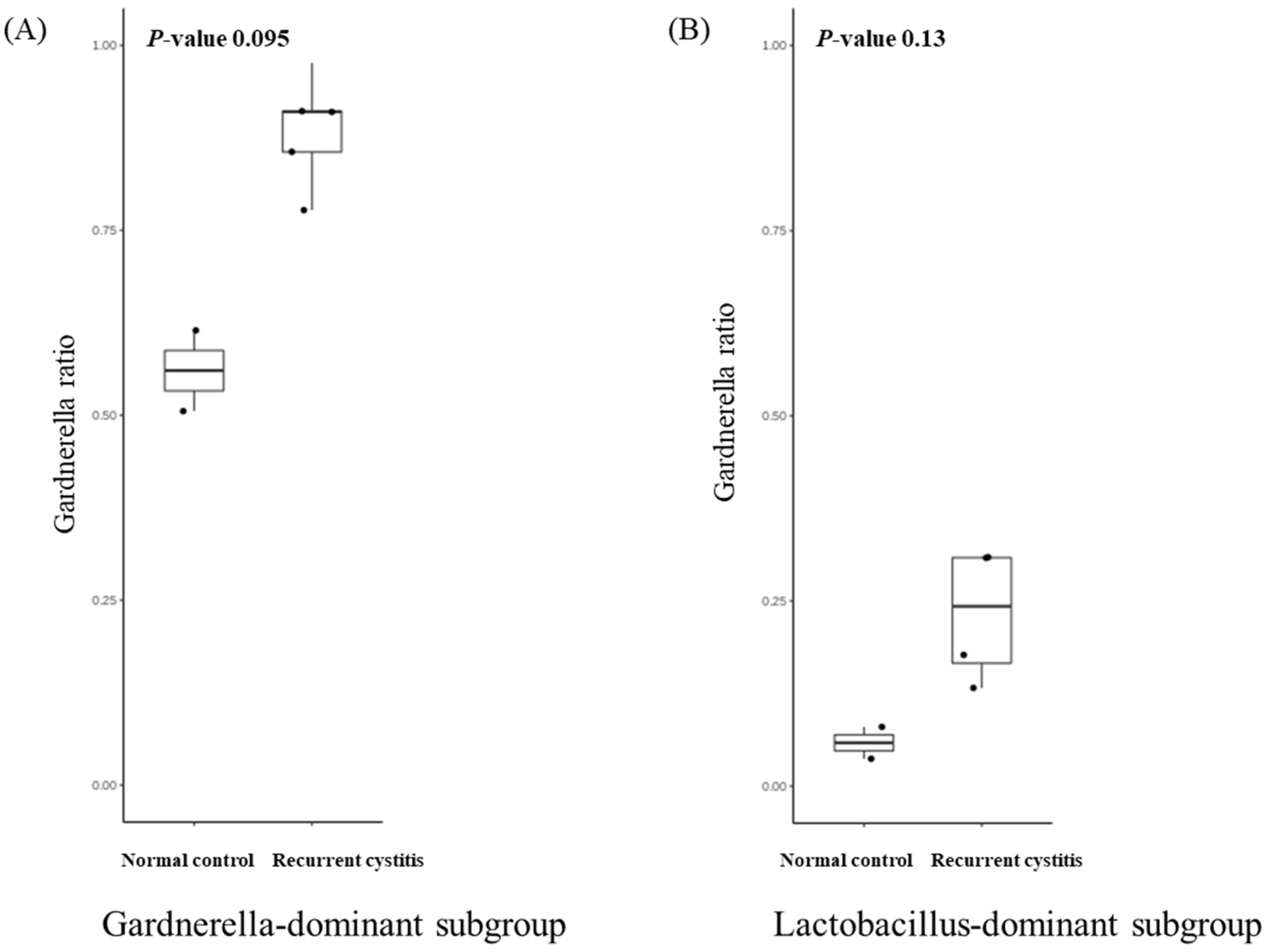
We investigated the presence of any patterns in the Gardnerella-positive group regardless of NC or rUTI status. The distribution of Gardnerella (+) urinary microbiota was analyzed with K-medoids clustering (Figure 3A), hierarchical clustering (Figure 3B), and a bar plot (Figure 3C), and was classified into three groups as follows (Figure 3D): Group 1 Escherichia urotype (Gardnerella is very few and Escherichia is dominant), Group 2 Gardnerella urotype (Gardnerella accounted for more than 50%), and Group 3 Lactobacillus urotype (Gardnerella is present in some cases, but Lactobacillus is predominant in more than 50%). All of Group 1 (Escherichia urotype) was associated with rUTI (5 rUTI, 0 NC). In Group 2 (Gardnerella urotype: 5 rUTI, 2 NC) or Group 3 (Lactobacillus urotype: 4 rUTI, 2 NC), both rUTI and asymptomatic NC were present (Figure 3A). However, in Group 2 and Group 3, the proportion of Gardnerella was higher in the rUTI group than the NC group, although not significant (Group 2, p = 0.095; Group 3, p = 0.13) (Figure 4).
