Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. siRNA Treatment
2.3. Real-Time Reverse Transcription–Polymerase Chain Reaction
2.4. Confocal Laser Scanning Microscopy Analysis
2.4.1. Single Immunofluorescence Staining
2.4.2. Double Immunofluorescence Staining
2.5. Western Blotting
2.6. Immunoprecipitation
2.7. Flow Cytometric Assay
2.8. Statistical Analysis
3. Results
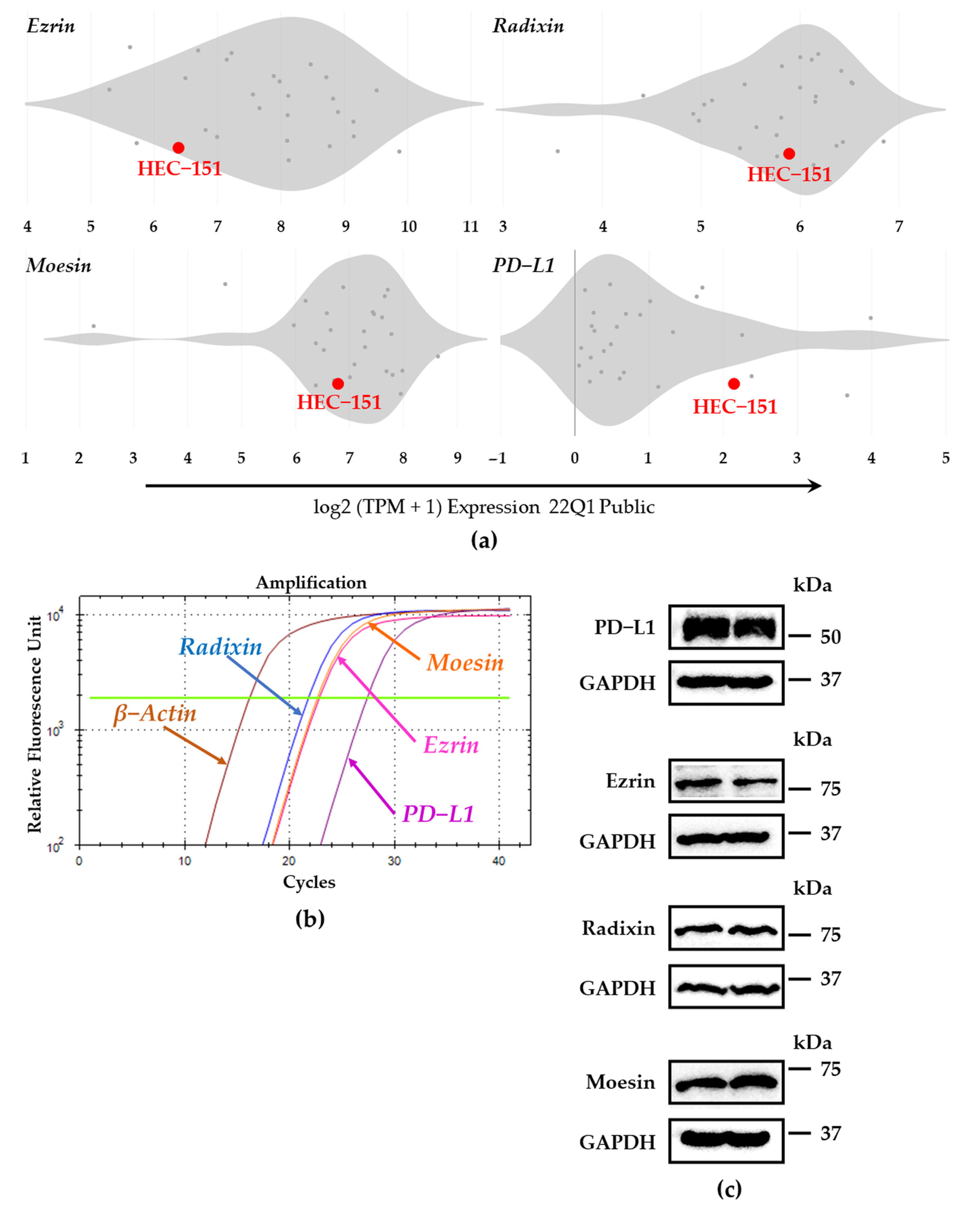
3.1. Expression Profiles of PD-L1 and Each ERM at mRNA and Protein Levels in HEC-151 Cells
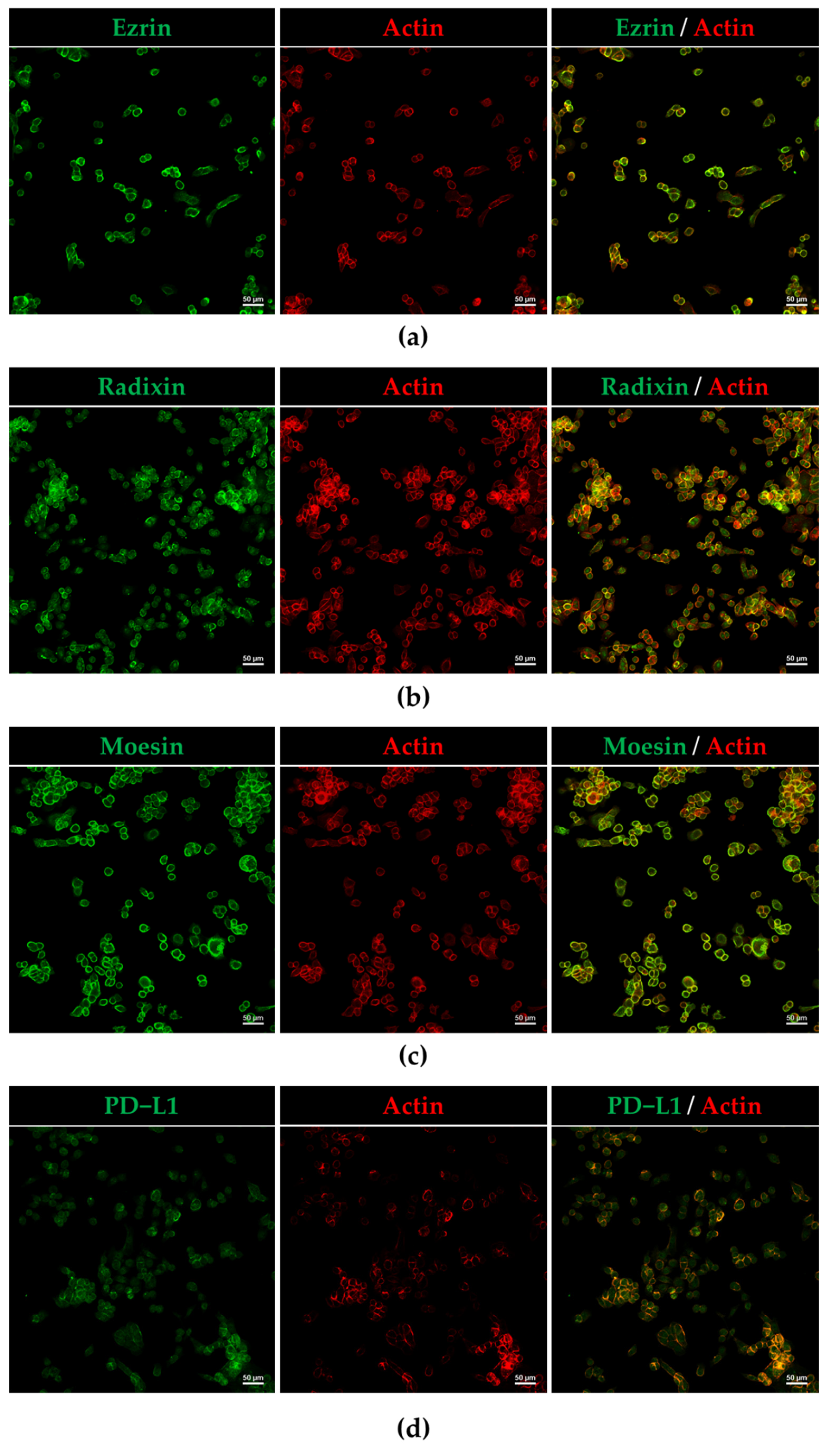
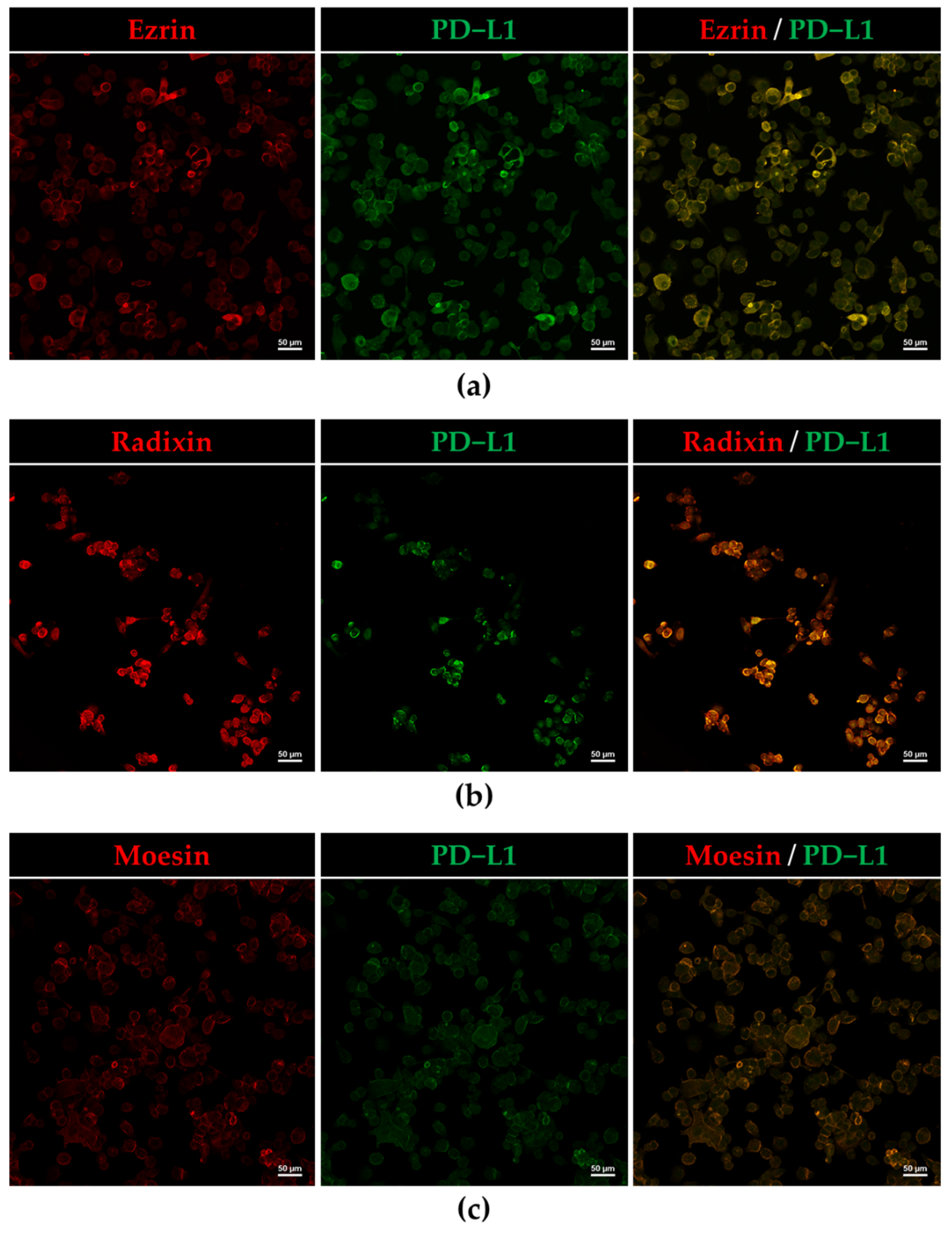
3.2. Plasma Membrane Localization of PD-L1 and ERM in HEC-151 Cells
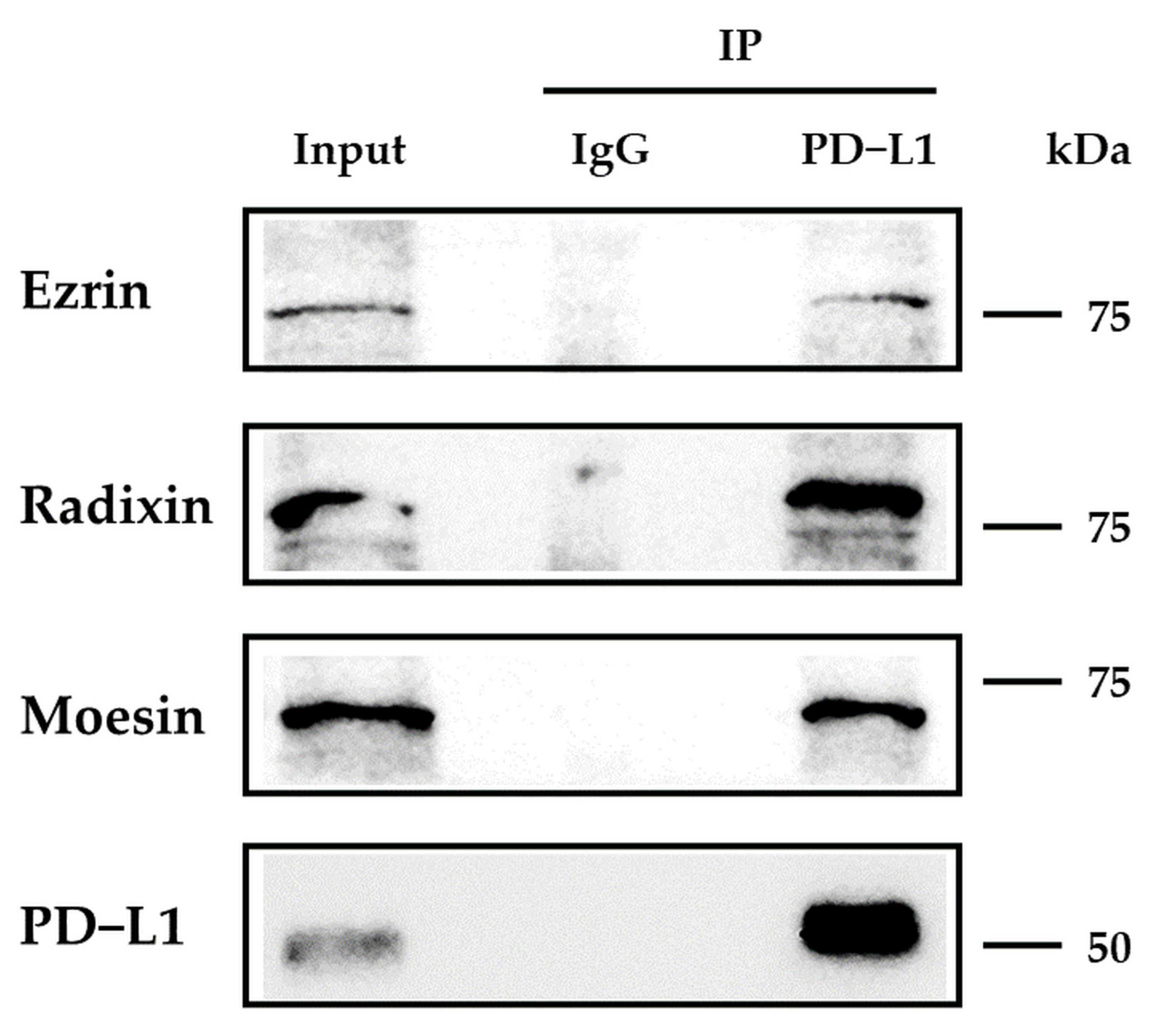
3.3. Protein–Protein Interaction between PD-L1 and ERM in HEC-151 Cells
3.4. Effect of siRNAs against ERM on the Expression Levels of PD-L1 in HEC-151 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Preti, E.; Landoni, F.; Carinelli, S.; Colombo, A.; Marini, C.; Sessa, C.; Group, E.G.W. Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013, 24 (Suppl. S6), vi33–vi38. [Google Scholar] [CrossRef] [PubMed]
- Koskas, M.; Amant, F.; Mirza, M.R.; Creutzberg, C.L. Cancer of the corpus uteri: 2021 update. Int. J. Gynaecol. Obstet. 2021, 155 (Suppl. S1), 45–60. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef]
- Melillo, G.; Chand, V.; Yovine, A.; Gupta, A.; Massacesi, C. Curative-Intent Treatment with Durvalumab in Early-Stage Cancers. Adv. Ther. 2021, 38, 2759–2778. [Google Scholar] [CrossRef]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Ohaegbulam, K.C.; Assal, A.; Lazar-Molnar, E.; Yao, Y.; Zang, X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 2015, 21, 24–33. [Google Scholar] [CrossRef]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef]
