Data-Driven Medicine in the Diagnosis and Treatment of Infertility
Abstract
1. Introduction
- A conceptual shift from a disease-centric to a health-centric model: How infertility care is moving beyond a disease-based reactive model to a pro-active model focused on enhancing patients’ health and well-being.
- Better prevention, diagnosis and treatments: Sophisticated big data analysis of cohorts have allowed for the development of better strategies for diagnosis and treatments.
- ML/AI in ART Treatments: How ML/AI are currently being used to improve IVF across almost all stages of the treatment process.
- The participatory citizen: How the individual is empowered to drive their own reproductive health and well-being [16].
2. A Conceptual Shift from a Disease-Centric to a Health-Centric Model
3. The Era of Big Data Is Enabling Better Prevention, Diagnosis and Treatments
3.1. Biomarkers and Screening Tests Can Guide Treatment Decisions
3.2. Mechanistic Understanding of Disease Can Help Stratify Patients for Treatment
3.3. Integrative Modelling of Non-Genetic Exposures Could Help Infertility Prevention Strategies
3.4. Genetic Data Can Be Used to Define Optimal Controlled Ovarian Hyperstimulation (COH) Dosing Regimens
3.5. The Microbiome as an Important Emerging Health Data Stream in Infertility
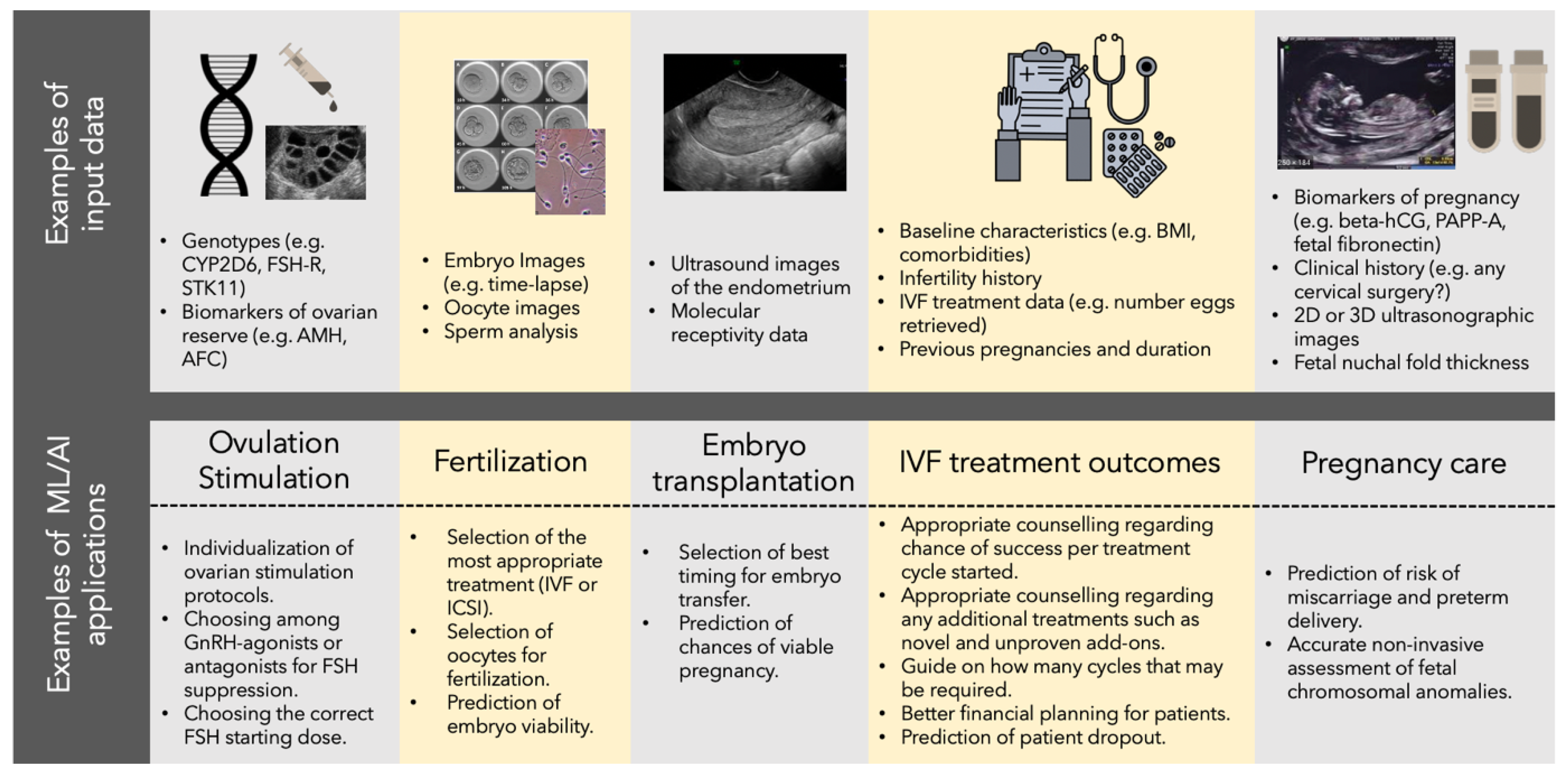
4. Machine Learning Is Aiding ART Treatments
5. The Participatory Citizen: From a Disease-Centric Model to Active Wellness
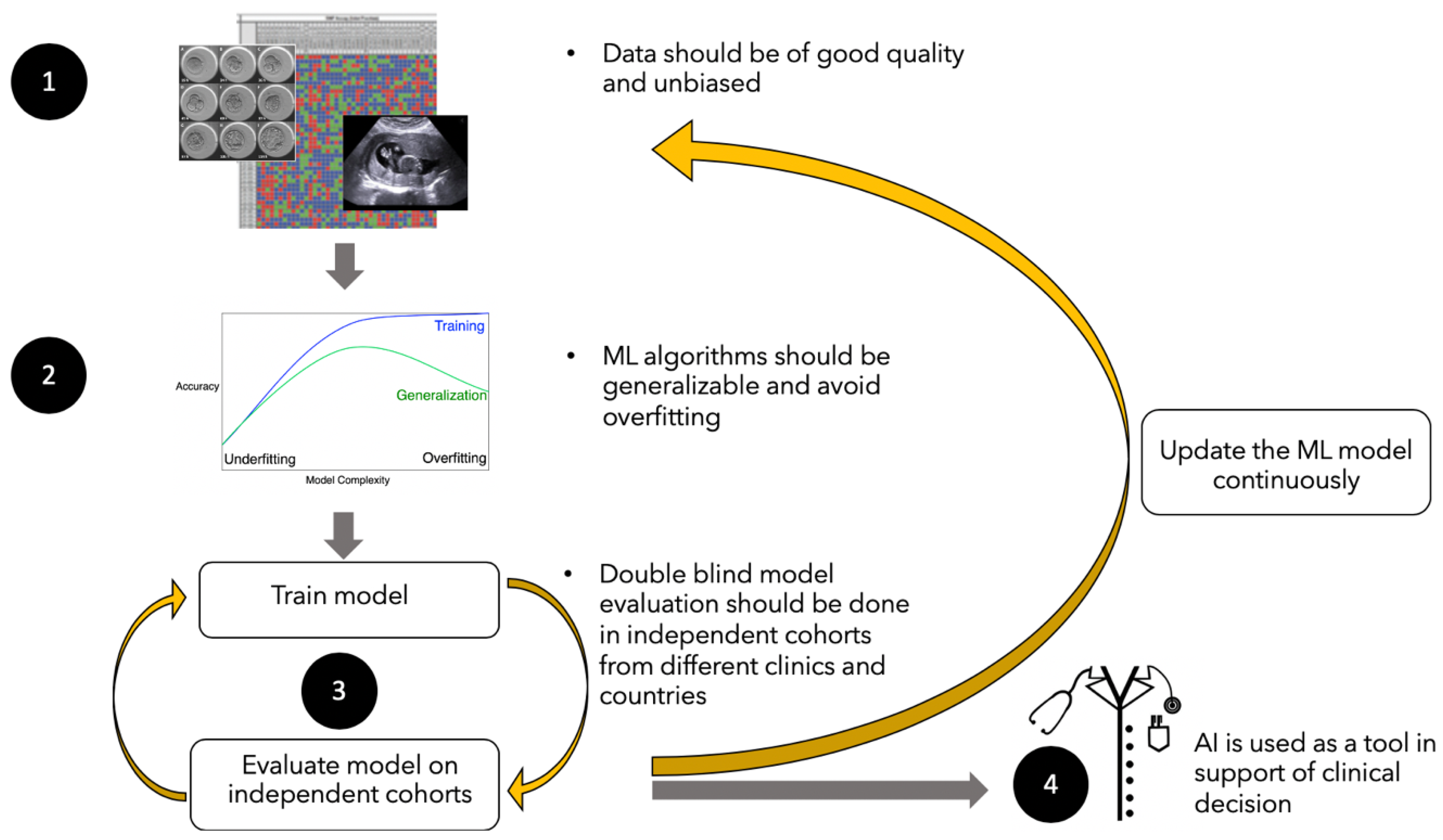
6. Challenges to the Use Machine Learning and Big Data in the Infertility Sector
6.1. High Quality and Quantities of Data
6.2. Generalizability of Learning
6.2.1. Data Biases Introduced by Population Heterogeneity
6.2.2. Non-Stationarity in Treatment Data and Historical Biases
6.3. Algorithm Validation Using Double-Blinded Datasets
6.4. The Challenge of Translation to Clinical Practice
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zegers-Hochschild, F.; Adamson, G.D.; de Mouzon, J.; Ishihara, O.; Mansour, R.; Nygren, K.; Sullivan, E.; van der Poel, S. The International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) Revised Glossary on ART Terminology, 2009. Hum. Reprod. 2009, 24, 2683–2687. [Google Scholar] [CrossRef]
- Healy, D.L.; Trounson, A.O.; Andersen, A. Female infertility: Causes and treatment. Lancet 1994, 343, 1539–1544. [Google Scholar] [CrossRef]
