Evaluating the Precision of Automatic Segmentation of Teeth, Gingiva and Facial Landmarks for 2D Digital Smile Design Using Real-Time Instance Segmentation Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Description
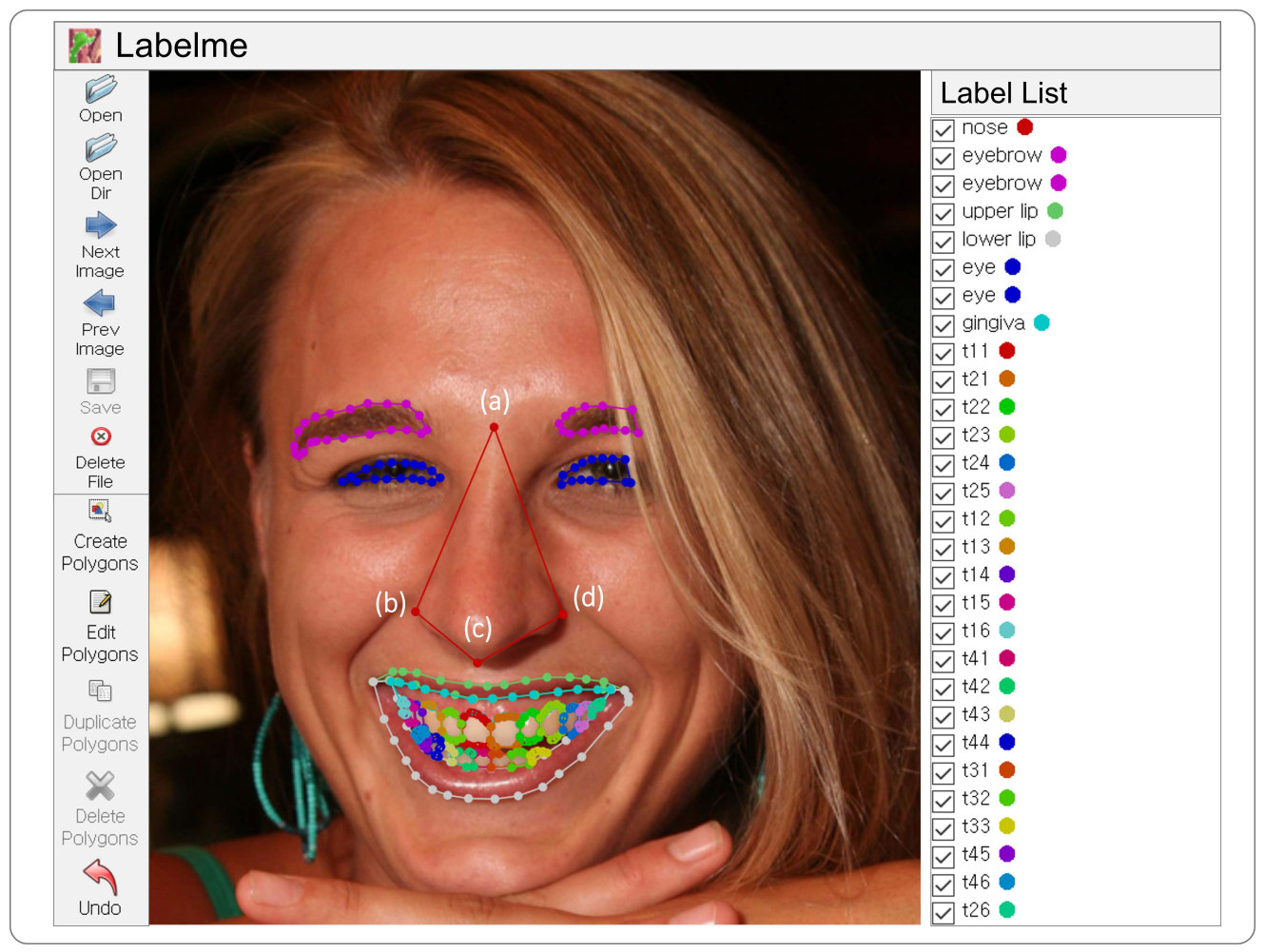
2.2. Dataset Annotation
- eye;
- eyebrow;
- nose;
- upper lip, lower lip;
- tragus;
- gingiva;
- buccal corridor;
- teeth (t11, t12, …, t18, t21, t22, …, t28, t31, …, t38, t41, …, t48).