- Tu, X.; Qin, B.; Zhang, Y.; Zhang, C.; Kahila, M.; Nowsheen, S.; Yin, P.; Yuan, J.; Pei, H.; Li, H.; et al. PD-L1 (B7-H1) Competes with the RNA Exosome to Regulate the DNA Damage Response and Can Be Targeted to Sensitize to Radiation or Chemotherapy. Mol. Cell 2019, 74, 1215–1226.e4. [Google Scholar] [CrossRef]
- Qin, W.; Hu, L.; Zhang, X.; Jiang, S.; Li, J.; Zhang, Z.; Wang, X. The Diverse Function of PD-1/PD-L Pathway Beyond Cancer. Front. Immunol. 2019, 10, 2298. [Google Scholar] [CrossRef] [PubMed]
- Makker, V.; Colombo, N.; Casado Herraez, A.; Santin, A.D.; Colomba, E.; Miller, D.S.; Fujiwara, K.; Pignata, S.; Baron-Hay, S.; Ray-Coquard, I.; et al. Lenvatinib plus Pembrolizumab for Advanced Endometrial Cancer. N. Engl. J. Med. 2022, 386, 437–448. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.M.; Bariani, G.M.; Cassier, P.A.; Marabelle, A.; Hansen, A.R.; De Jesus Acosta, A.; Miller, W.H., Jr.; Safra, T.; Italiano, A.; Mileshkin, L.; et al. Pembrolizumab in Patients with Microsatellite Instability-High Advanced Endometrial Cancer: Results from the KEYNOTE-158 Study. J. Clin. Oncol. 2022, 40, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Lee, H.H.; Hsu, J.L.; Yu, D.; Hung, M.C. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J. Biomed. Sci. 2020, 27, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, S.M.; Zhou, J.; Wang, Y.; Nie, R.C.; Chen, J.W.; Xie, D. Recent Findings in the Posttranslational Modifications of PD-L1. J. Oncol. 2020, 2020, 5497015. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef]