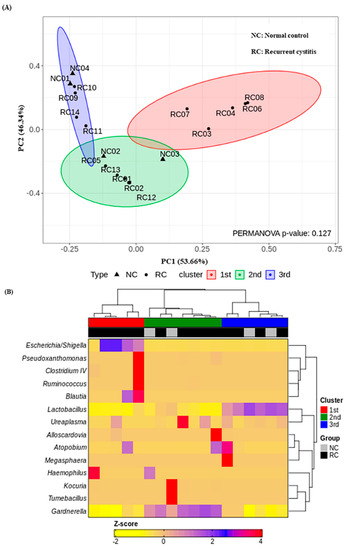
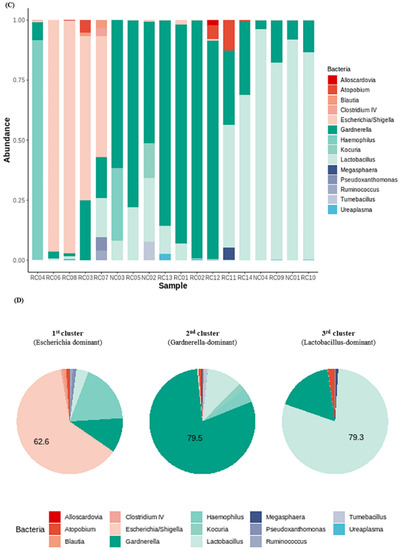
Figure 3.
Gardnerella (+) urinary microbiota revealed three distinct subgroups by (A) K-medoids clustering and (B) hierarchical clustering in R program version 4.1.2 (The R Foundation for Statistical Computing, Vienna, Austria; https://svn.r-project.org/R-packages/trunk/cluster, accessed on 17 March 2021), (C) bar plot, (D) pie chart.
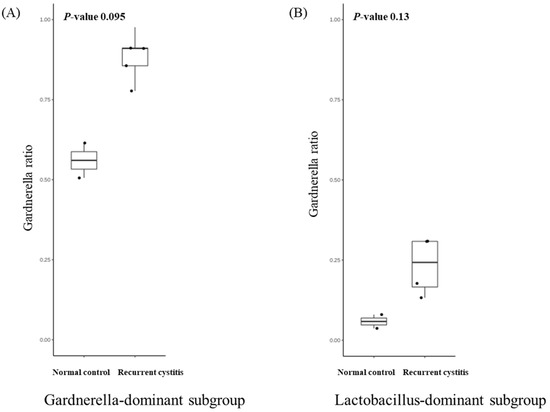
Figure 4.
Relative abundance of Gardnerella in (A) Gardnerella-dominant subgroup and (B) Lactobacillus-dominant subgroup.
4. Discussion
In this study, we found that there was no significant difference in urine microbiota results between the Gardnerella-positive NC group and Gardnerella-positive rUTI group. The Gardnerella-positive group could be divided into three urotypes: (1) Escherichia-dominant group, (2) Gardnerella-dominant group, and (3) Lactobacillus-dominant group. All Escherichia-dominant groups were associated with rUTI. In the Gardnerella-dominant and Lactobacillus-dominant groups, the NC and rUTI groups were mixed. In particular, bacterial vaginosis-associated strains such as Atopobium, Megasphaera, and Ureaplasma were detected only in the rUTI group.
