Modeling and Forecasting Monkeypox Cases Using Stochastic Models
Abstract
1. Introduction
2. Materials and Methods
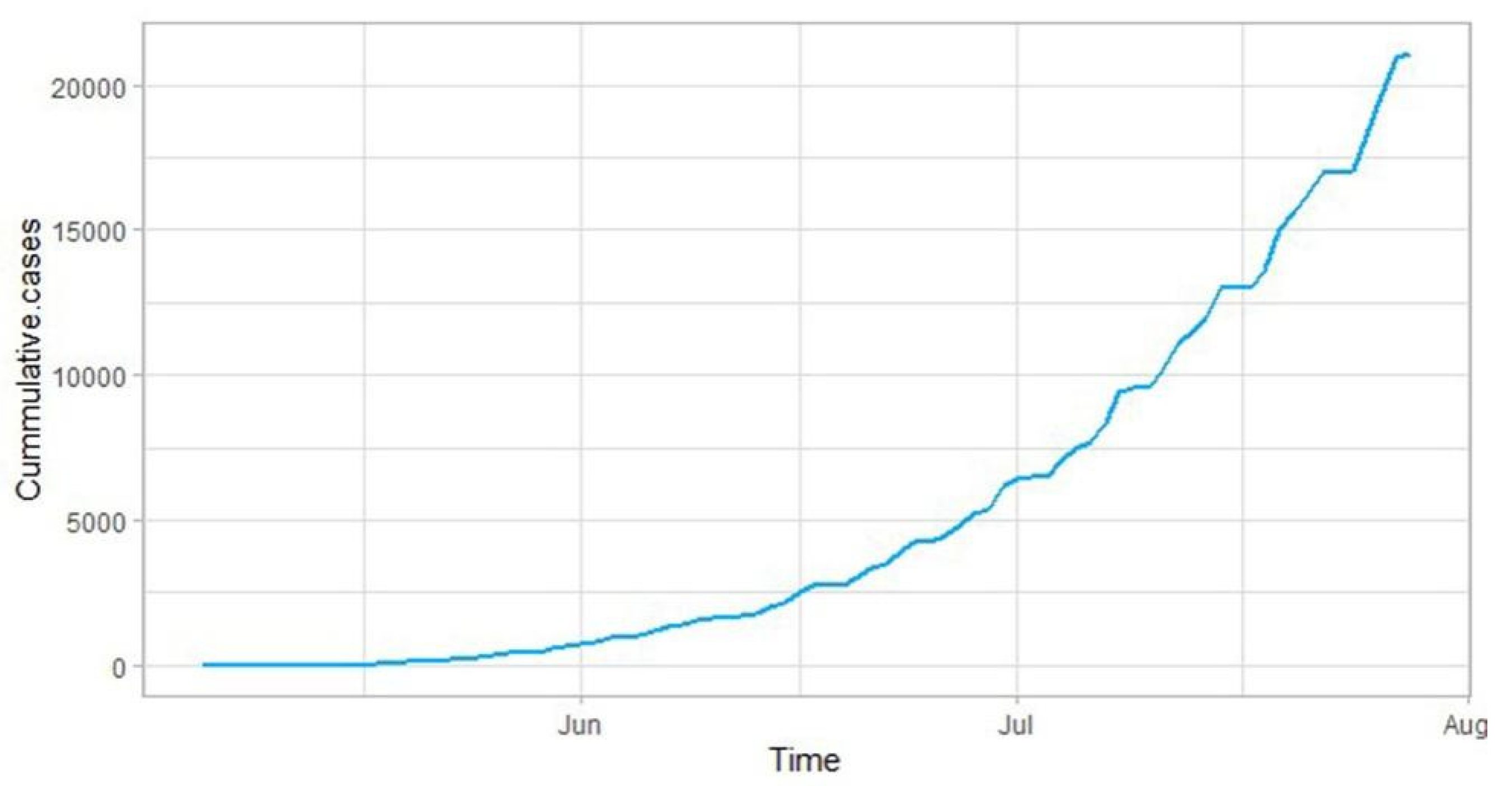
2.1. Study Area and Data Description
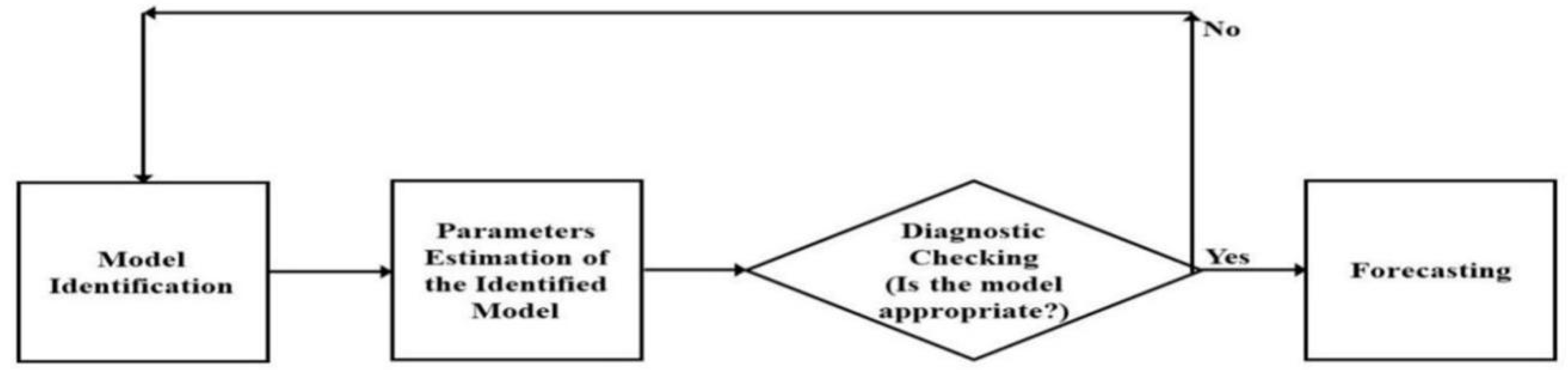
2.2. ARIMA Models
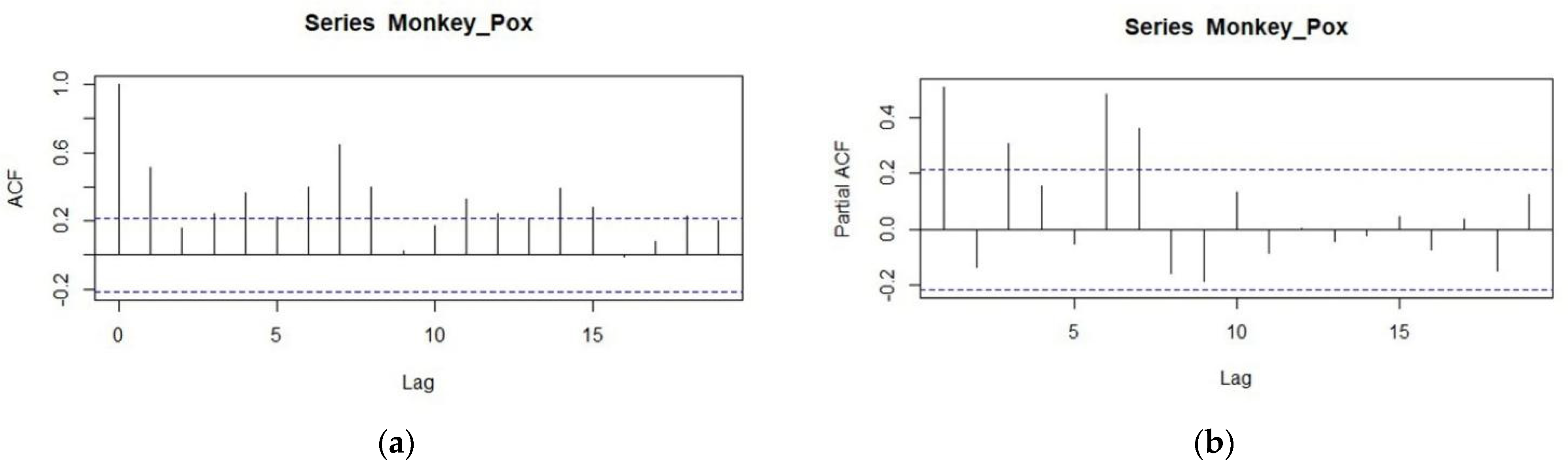
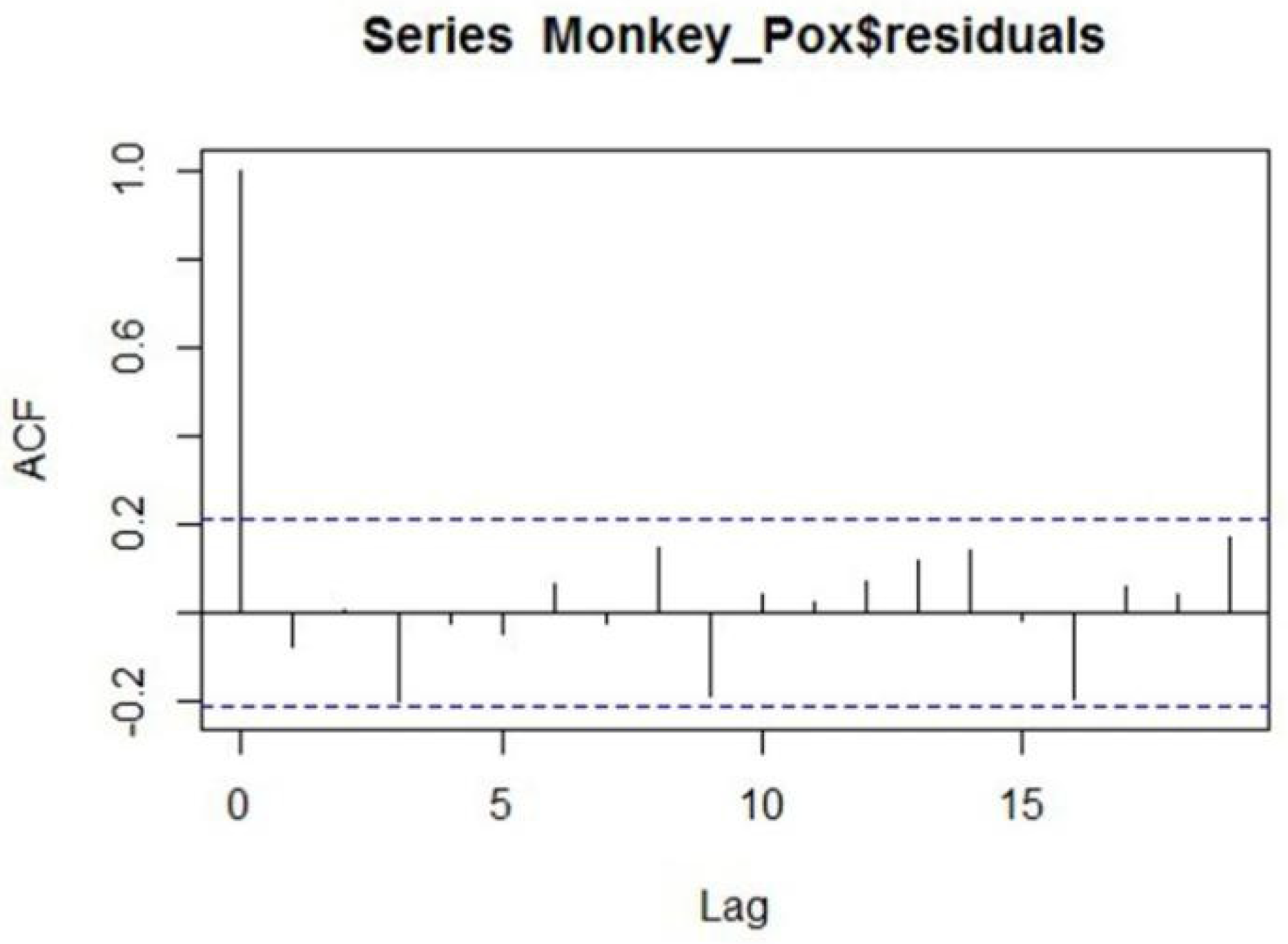
2.3. Methodology of ARIMA Models
2.4. Multilayer Perceptron Network (MLP)
2.5. Model Selection and Accuracy Measures
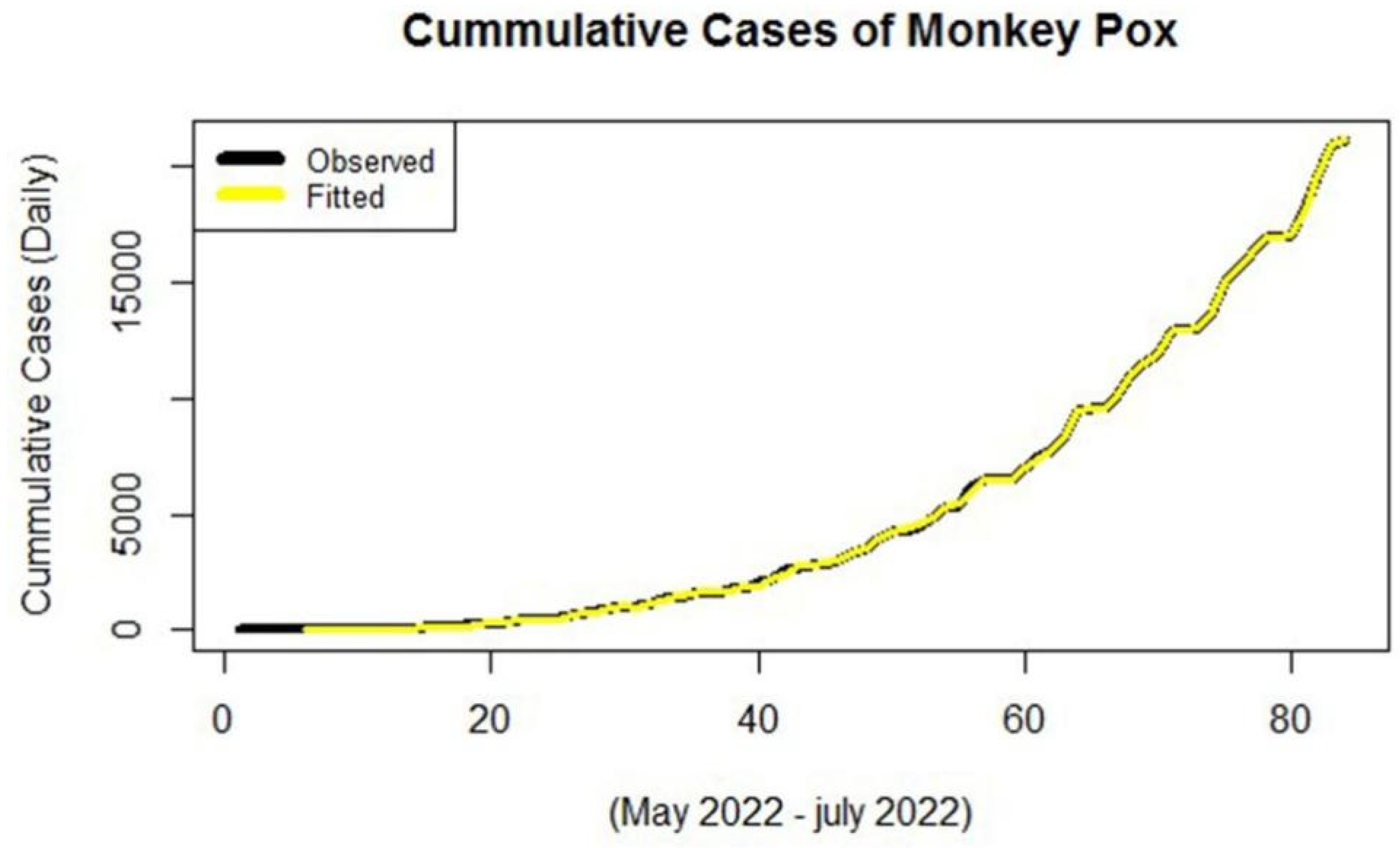
3. Results and Discussion
Multilayer Perceptron Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cho, C.T.; Wenner, H.A. Monkeypox virus. Bacteriol. Rev. 1973, 37, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Marennikova, S.; Šeluhina, E.M.; Mal’Ceva, N.; Čimiškjan, K.; Macevič, G. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull. World Health Organ. 1972, 46, 599. [Google Scholar] [PubMed]
- Bunge, E.M.; Hoet, B.; Chen, L.; Lienert, F.; Weidenthaler, H.; Baer, L.R.; Steffen, R. The changing epidemiology of human monkeypox—A potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022, 16, e0010141. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Perrichot, M.; Stemmler, M.; Emmerich, P.; Schmitz, H.; Varaine, F.; Shungu, R.; Tshioko, F.; Formenty, P. Outbreaks of disease suspected of being due to human monkeypox virus infection in the Democratic Republic of Congo in 2001. J. Clin. Microbiol. 2002, 40, 2919–2921. [Google Scholar] [CrossRef] [PubMed]
- Heymann, D.L.; Szczeniowski, M.; Esteves, K. Re-emergence of monkeypox in Africa: A review of the past six years. Br. Med. Bull. 1998, 54, 693–702. [Google Scholar] [CrossRef]