- Ogihara, T.; Mizoi, K.; Kamioka, H.; Yano, K. Physiological Roles of ERM Proteins and Transcriptional Regulators in Supporting Membrane Expression of Efflux Transporters as Factors of Drug Resistance in Cancer. Cancers 2020, 12, 3352. [Google Scholar] [CrossRef]
- Luciani, F.; Molinari, A.; Lozupone, F.; Calcabrini, A.; Lugini, L.; Stringaro, A.; Puddu, P.; Arancia, G.; Cianfriglia, M.; Fais, S. P-glycoprotein-actin association through ERM family proteins: A role in P-glycoprotein function in human cells of lymphoid origin. Blood 2002, 99, 641–648. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Yoshida, S.; Hatano, R.; Asano, S. Pathophysiological Roles of Ezrin/Radixin/Moesin Proteins. Biol. Pharm. Bull. 2017, 40, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Asp, N.; Kvalvaag, A.; Sandvig, K.; Pust, S. Regulation of ErbB2 localization and function in breast cancer cells by ERM proteins. Oncotarget 2016, 7, 25443–25460. [Google Scholar] [CrossRef] [PubMed]
- Rudnicka, D.; Oszmiana, A.; Finch, D.K.; Strickland, I.; Schofield, D.J.; Lowe, D.C.; Sleeman, M.A.; Davis, D.M. Rituximab causes a polarization of B cells that augments its therapeutic function in NK-cell-mediated antibody-dependent cellular cytotoxicity. Blood 2013, 121, 4694–4702. [Google Scholar] [CrossRef] [PubMed]
- Clucas, J.; Valderrama, F. ERM proteins in cancer progression. J. Cell Sci. 2014, 127, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Faure, C.; Djerbi-Bouillie, R.; Domingot, A.; Bouzinba-Segard, H.; Taouji, S.; Saidi, Y.; Bernard, S.; Carallis, F.; Rothe-Walther, R.; Lenormand, J.L.; et al. Allosteric Inhibition of HER2 by Moesin-Mimicking Compounds Targets HER2-Positive Cancers and Brain Metastases. Cancer Res. 2021, 81, 5464–5476. [Google Scholar] [CrossRef]