Our research group has completed two papers related to the urine microbiome. The core of the first paper was that E. coli was the most causative strain in acute and recurrent cystitis, but the base from which E. coli grew, i.e., the bladder condition (commensal or pathogenic organism), was completely different [14]. In other words, the first paper was a study on the E. coli-dominant urotype. In this paper, the second study, we found the possibility that Gardnerella in the bladder could influence the uncertain pathophysiology of the dominant urotype of recurrent urinary tract infection.
A recent UTI guideline is based on antibiotic treatment based on urine culture for rUTI. In particular, continuous low-dose antimicrobial prophylaxis is recommended if rUTI persists after behavioral interventions have failed. In reality, 75% of the patients in clinical practice with rUTI are taking empirical antibiotics without undergoing tests [27]. Consequently, antibiotic resistance has increased while rUTI prevalence has not decreased [28].
Recently, through the new bacterial technique (16s RNA sequencing, EQUC), the classical perception of the bladder microbiome is being reconsidered, and the role of various microorganisms in urinary tract infection needs to be re-established [7,14]. According to a previous classical urine culture-based study, the most common causative bacteria of uncomplicated UTI were E. coli (58%), mixed flora (13.4%), and K. pneumoniae (6.5%) [29]. However, the pattern was different in our group’s previous pilot study [14]. This study, based on NGS, additionally discovered Sphingomonas, Staphylococcus, Streptococcus, and Rothia spp., which were not found in the existing culture, especially for the rUTI group. Although the NGS test has better diagnostic yield than the existing tests, it should also be noted that there are several drawbacks of the NGS test. First of all, NGS is not yet a widely used standard test method, as with urine culture, in clinical practice, and it is mainly used for research purposes [29]. In addition, the cost of NGS is more expensive than urine culture or EQUC, and when various bacteria are identified, it is difficult to interpret the results as to which bacteria are the actual pathogens [30]. Lastly, the lack of primer universality can be a major problem compared with urine culture or EQUC [13].
With the accumulation of the knowledge regarding this microbiome, the importance of the gut–vagina–bladder axis for rUTI has been increasingly emphasized. Clinical evidence of an association between the vagina and bladder is as follows. First, bacterial vaginosis is a risk factor for UTI [31]; second, UTI decreases when hormone treatment such as vaginal estrogen is administered [32,33], and lastly, there are many patients who complain of frequent UTI after sex [34]. On a microbiological basis, it has been reported that around two thirds of the bladder microbiota overlaps with the gut microbiota, and around one third of the bladder microbiota exists only in the vagina [35].
The first finding of our study was that there was no difference in the detection rate of Gardnerella between the NC and rUTI groups. Similar to our result, a previous study reported that Gardnerella was detected in 27% of the normal population, and this ratio was not significantly different from patients with urinary symptoms [10]. Gardnerella is often detected even in normal asymptomatic individuals. However, in the absence of symptoms due to a host–pathogen immune response, Gardnerella does not cause disease. Therefore, not only the detection rate of Gardnerella but also symptoms are important for interpretation. Taken together, Gardnerella can influence the uncertain pathophysiology of the dominant urotype in rUTI under the assumption that Gardnerella causes symptoms. In our study, in the normal group, E. coli did not appear even if Gardnerella was detected, but in the rUTI group, 22.57% of patients with Gardnerella were detected (Table 3). Although the clinical significance cannot be confirmed due to the small number of subjects, it shows abnormal diversity in the rUTI group. This means that bladder dysbiosis may present a differential immune response to bacterial colonization and different symptoms. In particular, Atopobium, Megasphaera, and Ureaplasma, known to be associated with bacterial vaginosis, were detected only in the rUTI group.