- Kumar, N.; Acharya, A.; Gendelman, H.E.; Byrareddy, S.N. The 2022 outbreak and the pathobiology of the monkeypox virus. J. Autoimmun. 2022, 131, 102855. [Google Scholar] [CrossRef]
- Vivancos, R.; Anderson, C.; Blomquist, P.; Balasegaram, S.; Bell, A.; Bishop, L.; Brown, C.S.; Chow, Y.; Edeghere, O.; Florence, I. Community transmission of monkeypox in the United Kingdom, April to May 2022. Eurosurveillance 2022, 27, 2200422. [Google Scholar] [CrossRef]
- WHO. 2022 Monkeypox Outbreak Global Map. Available online: https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html (accessed on 27 July 2022).
- Wang, Y.-W.; Shen, Z.-Z.; Jiang, Y. Comparison of ARIMA and GM (1, 1) models for prediction of hepatitis B in China. PLoS ONE 2018, 13, e0201987. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Zheng, Y.; Wang, K.; Zhang, X.; Zheng, Y. Time prediction models for echinococcosis based on gray system theory and epidemic dynamics. Int. J. Environ. Res. Public Health 2017, 14, 262. [Google Scholar] [CrossRef]
- Kandula, S.; Yamana, T.; Pei, S.; Yang, W.; Morita, H.; Shaman, J. Evaluation of mechanistic and statistical methods in forecasting influenza-like illness. J. R. Soc. Interface 2018, 15, 20180174. [Google Scholar] [CrossRef]
- Ren, H.; Li, J.; Yuan, Z.-A.; Hu, J.-Y.; Yu, Y.; Lu, Y.-H. The development of a combined mathematical model to forecast the incidence of hepatitis E in Shanghai, China. BMC Infect. Dis. 2013, 13, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Yang, M.; Zhang, T.; Young, A.A.; Li, X. Comparative study of four time series methods in forecasting typhoid fever incidence in China. PLoS ONE 2013, 8, e63116. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.C.; Molesworth, A.M.; Djingarey, M.H.; Yameogo, K.; Belanger, F.; Cuevas, L.E.J.T.M. Potential of environmental models to predict meningitis epidemics in Africa. Trop. Med. Int. Health 2006, 11, 781–788. [Google Scholar] [CrossRef]
- Orbann, C.; Sattenspiel, L.; Miller, E.; Dimka, J. Defining epidemics in computer simulation models: How do definitions influence conclusions? Epidemics 2017, 19, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Kurbalija, V.; Radovanović, M.; Ivanović, M.; Schmidt, D.; von Trzebiatowski, G.L.; Burkhard, H.-D.; Hinrichs, C. Time-series analysis in the medical domain: A study of Tacrolimus administration and influence on kidney graft function. Comput. Biol. Med. 2014, 50, 19–31. [Google Scholar] [CrossRef]
- Bernal, J.L.; Cummins, S.; Gasparrini, A. Interrupted time series regression for the evaluation of public health interventions: A tutorial. Int. J. Epidemiol. 2017, 46, 348–355. [Google Scholar] [CrossRef]
- Bernal, J.L.; Soumerai, S.; Gasparrini, A. A methodological framework for model selection in interrupted time series studies. J. Clin. Epidemiol. 2018, 103, 82–91. [Google Scholar] [CrossRef]
- Polwiang, S. The time series seasonal patterns of dengue fever and associated weather variables in Bangkok (2003–2017). BMC Infect. Dis. 2020, 20, 208. [Google Scholar] [CrossRef]
- Gaudart, J.; Touré, O.; Dessay, N.; Dicko, A.L.; Ranque, S.; Forest, L.; Demongeot, J.; Doumbo, O.K. Modelling malaria incidence with environmental dependency in a locality of Sudanese savannah area, Mali. Malar. J. 2009, 8, 61. [Google Scholar] [CrossRef]