- Kamata, Y.; Watanabe, J.; Hata, H.; Hamano, M.; Kuramoto, H. Quantitative study on the correlation between p53 gene mutation and its expression in endometrial carcinoma cell lines. Eur. J. Gynaecol. Oncol. 2004, 25, 55–60. [Google Scholar]
- Kobori, T.; Doukuni, R.; Ishikawa, H.; Ito, Y.; Okada, R.; Tanaka, C.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Ezrin and Radixin Differentially Modulate Cell Surface Expression of Programmed Death Ligand-1 in Human Pancreatic Ductal Adenocarcinoma KP-2 Cells. Immuno 2022, 2, 6. [Google Scholar] [CrossRef]
- Tanaka, C.; Kobori, T.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Ezrin Modulates the Cell Surface Expression of Programmed Cell Death Ligand-1 in Human Cervical Adenocarcinoma Cells. Molecules 2021, 26, 5648. [Google Scholar] [CrossRef]
- Kobori, T.; Tanaka, C.; Tameishi, M.; Urashima, Y.; Ito, T.; Obata, T. Role of Ezrin/Radixin/Moesin in the Surface Localization of Programmed Cell Death Ligand-1 in Human Colon Adenocarcinoma LS180 Cells. Pharmaceuticals 2021, 14, 864. [Google Scholar] [CrossRef]
- Tameishi, M.; Kobori, T.; Tanaka, C.; Urashima, Y.; Ito, T.; Obata, T. Contribution of Ezrin on the Cell Surface Plasma Membrane Localization of Programmed Cell Death Ligand-1 in Human Choriocarcinoma JEG-3 Cells. Pharmaceuticals 2021, 14, 963. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011, 13, R45. [Google Scholar] [CrossRef] [PubMed]
- Mhawech-Fauceglia, P.; Wang, D.; Lele, S.; Frederick, P.J.; Pejovic, T.; Liu, S. Claudin7 and moesin in endometrial Adenocarcinoma; a retrospective study of 265 patients. BMC Res. Notes 2012, 5, 65. [Google Scholar] [CrossRef]