Second, our study suggested three patterns of Gardnerella-positive patients. First, in the E. coli-dominant group, this is the first study of humans showing that Gardnerella might be a covert pathogen that activates E. coli. It has already been shown that the vagina acts as a reservoir for uropathogens such as E. coli. Moreover, even short exposure of the bladder to Gardnerella caused bladder cell damage such as urothelial exfoliation and urothelial apoptosis in an animal study [17]. The second group (Gardnerella-dominant group) provides clues that increased amounts of Gardnerella itself may be associated with rUTI. Although rare, it has been reported that Gardnerella acts as a causative agent of UTIs [31]. Considering the low culture-positive rate of Gardnerella, the clinical significance is likely to be higher in practice. The clues from the study that the increased amount of Gardnerella in the Gardnerella urotype in the bladder may be related to rUTI are as follows. First, in these patients, no rUTI-causing bacteria were detected other than Gardnerella, and secondly, in this case, other vaginitis-causing bacteria were also accompanied. Taken together, we interpreted that dysbiosis caused by vaginitis strains, especially Gardnerella, induced symptomatic rUTI. In the case of the third Lactobacillus-dominant group, since the protectivity effect of Lactobacillus is different depending on the type of Lactobacillus strain, cystitis can occur even with a small percentage of Gardnerella in Lactobacillus strains with poor protectivity. However, in this study, the Lactobacillus strain was not analyzed, and thus further follow-up studies are required to investigate the difference between normal and rUTI strains.
Considering that both NC and rUTI existed in Group 2 (Gardnerella-dominant) and Group 3 (Lactobacillus-dominant), we can make the following inferences. Asymptomatic Gardnerella infection is present in the vagina, and some are self-treated [36]. Likewise, the presence of Gardnerella does not necessarily cause rUTI, just as the presence of Gardnerella does not necessarily cause bacterial vaginosis. Although Gardnerella itself does not have high virulence, it is affected by other risk factors for causing symptoms. Risk factors such as host immunity, Gardnerella proliferation, the microbiome environment, and the residence environment seem to influence the development of the phenotype (asymptomatic or rUTI). For example, it seems that recurrent cystitis tends to occur when Gardnerella and other bacterial vaginosis strains are co-present, which can be interpreted as the influence of the microbiome environment. In addition, the fact that the proportion of Gardnerella was higher in rUTI patients (although not significant) could be evidence that the amount of Gardnerella proliferation had an effect on phenotype expression. Overall, our results provide clues for a new pathophysiology of rUTI. The usage of antibiotics based on traditional uropathogens in Group 2 or Group 3 has weak clinical effects and may cause side effects of antibiotic resistance to occur.
We suppose that the increased number of UTIs in menopause might be caused by decreased estrogen levels and associated physical changes. Estrogen helps the growth of Lactobacillus bacteria in the vagina and bladder, and the proliferation of Lactobacillus bacteria reduces urinary tract infections. After menopause, estrogen reduction and changes in Lactobacillus are thought to promote UTI. In fact, estrogen replacement hormone therapy in postmenopausal women reduced cystitis by providing a microenvironment that improved the growth of normal Lactobacillus in the vagina [37,38,39,40,41,42]. However, in this study, it was not possible to analyze the effect of estrogen replacement, because prescription of vaginal hormones was impossible during the study period due to the shortage of medication.
Our study has several advantages. First, we demonstrated the clinical importance of the gut–vagina–bladder axis in humans, focusing on Gardnerella for the first time. The gut–vagina–bladder axis has been explained to some extent through microbiological and animal experiments, but studies on humans are still lacking [8,17,35]. Second, in our study, the specimen was collected by transurethral catheterization, so the bladder microbiome was well reflected without contamination. Finally, our study suggested several types and novel mechanisms of rUTI that had not been previously elucidated.