- Wei, W.; Jiang, J.; Liang, H.; Gao, L.; Liang, B.; Huang, J.; Zang, N.; Liao, Y.; Yu, J.; Lai, J.; et al. Application of a Combined Model with Autoregressive Integrated Moving Average (ARIMA) and Generalized Regression Neural Network (GRNN) in Forecasting Hepatitis Incidence in Heng County, China. PLoS ONE 2016, 11, e0156768. [Google Scholar] [CrossRef]
- Zheng, Y.L.; Zhang, L.P.; Zhang, X.L.; Wang, K.; Zheng, Y.J. Forecast model analysis for the morbidity of tuberculosis in Xinjiang, China. PLoS ONE 2015, 10, e0116832. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Tao, H. Epidemiology and ARIMA model of positive-rate of influenza viruses among children in Wuhan, China: A nine-year retrospective study. Int. J. Infect. Dis. 2018, 74, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Roser, M.; Ritchie, H.; Ortiz-Ospina, E.; Hasell, J. Coronavirus Pandemic (COVID-19). Our World Data 2020, 4. [Google Scholar]
- Bartholomew, D. Time Series Analysis Forecasting and Control; Wiley: Hoboken, NJ, USA, 1971; Volume 22, pp. 199–201. [Google Scholar]
- Minhaj, F.S.; Ogale, Y.P.; Whitehill, F.; Schultz, J.; Foote, M.; Davidson, W.; Hughes, C.M.; Wilkins, K.; Bachmann, L.; Chatelain, R. Monkeypox outbreak—Nine states, May 2022. Morb. Mortal. Wkly. Rep. 2022, 71, 764. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, J.P.; von Elm, E.; Altman, D.G.; Gøtzsche, P.C.; Mulrow, C.D.; Pocock, S.J.; Poole, C.; Schlesselman, J.J.; Egger, M.; STROBE Initiative. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and elaboration. Ann. Intern. Med. 2007, 147, W-163–W-194. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, X.; Jiang, B.; Yang, W. Forecasting incidence of hemorrhagic fever with renal syndrome in China using ARIMA model. BMC Infect. Dis. 2011, 11, 218. [Google Scholar] [CrossRef]