- Pluciennik, E.; Nowakowska, M.; Galdyszynska, M.; Popeda, M.; Bednarek, A.K. The influence of the WWOX gene on the regulation of biological processes during endometrial carcinogenesis. Int. J. Mol. Med. 2016, 37, 807–815. [Google Scholar] [CrossRef]
- Wang, D.; Tian, Y.; Feng, W.; Zhao, L.; Zhao, M.; Liu, J.; Wang, Q. Pseudolaric acid B induces endometrial cancer Ishikawa cell apoptosis and inhibits metastasis through AKT-GSK-3beta and ERK1/2 signaling pathways. Anticancer Drugs 2017, 28, 603–612. [Google Scholar] [CrossRef]
- Williams, E.; Villar-Prados, A.; Bowser, J.; Broaddus, R.; Gladden, A.B. Loss of polarity alters proliferation and differentiation in low-grade endometrial cancers by disrupting Notch signaling. PLoS ONE 2017, 12, e0189081. [Google Scholar] [CrossRef]
- Yan, H.; Sun, B.M.; Zhang, Y.Y.; Li, Y.J.; Huang, C.X.; Feng, F.Z.; Li, C. Upregulation of miR-183–5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin. Int. J. Mol. Med. 2018, 42, 2469–2480. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, X.; Xia, H.; Wang, L.; Zhao, H.; Xu, B.; Zhang, A.; Zhang, W. Promotion of the occurrence of endometrioid carcinoma by S100 calcium binding protein P. BMC Cancer 2020, 20, 845. [Google Scholar] [CrossRef]
- Xu, D.; Dong, P.; Xiong, Y.; Chen, R.; Konno, Y.; Ihira, K.; Yue, J.; Watari, H. PD-L1 Is a Tumor Suppressor in Aggressive Endometrial Cancer Cells and Its Expression Is Regulated by miR-216a and lncRNA MEG3. Front. Cell Dev. Biol. 2020, 8, 598205. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Li, L.; Li, N.; Li, F.; Qin, X.; Li, T.; Liu, M. Metformin suppresses cancer cell growth in endometrial carcinoma by inhibiting PD-L1. Eur. J. Pharmacol. 2019, 859, 172541. [Google Scholar] [CrossRef] [PubMed]