- (a)
- a point between eyebrows;
- (b)
- a point located at the left ala of nose;
- (c)
- a point falling to the pharynx;
- (d)
- a point located at the right ala of nose.
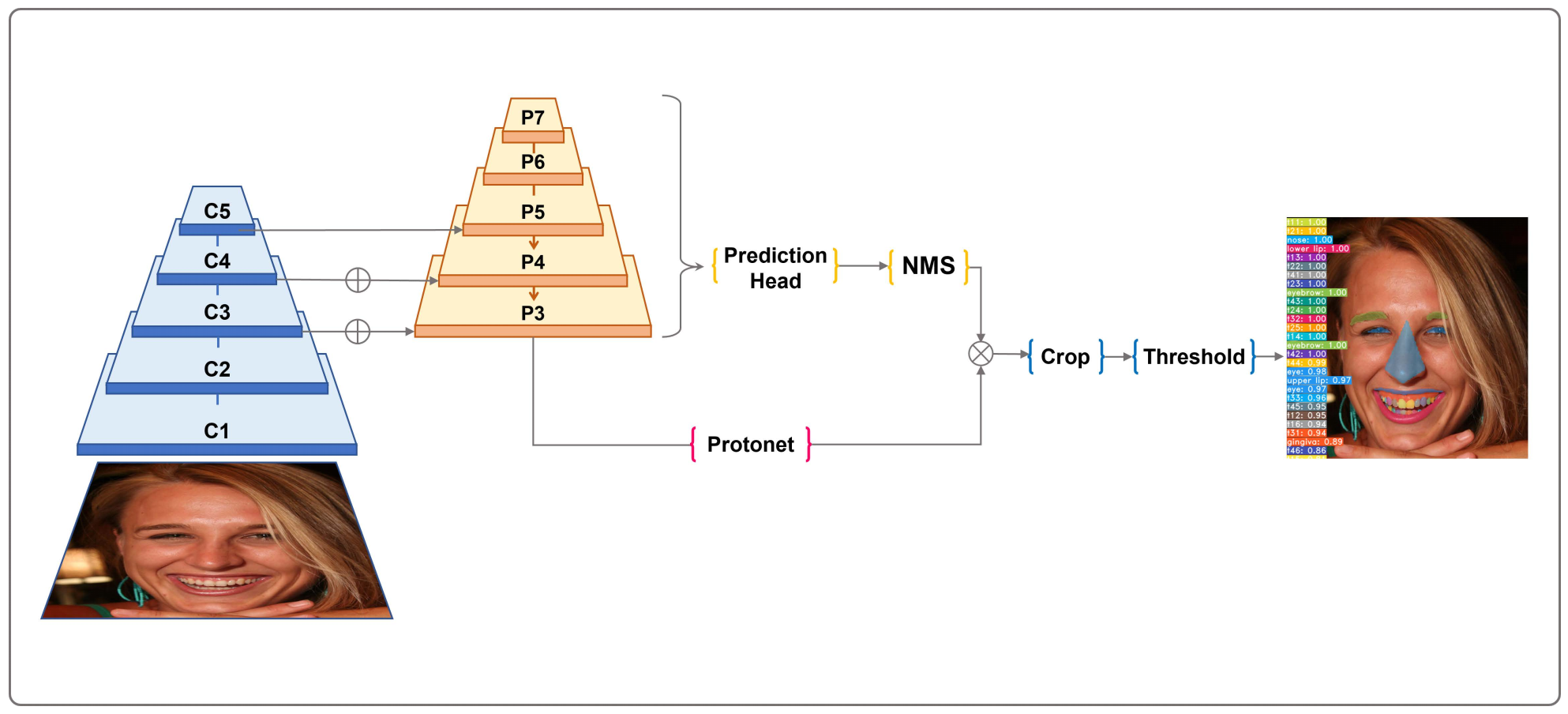
2.3. Deep Learning Model
2.4. Model Training
2.5. Statistical Analysis
3. Results
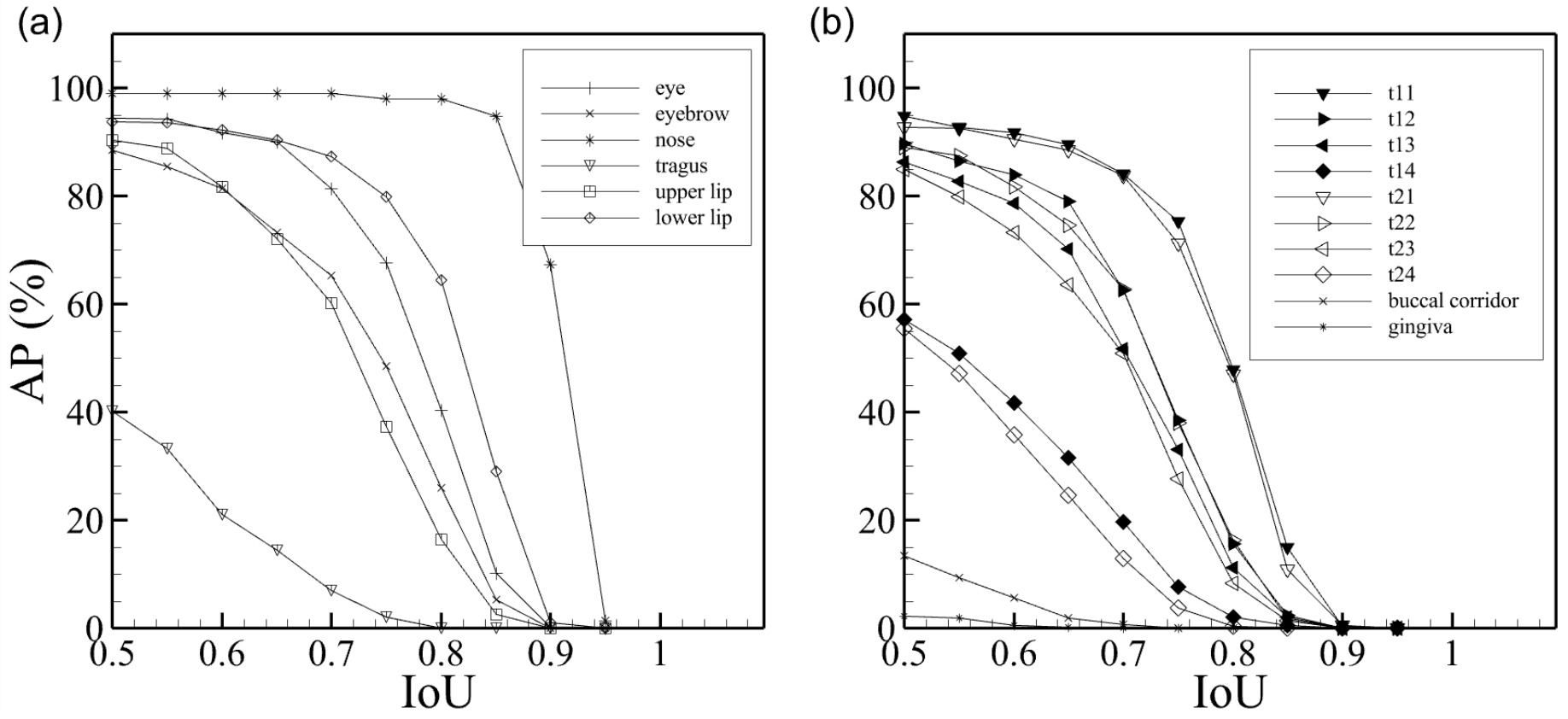
3.1. Quantitative Segmentation Results
3.2. Qualitative Segmentation Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raj, V. Esthetic Paradigms in the Interdisciplinary Management of Maxillary Anterior Dentition-A Review. J. Esthet. Restor. Dent. 2013, 25, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Hasanreisoglu, U.; Berksun, S.; Aras, K.; Arslan, I. An analysis of maxillary anterior teeth: Facial and dental proportions. J. Prosthet. Dent. 2005, 94, 530–538. [Google Scholar] [CrossRef]
- Rifkin, R. Facial analysis: A comprehensive approach to treatment planning in aesthetic dentistry. Pract. Periodontics Aesthetic Dent. 2000, 12, 865–871. [Google Scholar]
- Paul, S.J. Smile analysis and face-bow transfer: Enhancing aesthetic restorative treatment. Pract. Proced. Aesthetic Dent. 2001, 13, 217–222. [Google Scholar]
- Hatzi, P.; Millstein, P.; Maya, A. Determining the accuracy of articulator interchangeability and hinge axis reproducibility. J. Prosthet. Dent. 2001, 85, 236–245. [Google Scholar] [CrossRef]
- Borgh, O.; Posselt, U. Hinge axis registration: Experiments on the articulator. J. Prosthet. Dent. 1958, 8, 35–40. [Google Scholar] [CrossRef]
- Jafri, Z.; Ahmad, N.; Sawai, M.; Sultan, N.; Bhardwaj, A. Digital Smile Design-An innovative tool in aesthetic dentistry. J. Oral Biol. Craniofacial Res. 2020, 10, 194–198. [Google Scholar] [CrossRef]
- Coachman, C.; Calamita, M.A.; Sesma, N. Dynamic documentation of the smile and the 2D/3D digital smile design process. Int. J. Periodontics Restor. Dent. 2017, 37, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Romeo, G.; Bresciano, M. Diagnostic and Technical Approach to Esthetic Rehabilitations. J. Esthet. Restor. Dent. 2003, 15, 204–216. [Google Scholar] [CrossRef]