- Petraglia, F.; Serour, G.I.; Chapron, C. The changing prevalence of infertility. Int. J. Gynecol. Obstet. 2013, 123, S4–S8. [Google Scholar] [CrossRef]
- Mascarenhas, M.N.; Flaxman, S.R.; Boerma, T.; Vanderpoel, S.; Stevens, G.A. National, Regional, and Global Trends in Infertility Prevalence Since 1990: A Systematic Analysis of 277 Health Surveys. PLoS Med. 2012, 9, e1001356. [Google Scholar] [CrossRef]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Mindlis, I.; Pinotti, R.; Swan, S.H. Temporal trends in sperm count: A systematic review and meta-regression analysis. Hum. Reprod. Update 2017, 23, 646–659. [Google Scholar] [CrossRef]
- Quaas, A.; Dokras, A. Diagnosis and treatment of unexplained infertility. Rev. Obstet. Gynecol. 2008, 1, 69–76. [Google Scholar]
- Baird, D.T.; Collins, J.; Egozcue, J.; Evers, L.H.; Gianaroli, L.; Leridon, H.; Sunde, A.; Templeton, A.; Van Steirteghem, A.; Cohen, J.; et al. Fertility and ageing. Hum. Reprod. Update 2005, 11, 261–276. [Google Scholar] [CrossRef]
- OECD. Age of Mothers at Childbirth and Age-Specific Fertility. OECD—Social Policy Division—Directorate of Employment, Labour and Social Affairs. 2020. Available online: http://www.oecd.org/els/soc/SF_2_3_Age_mothers_childbirth.pdf (accessed on 18 May 2020).
- De Geyter, C.; Calhaz-Jorge, C.; Kupka, M.S.; Wyns, C.; Mocanu, E.; Motrenko, T.; Scaravelli, G.; Smeenk, J.; Vidakovic, S.; Goossens, V.; et al. ART in Europe, 2014: Results generated from European registries by ESHRE: The European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). Hum. Reprod. 2018, 33, 1586–1601. [Google Scholar] [CrossRef] [PubMed]
- Evers, J.L. Female subfertility. Lancet 2002, 360, 151–159. [Google Scholar] [CrossRef]
- Palermo, G.; Joris, H.; Devroey, P.; Van Steirteghem, A.C. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992, 340, 17–18. [Google Scholar] [CrossRef]
- Handyside, A.H.; Kontogianni, E.H.; Hardy, K.; Winston, R.M.L. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 1990, 344, 768–770. [Google Scholar] [CrossRef] [PubMed]