- Kharma, B.; Baba, T.; Matsumura, N.; Kang, H.S.; Hamanishi, J.; Murakami, R.; McConechy, M.M.; Leung, S.; Yamaguchi, K.; Hosoe, Y.; et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 2014, 74, 6519–6530. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Tang, R.; Zhang, R.; Wang, H.; Ji, Z.; Shao, Y.; Wang, S.; Zhong, T.; Gu, Y.; Meng, J. A comprehensive analysis of IDO1 expression with tumour-infiltrating immune cells and mutation burden in gynaecologic and breast cancers. J. Cell. Mol. Med. 2020, 24, 5238–5248. [Google Scholar] [CrossRef]
- Tamura, K.; Hasegawa, K.; Katsumata, N.; Matsumoto, K.; Mukai, H.; Takahashi, S.; Nomura, H.; Minami, H. Efficacy and safety of nivolumab in Japanese patients with uterine cervical cancer, uterine corpus cancer, or soft tissue sarcoma: Multicenter, open-label phase 2 trial. Cancer Sci. 2019, 110, 2894–2904. [Google Scholar] [CrossRef]
- Hu, X.; Lin, Z.; Wang, Z.; Zhou, Q. Emerging role of PD-L1 modification in cancer immunotherapy. Am. J. Cancer Res. 2021, 11, 3832–3840. [Google Scholar]
- Meng, F.; Su, Y.; Xu, B. Rho-associated protein kinase-dependent moesin phosphorylation is required for PD-L1 stabilization in breast cancer. Mol. Oncol. 2020, 14, 2701–2712. [Google Scholar] [CrossRef]
- Chan, L.C.; Li, C.W.; Xia, W.; Hsu, J.M.; Lee, H.H.; Cha, J.H.; Wang, H.L.; Yang, W.H.; Yen, E.Y.; Chang, W.C.; et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J. Clin. Investig. 