- Benvenuto, D.; Giovanetti, M.; Vassallo, L.; Angeletti, S.; Ciccozzi, M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief. 2020, 29, 105340. [Google Scholar] [CrossRef]
- Priebe, S.; Huxley, P.; Knight, S.; Evans, S. Application and Results of the Manchester Short Assessment of Quality of Life (Mansa). Int. J. Soc. Psychiatry 1999, 45, 7–12. [Google Scholar] [CrossRef]
- Pankratz, A. Forecasting with Univariate Box-Jenkins Models: Concepts and Cases; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 224. [Google Scholar]
- Ljung, G.M.; Box, G.E.J.B. On a measure of lack of fit in time series models. Biometrika 1978, 65, 297–303. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]

| Country | Cases | Category |
|---|---|---|
| United States | 4638 | Has not historically reported monkeypox |
| Spain | 3738 | Has not historically reported monkeypox |
| Germany | 2459 | Has not historically reported monkeypox |
| United Kingdom | 2432 | Has not historically reported monkeypox |
| France | 1837 | Has not historically reported monkeypox |
| Netherlands | 818 | Has not historically reported monkeypox |
| Canada | 745 | Has not historically reported monkeypox |
| Brazil | 696 | Has not historically reported monkeypox |
| Portugal | 588 | Has not historically reported monkeypox |
| Italy | 426 | Has not historically reported monkeypox |
| Belgium | 393 | Has not historically reported monkeypox |