However, our study also has several limitations. First, since this study was conducted in a retrospective manner, selection bias is inevitable, especially regarding the NC group. Moreover, we could not match the cases and controls regarding their estrogen hormonal status. Due to the limitations of the retrospective study, the number of subjects corresponding to the normal control group was relatively small. In particular, in our study, normal controls were selected from those who had undergone a health check-up, and it was rare for premenopausal women to even receive a urine NGS test during a health check-up. In Korea, if a patient has urinary symptoms, the NGS test is reimbursed by the government and the cost of the NGS is low at USD 160. On the other hand, if an asymptomatic subject undergoes an NGS test for health screening purposes, the cost of the NGS is between USD 350 and 450. Due to the burden of testing costs, most asymptomatic subjects do not frequently undergo NGS testing. For this reason, it was difficult to secure additional normal controls in this study. We are aware of these limitations and are working to increase the NGS test rate in the normal population by securing research funds. Second, our study revealed the importance of Gardnerella infection as a covert pathogen, but did not provide a cut-off for the required amount of infection for clinical symptoms to appear. Third, the difference between the Lactobacillus strains in the NC group and the rUTI group could not be suggested. This requires follow-up studies, and it is believed that it will provide clues about Lactobacillus prophylaxis in patients with rUTI in the future. Fourth, this study did not suggest whether treating Gardnerella could actually improve the clinical symptoms of rUTI. In general, antibiotics used for bacterial vaginosis and those used for rUTI are different [43]. The choice of antibiotics for UTIs can also affect the vaginal microbiome. For example, the use of beta-lactam antibiotics is less effective against vaginally colonized E. coli, and recurrent UTIs caused by vaginal E. coli easily occur with such beta-lactam antibiotics [9,44]. Furthermore, Gardnerella is difficult to treat due to biofilm formation [45]. Metronidazole or tobramycin, which were previously recommended as therapeutic agents for Gardnerella, can prevent the formation of a new biofilm, but are known to have less effect on the previously formed biofilm [45]. Therefore, further studies are needed to determine which antibiotic is most suitable for rUTI caused by Gardnerella. Fifth, this study was able to analyze the results up to the genus level because the partial V4 region of 16S rDNA was analyzed as a target. Further follow-up studies up to the species level will be needed.
5. Conclusions
In summary, if conventional uropathogens are not detected in patients with rUTI, Gardnerella infection should be considered. Asymptomatic urine Gardnerella (asymptomatic bacteriuria) does not require treatment, but symptomatic patients with Gardnerella found in the urine may need treatment, considering the possibility of the causative organism. Moreover, even for asymptomatic cases, treatment should be considered if E. coli or the causative agent of bacterial vaginosis is detected in addition to Gardnerella. Overall, our study suggest that dysbiosis of the bladder microbiome may show different colonization and different symptoms.
Author Contributions
Conceptualization, S.R., J.H.K. and Y.H.K.; formal analysis, E.J.S., H.-Y.S., S.S.K., J.C.C., J.C.K., B.C.Y., E.-S.L. and S.S.J.; investigation, J.Y., H.B.S., M.-A.J. and C.B.R.; data curation, all authors; writing—original draft preparation, J.-J.Y. and J.S.S.; writing—review and editing, J.-J.Y., W.B.K. and J.S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2021R1G1A1007886, 2021R1G1A1010833), and in part by the Soonchunhyang University Research Fund.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Soonchunhyang University Bucheon Hospital (IRB number SCHBC 2021-10-011-01).
Informed Consent Statement