- Derbabian, K.; Marzola, R.; Donovan, T.E.; Arcidiacono, A. The Science of Communicating the Art of Esthetic Dentistry. Part III: Precise Shade Communication. J. Esthet. Restor. Dent. 2001, 13, 154–162. [Google Scholar] [CrossRef]
- Kahng, L.S. Patient–Dentist–Technician Communication within the Dental Team: Using a Colored Treatment Plan Wax-Up. J. Esthet. Restor. Dent. 2006, 18, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Charavet, C.; Bernard, J.C.; Gaillard, C.; Gall, M.L. Benefits of Digital Smile Design (DSD) in the conception of a complex orthodontic treatment plan: A case report-proof of concept. Int. Orthod. 2019, 17, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.P.; da Costa, R.G.; Calgaro, M.; Ritter, A.V.; Correr, G.M.; da Cunha, L.F.; Gonzaga, C.C. Digital smile design and mock-up technique for esthetic treatment planning with porcelain laminate veneers. J. Conserv. Dent. 2018, 21, 455. [Google Scholar] [CrossRef]
- Mahn, E.; Sampaio, C.S.; da Silva, B.P.; Stanley, K.; Valdés, A.M.; Gutierrez, J.; Coachman, C. Comparing the use of static versus dynamic images to evaluate a smile. J. Prosthet. Dent. 2020, 123, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wang, K.P.; Liu, Y.; Liu, Y.; Zhou, Y. Four-dimensional digital prediction of the esthetic outcome and digital implementation for rehabilitation in the esthetic zone. J. Prosthet. Dent. 2020, 123, 557–563. [Google Scholar] [CrossRef]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Hung, K.; Montalvao, C.; Tanaka, R.; Kawai, T.; Bornstein, M.M. The use and performance of artificial intelligence applications in dental and maxillofacial radiology: A systematic review. Dentomaxillofac. Radiol. 2020, 49, 20190107. [Google Scholar] [CrossRef]
- Chouhan, N.; Khan, A.; Shah, J.Z.; Hussnain, M.; Khan, M.W. Deep convolutional neural network and emotional learning based breast cancer detection using digital mammography. Comput. Biol. Med. 2021, 132, 104318. [Google Scholar] [CrossRef]
- Shanthi, T.; Sabeenian, R.; Anand, R. Automatic diagnosis of skin diseases using convolution neural network. Microprocess. Microsyst. 2020, 76, 103074. [Google Scholar] [CrossRef]