| Switzerland | 251 | Has not historically reported monkeypox |
| Peru | 224 | Has not historically reported monkeypox |
| The Democratic Republic of the Congo | 163 | Has historically reported monkeypox |
| Israel | 121 | Has not historically reported monkeypox |
| Nigeria | 117 | Has historically reported monkeypox |
| Austria | 115 | Has not historically reported monkeypox |
| Ireland | 85 | Has not historically reported monkeypox |
| Sweden | 81 | Has not historically reported monkeypox |
| Denmark | 71 | Has not historically reported monkeypox |
| Mexico | 59 | Has not historically reported monkeypox |
| Min | 1st Quartile | Median | Mode | 3rd Quartile | Max |
|---|---|---|---|---|---|
| 1 | 401 | 2654 | 5218 | 8657 | 21,099 |
| Candidate Model | MSE | RMSE | MAE | MAPE |
|---|---|---|---|---|
| ARIMA (5,1,5) | 38,549.4 | 196.34 | 118.05 | 6.52 |
| ARIMA (6,1,5) | 25,766.67 | 160.52 | 94.55 | 6.29 |
| ARIMA (7,1,7) | 22,734.61 | 150.78 | 88.65 | 5.72 |
| Serial No | Forecasted Values | Upper 95% C. I | Lower 95% C. I |
|---|---|---|---|
| 1 | 21,516.89 | 21,845.83 | 21,187.94 |
| 2 | 21,667.12 | 22,147.57 | 21,186.67 |
| 3 | 22,137.39 | 22,724.06 | 21,550.72 |
| 4 | 23,283.64 | 23,977.30 | 22,589.98 |
| 5 | 24,843.72 | 25,670.73 | 24,016.71 |
| 6 | 25,930.43 | 26,834.66 | 25,026.20 |
| 7 | 25,916.84 | 26,834.66 | 24,902.92 |
| 8 | 26,021.02 | 26,930.75 | 24,738.57 |
| 9 | 26,474.18 | 27,303.47 | 24,930.92 |
| 10 | 27,300.65 | 28,017.44 | 25,559.52 |
| Candidate Model | MSE | RMSE | MAE | MAPE |
|---|---|---|---|---|
| With 5 hidden neurons | 6964.31 | 83.45 | 56.70 | 0.27 |
| With 7 hidden neurons | 3895.64 | 62.41 | 41.66 | 0.19 |