Informed consent was waived by the IRB of Soonchunhyang University Bucheon Hospital due to the retrospective design of this study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cox, C.E.; Lacy, S.S.; Hinman, F., Jr. The urethra and its relationship to urinary tract infection. II. The urethral flora of the female with recurrent urinary infection. J. Urol. 1968, 99, 632–638. [Google Scholar] [CrossRef]
- Al-Badr, A.; Al-Shaikh, G. Recurrent Urinary Tract Infections Management in Women: A review. Sultan Qaboos Univ. Med. J. 2013, 13, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am. J. Med. 2002, 113 (Suppl. S1), 5S–13S. [Google Scholar] [CrossRef]
- Kalra, O.P.; Raizada, A. Approach to a patient with urosepsis. J. Glob. Infect. Dis. 2009, 1, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Renard, J.; Ballarini, S.; Mascarenhas, T.; Zahran, M.; Quimper, E.; -Choucair, J.; Iselin, C.E. Recurrent Lower Urinary Tract Infections Have a Detrimental Effect on Patient Quality of Life: A Prospective, Observational Study. Infect. Dis. Ther. 2014, 4, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josephs-Spaulding, J.; Krogh, T.J.; Rettig, H.C.; Lyng, M.; Chkonia, M.; Waschina, S.; Graspeuntner, S.; Rupp, J.; Moller-Jensen, J.; Kaleta, C. Recurrent Urinary Tract Infections: Unraveling the Complicated Environment of Uncomplicated rUTIs. Front. Cell. Infect. Microbiol. 2021, 11, 562525. [Google Scholar] [CrossRef]
- Mestrovic, T.; Matijasic, M.; Peric, M.; Cipcic Paljetak, H.; Baresic, A.; Verbanac, D. The Role of Gut, Vaginal, and Urinary Microbiome in Urinary Tract Infections: From Bench to Bedside. Diagnostics 2020, 11, 7. [Google Scholar] [CrossRef]
- Stapleton, A.E. The Vaginal Microbiota and Urinary Tract Infection. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.H.; Birch, D.F.; Fairley, K.F. Prevalence of Gardnerella vaginalis in the urinary tract. J. Clin. Microbiol. 1988, 26, 1130–1133. [Google Scholar] [CrossRef] [Green Version]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Gasiorek, M.; Hsieh, M.H.; Forster, C.S. Utility of DNA Next-Generation Sequencing and Expanded Quantitative Urine Culture in Diagnosis and Management of Chronic or Persistent Lower Urinary Tract Symptoms. J. Clin. Microbiol. 2019, 58, e00204-19. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.J.; Shin, H.B.; Song, J.S.; Kim, M.; Yun, J.; Kim, Z.; Lee, Y.M.; Lee, S.W.; Lee, K.W.; Kim, W.B.; et al. Urinary Microbiome Characteristics in Female Patients with Acute Uncomplicated Cystitis and Recurrent Cystitis. J. Clin. Med. 2021, 10, 1097. [Google Scholar] [CrossRef]
- Lewis, A.L.; Gilbert, N.M. Roles of the vagina and the vaginal microbiota in urinary tract infection: Evidence from clinical correlations and experimental models. GMS Infect. Dis. 2020, 8, Doc02. [Google Scholar] [CrossRef]
- Komesu, Y.M.; Dinwiddie, D.L.; Richter, H.E.; Lukacz, E.S.; Sung, V.W.; Siddiqui, N.Y.; Zyczynski, H.M.; Ridgeway, B.; Rogers, R.G.; Arya, L.A.; et al. Defining the relationship between vaginal and urinary microbiomes. Am. J. Obstet. Gynecol. 2020, 222, 154.e1–154.e10. [Google Scholar] [CrossRef]
- Gilbert, N.M.; O’Brien, V.P.; Lewis, A.L. Transient microbiota exposures activate dormant Escherichia coli infection in the bladder and drive severe outcomes of recurrent disease. PLoS Pathog. 2017, 13, e1006238. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [Green Version]
- Kaehler, B.D.; Bokulich, N.A.; McDonald, D.; Knight, R.; Caporaso, J.G.; Huttley, G.A. Species abundance information improves sequence taxonomy classification accuracy. Nat. Commun. 2019, 10, 4643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. microDecon: A highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Tapiainen, T.; Paalanne, N.; Tejesvi, M.V.; Koivusaari, P.; Korpela, K.; Pokka, T.; Salo, J.; Kaukola, T.; Pirttila, A.M.; Uhari, M.; et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 2018, 84, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Delbeke, H.; Younas, S.; Casteels, I.; Joossens, M. Current knowledge on the human eye microbiome: A systematic review of available amplicon and metagenomic sequencing data. Acta Ophthalmol. 2021, 99, 16–25. [Google Scholar] [CrossRef]