2019, 129, 3324–3338. [Google Scholar] [CrossRef]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 expression in the tumor microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Takamatsu, H.; Espinoza, J.L.; Lu, X.; Qi, Z.; Okawa, K.; Nakao, S. Anti-moesin antibodies in the serum of patients with aplastic anemia stimulate peripheral blood mononuclear cells to secrete TNF-alpha and IFN-gamma. J. Immunol. 2009, 182, 703–710. [Google Scholar] [CrossRef]
- Suzuki, K.; Nagao, T.; Itabashi, M.; Hamano, Y.; Sugamata, R.; Yamazaki, Y.; Yumura, W.; Tsukita, S.; Wang, P.C.; Nakayama, T.; et al. A novel autoantibody against moesin in the serum of patients with MPO-ANCA-associated vasculitis. Nephrol. Dial. Transplant. 2014, 29, 1168–1177. [Google Scholar] [CrossRef] [PubMed][Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, C.; Kobori, T.; Okada, R.; Doukuni, R.; Tameishi, M.; Urashima, Y.; Ito, T.; Takagaki, N.; Obata, T. Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells. J. Clin. Med. 2022, 11, 2226. https://doi.org/10.3390/jcm11082226
Tanaka C, Kobori T, Okada R, Doukuni R, Tameishi M, Urashima Y, Ito T, Takagaki N, Obata T. Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells. Journal of Clinical Medicine. 2022; 11(8):2226. https://doi.org/10.3390/jcm11082226
Chicago/Turabian StyleTanaka, Chihiro, Takuro Kobori, Rie Okada, Rina Doukuni, Mayuka Tameishi, Yoko Urashima, Takuya Ito, Nobumasa Takagaki, and Tokio Obata. 2022. "Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells" Journal of Clinical Medicine 11, no. 8: 2226. https://doi.org/10.3390/jcm11082226
APA StyleTanaka, C., Kobori, T., Okada, R., Doukuni, R., Tameishi, M., Urashima, Y., Ito, T., Takagaki, N., & Obata, T. (2022). Ezrin Regulates the Cell Surface Localization of PD-L1 in HEC-151 Cells. Journal of Clinical Medicine, 11(8), 2226. https://doi.org/10.3390/jcm11082226