- Swain, J.; VerMilyea, M.T.; Meseguer, M.; Ezcurra, D.; Letterie, G.; Sánchez, P.; Trew, G.; Nayot, D.; Campbell, A.; Huangv, I.; et al. AI in the treatment of fertility: Key considerations. J. Assist. Reprod. Genet. 2020, 37, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Curchoe, C.L.; Malmsten, J.; Bormann, C.; Shafiee, H.; Farias, A.F.-S.; Mendizabal, G.; Chavez-Badiola, A.; Sigaras, A.; Alshubbar, H.; Chambost, J.; et al. Predictive modeling in reproductive medicine: Where will the future of artificial intelligence research take us? Fertil. Steril. 2020, 114, 934–940. [Google Scholar] [CrossRef]
- Hood, L. Systems biology: Integrating technology, biology, and computation. Mech. Ageing Dev. 2002, 124, 9–16. [Google Scholar] [CrossRef]
- Kaye, J.; Curren, L.; Anderson, N.; Edwards, K.; Fullerton, S.; Kanellopoulou, N.; Lund, D.; MacArthur, D.G.; Mascalzoni, D.; Shepherd, J.; et al. From patients to partners: Participant-centric initiatives in biomedical research. Nat. Rev. Genet. 2012, 13, 371–376. [Google Scholar] [CrossRef]
- Spandorfer, S.D.; Avrech, O.M.; Colombero, L.T.; Palermo, G.D.; Rosenwaks, Z. Effect of parental age on fertilization and pregnancy characteristics in couples treated by intracytoplasmic sperm injection. Hum. Reprod. 1998, 13, 334–338. [Google Scholar] [CrossRef]
- Westrom, L. Effect of pelvic inflammatory disease on fertility. Venereol. Off. Publ. Natl. Venereol. Counc. Aust. 1995, 8, 219–222. [Google Scholar]
- Tsevat, D.G.; Wiesenfeld, H.C.; Parks, C.; Peipert, J.F. Sexually transmitted diseases and infertility. Am. J. Obstet. Gynecol. 2017, 216, 1–9. [Google Scholar] [CrossRef]
- Norman, R.J.; Noakes, M.; Wu, R.; Davies, M.J.; Moran, L.; Wang, J.X. Improving reproductive performance in overweight/obese women with effective weight management. Hum. Reprod. Update 2004, 10, 267–280. [Google Scholar] [CrossRef]
- Balen, A.H.; Dresner, M.; Scott, E.M.; Drife, J.O. Should obese women with polycystic ovary syndrome receive treatment for infertility? BMJ 2006, 332, 434–435. [Google Scholar] [CrossRef]
- HFEA. Fertility Treatment 2018: Trends and Figures. HFEA. 2022. Available online: https://www.hfea.gov.uk/about-us/publications/research-and-data/fertility-treatment-2018-trends-and-figures/#storage (accessed on 17 October 2021).
- Egg Freezing in Fertility Treatment Trends and Figures: 2010–2016. HFEA. Available online: https://www.hfea.gov.uk/media/2656/egg-freezing-in-fertility-treatment-trends-and-figures-2010-2016-final.pdf (accessed on 17 October 2021).