- Park, H.-S.; Jun, C.-H. A simple and fast algorithm for K-medoids clustering. Expert Syst. Appl. 2009, 36, 3336–3341. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Schauberger, C.W.; Merkitch, K.W.; Prell, A.M. Acute cystitis in women: Experience with a telephone-based algorithm. WMJ Off. Publ. State Med Soc. Wis. 2007, 106, 326–329. [Google Scholar]
- Lee, D.S.; Lee, S.J.; Choe, H.S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. Biomed Res. Int. 2018, 2018, 7656752. [Google Scholar] [CrossRef] [Green Version]
- Muhamad Rizal, N.S.; Neoh, H.M.; Ramli, R.; PR, A.L.K.P.; Hanafiah, A.; Abdul Samat, M.N.; Tan, T.L.; Wong, K.K.; Nathan, S.; Chieng, S.; et al. Advantages and Limitations of 16S rRNA Next-Generation Sequencing for Pathogen Identification in the Diagnostic Microbiology Laboratory: Perspectives from a Middle-Income Country. Diagnostics 2020, 10, 816. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez, R.L.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef] [Green Version]
- Harmanli, O.H.; Cheng, G.Y.; Nyirjesy, P.; Chatwani, A.; Gaughan, J.P. Urinary tract infections in women with bacterial vaginosis. Obstet. Gynecol. 2000, 95, 710–712. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, S.; Pastijn, A.; Gevers, R.; Murillo, D. Estrogen therapy in older patients with recurrent urinary tract infections: A review. Int. J. Fertil. Womens Med. 2004, 49, 71–74. [Google Scholar] [PubMed]
- Krause, M.; Wheeler, T.L., 2nd; Snyder, T.E.; Richter, H.E. Local Effects of Vaginally Administered Estrogen Therapy: A Review. J. Pelvic Med. Surg. 2009, 15, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.E.; Hawes, S.E.; Scholes, D.; Boyko, E.J.; Hughes, J.P.; Fihn, S.D. Sexual intercourse and risk of symptomatic urinary tract infection in post-menopausal women. J. Gen. Intern. Med. 2008, 23, 595–599. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, A.J.; Brubaker, L. Urobiome updates: Advances in urinary microbiome research. Nat. Rev. Urol. 2019, 16, 73–74. [Google Scholar] [CrossRef]
- Paladine, H.L.; Desai, U.A. Vaginitis: Diagnosis and Treatment. Am. Fam. Physician 2018, 97, 321–329. [Google Scholar]
- Heinemann, C.; Reid, G. Vaginal microbial diversity among postmenopausal women with and without hormone replacement therapy. Can. J. Microbiol. 2005, 51, 777–781. [Google Scholar] [CrossRef]
- Raz, R. Urinary tract infection in postmenopausal women. Korean J. Urol. 2011, 52, 801–808. [Google Scholar] [CrossRef] [Green Version]
- Raz, R. Hormone replacement therapy or prophylaxis in postmenopausal women with recurrent urinary tract infection. J. Infect. Dis. 2001, 183, S74–S76. [Google Scholar] [CrossRef] [Green Version]
- Raz, R.; Stamm, W.E. A controlled trial of intravaginal estriol in postmenopausal women with recurrent urinary tract infections. N. Engl. J. Med. 1993, 329, 753–756. [Google Scholar] [CrossRef] [Green Version]
- Cauci, S.; Driussi, S.; De Santo, D.; Penacchioni, P.; Iannicelli, T.; Lanzafame, P.; De Seta, F.; Quadrifoglio, F.; de Aloysio, D.; Guaschino, S. Prevalence of bacterial vaginosis and vaginal flora changes in peri- and postmenopausal women. J. Clin. Microbiol. 2002, 40, 2147–2152. [Google Scholar] [CrossRef] [Green Version]
- Eriksen, B. A randomized, open, parallel-group study on the preventive effect of an estradiol-releasing vaginal ring (Estring) on recurrent urinary tract infections in postmenopausal women. Am. J. Obstet. Gynecol. 1999, 180, 1072–1079. [Google Scholar] [CrossRef]
- Pedraza-Aviles, A.G.; Zaragoza, M.C.; Mota-Vazquez, R.; Hernandez-Soto, C.; Ramirez-Santana, M.; Terrazas-Maldonado, M.L. Treatment of urinary tract infection by Gardnerella vaginalis: A comparison of oral metronidazole versus ampicillin. Rev. Latinoam. Microbiol. 2001, 43, 65–69. [Google Scholar] [PubMed]
- Hooton, T.M.; Scholes, D.; Gupta, K.; Stapleton, A.E.; Roberts, P.L.; Stamm, W.E. Amoxicillin-clavulanate vs ciprofloxacin for the treatment of uncomplicated cystitis in women: A randomized trial. JAMA 2005, 293, 949–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottschick, C.; Szafranski, S.P.; Kunze, B.; Sztajer, H.; Masur, C.; Abels, C.; Wagner-Dobler, I. Screening of Compounds against Gardnerella vaginalis Biofilms. PLoS ONE 2016, 11, e0154086. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).