- Xu, K.; Feng, D.; Mi, H. Deep Convolutional Neural Network-Based Early Automated Detection of Diabetic Retinopathy Using Fundus Image. Molecules 2017, 22, 2054. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, D.H.; Jeong, S.N.; Choi, S.H. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J. Dent. 2018, 77, 106–111. [Google Scholar] [CrossRef]
- Krois, J.; Ekert, T.; Meinhold, L.; Golla, T.; Kharbot, B.; Wittemeier, A.; Dörfer, C.; Schwendicke, F. Deep Learning for the Radiographic Detection of Periodontal Bone Loss. Sci. Rep. 2019, 9, 8495. [Google Scholar] [CrossRef] [PubMed]
- Tuzoff, D.V.; Tuzova, L.N.; Bornstein, M.M.; Krasnov, A.S.; Kharchenko, M.A.; Nikolenko, S.I.; Sveshnikov, M.M.; Bednenko, G.B. Tooth detection and numbering in panoramic radiographs using convolutional neural networks. Dentomaxillofac. Radiol. 2019, 48, 20180051. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Kim, D.; Jeong, H.; Yoon, S.J.; Youm, S. Automatic Tooth Detection and Numbering Using a Combination of a CNN and Heuristic Algorithm. Appl. Sci. 2020, 10, 5624. [Google Scholar] [CrossRef]
- Ekert, T.; Krois, J.; Meinhold, L.; Elhennawy, K.; Emara, R.; Golla, T.; Schwendicke, F. Deep Learning for the Radiographic Detection of Apical Lesions. J. Endod. 2019, 45, 917–922.e5. [Google Scholar] [CrossRef] [PubMed]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 4396–4405. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Honolulu, HI, USA, 21–26 July 2017; pp. 2980–2988. [Google Scholar] [CrossRef]
- Li, Y.; Qi, H.; Dai, J.; Ji, X.; Wei, Y. Fully Convolutional Instance-Aware Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honoullu, HI, USA, 21–26 July 2017; pp. 4438–4446. [Google Scholar] [CrossRef] [Green Version]
- Girshick, R. Fast R-CNN. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar] [CrossRef]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-fcn: Object detection via region-based fully convolutional networks. In Proceedings of the NIPS’16: Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 379–387. Available online: https://proceedings.neurips.cc/paper/2016/file/577ef1154f3240ad5b9b413aa7346a1e-Paper.pdf (accessed on 7 January 2022).