| With 10 hidden neurons | 2960.29 | 54.40 | 32.59 | 0.12 |
| Serial No | Forecasted Values | Upper 95% C. I | Lower 95% C. I |
|---|---|---|---|
| 1 | 21,124.99 | 21,960.54 | 21,222.59 |
| 2 | 21,856.00 | 22,859.63 | 22,454.63 |
| 3 | 21,830.08 | 23,597.83 | 23,182.77 |
| 4 | 21,926.20 | 24,765.09 | 24,295.99 |
| 5 | 21,704.02 | 24,806.16 | 25,108.36 |
| 6 | 22,317.85 | 25,757.07 | 25,167.07 |
| 7 | 22,507.93 | 25,046.78 | 24,846.67 |
| 8 | 22,722.82 | 26,909.01 | 26,709.41 |
| 9 | 22,950.31 | 27,886.04 | 27,186.14 |
| 10 | 24,269.96 | 27,995.00 | 27,885.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qureshi, M.; Khan, S.; Bantan, R.A.R.; Daniyal, M.; Elgarhy, M.; Marzo, R.R.; Lin, Y. Modeling and Forecasting Monkeypox Cases Using Stochastic Models. J. Clin. Med. 2022, 11, 6555. https://doi.org/10.3390/jcm11216555
Qureshi M, Khan S, Bantan RAR, Daniyal M, Elgarhy M, Marzo RR, Lin Y. Modeling and Forecasting Monkeypox Cases Using Stochastic Models. Journal of Clinical Medicine. 2022; 11(21):6555. https://doi.org/10.3390/jcm11216555
Chicago/Turabian StyleQureshi, Moiz, Shahid Khan, Rashad A. R. Bantan, Muhammad Daniyal, Mohammed Elgarhy, Roy Rillera Marzo, and Yulan Lin. 2022. "Modeling and Forecasting Monkeypox Cases Using Stochastic Models" Journal of Clinical Medicine 11, no. 21: 6555. https://doi.org/10.3390/jcm11216555
APA StyleQureshi, M., Khan, S., Bantan, R. A. R., Daniyal, M., Elgarhy, M., Marzo, R. R., & Lin, Y. (2022). Modeling and Forecasting Monkeypox Cases Using Stochastic Models. Journal of Clinical Medicine, 11(21), 6555. https://doi.org/10.3390/jcm11216555