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. YOLACT++: Better Real-time Instance Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 1108–1121. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Proceedings of the Computer Vision—ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; pp. 21–37. [Google Scholar] [CrossRef] [Green Version]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honoullu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Bottou, L. Stochastic gradient learning in neural networks. Proc. Neuro-Nımes 1991, 91, 12. [Google Scholar]
- Bottou, L. Large-Scale Machine Learning with Stochastic Gradient Descent. In Proceedings of the COMPSTAT’2010, Physica-Verlag HD, Paris, France, 22–27 August 2010; pp. 177–186. [Google Scholar] [CrossRef] [Green Version]
- Rakhlin, A.; Shamir, O.; Sridharan, K. Making gradient descent optimal for strongly convex stochastic optimization. arXiv 2011, arXiv:1109.5647. [Google Scholar]
- Hoiem, D.; Divvala, S.K.; Hays, J.H. Pascal VOC 2008 challenge. World Lit. Today 2009, 24. Available online: http://host.robots.ox.ac.uk/pascal/VOC/voc2008/htmldoc/voc.html (accessed on 5 January 2022).
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Lee, K.; Shin, E.; Choy, G.; Do, S. How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? arXiv 2015, arXiv:1511.06348. [Google Scholar]
- Boiko, O.; Hyttinen, J.; Fält, P.; Jäsberg, H.; Mirhashemi, A.; Kullaa, A.; Hauta-Kasari, M. Deep Learning for Dental Hyperspectral Image Analysis. Color Imaging Conf. 2019, 2019, 295–299. [Google Scholar] [CrossRef]


| Type | () | () | () | () | () |
|---|---|---|---|---|---|
| Box | 0.341 (0.635) | 0.621 (0.946) | 0.879 (0.990) | 0.645 (0.942) | 0.303 (0.604) |
| Mask | 0.229 (0.472) | 0.570 (0.945) | 0.855 (0.990) | 0.541 (0.921) | 0.175 (0.411) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Kim, J.-E. Evaluating the Precision of Automatic Segmentation of Teeth, Gingiva and Facial Landmarks for 2D Digital Smile Design Using Real-Time Instance Segmentation Network. J. Clin. Med. 2022, 11, 852. https://doi.org/10.3390/jcm11030852
Lee S, Kim J-E. Evaluating the Precision of Automatic Segmentation of Teeth, Gingiva and Facial Landmarks for 2D Digital Smile Design Using Real-Time Instance Segmentation Network. Journal of Clinical Medicine. 2022; 11(3):852. https://doi.org/10.3390/jcm11030852
Chicago/Turabian StyleLee, Seulgi, and Jong-Eun Kim. 2022. "Evaluating the Precision of Automatic Segmentation of Teeth, Gingiva and Facial Landmarks for 2D Digital Smile Design Using Real-Time Instance Segmentation Network" Journal of Clinical Medicine 11, no. 3: 852. https://doi.org/10.3390/jcm11030852
APA StyleLee, S., & Kim, J.-E. (2022). Evaluating the Precision of Automatic Segmentation of Teeth, Gingiva and Facial Landmarks for 2D Digital Smile Design Using Real-Time Instance Segmentation Network. Journal of Clinical Medicine, 11(3), 852. https://doi.org/10.3390/jcm11030852