- Goldman, R.H.; Racowsky, C.; Farland, L.V.; Munné, S.; Ribustello, L.; Fox, J.H. Predicting the likelihood of live birth for elective oocyte cryopreservation: A counseling tool for physicians and patients. Hum. Reprod. 2017, 32, 853–859. [Google Scholar] [CrossRef] [PubMed]
- HFEA. Egg Freezing. 2020. Available online: https://www.hfea.gov.uk/treatments/fertility-preservation/egg-freezing/ (accessed on 17 October 2021).
- Tüttelmann, F.; Nieschlag, E. Classification of andrological disorders. In Andrology: Male Reproductive Health and Dysfunction; Nieschlag, E., Behre, H.M., Nieschlag, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 87–92. [Google Scholar]
- Yan, S.; Shabbir, M.; Yap, T.; Homa, S.; Ramsay, J.; McEleny, K.; Minhas, S. Should the current guidelines for the treatment of varicoceles in infertile men be re-evaluated? Hum. Fertil. 2019, 24, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Mansour Ghanaie, M.; Asgari, S.A.; Dadrass, N.; Allahkhah, A.; Iran-Pour, E.; Safarinejad, M.R. Effects of varicocele repair on spontaneous first trimester miscarriage: A randomized clinical trial. Urol. J. 2012, 9, 505–513. [Google Scholar] [PubMed]
- Pastuszak, A.W.; Wang, R. Varicocele and testicular function. Asian J. Androl. 2015, 17, 659. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Deepinder, F.; Cocuzza, M.; Agarwal, R.; Short, R.A.; Sabanegh, E.; Marmar, J.L. Efficacy of Varicocelectomy in Improving Semen Parameters: New Meta-analytical Approach. Urology 2007, 70, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Leng, P.; Wang, J.; Yang, G.; Zu, R.; Jia, X.; Zhang, K.; Mengesha, B.A.; Huang, J.; Wang, D.; et al. Clinlabomics: Leveraging clinical laboratory data by data mining strategies. BMC Bioinform. 2022, 23, 387. [Google Scholar] [CrossRef] [PubMed]
- Curchoe, C.L.; Bormann, C.L. Artificial intelligence and machine learning for human reproduction and embryology presented at ASRM and ESHRE 2018. J. Assist. Reprod. Genet. 2019, 36, 591–600. [Google Scholar] [CrossRef]
- Armstrong, S.; Bhide, P.; Jordan, V.; Pacey, A.; Marjoribanks, J.; Farquhar, C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst. Rev. 2019, 5, CD011320. [Google Scholar] [CrossRef]
- Bhide, P.; Srikantharajah, A.; Lanz, D.; Dodds, J.; Collins, B.; Zamora, J.; Chan, D.; Thangaratinam, S.; Khan, K.S. TILT: Time-Lapse Imaging Trial—A pragmatic, multi-centre, three-arm randomised controlled trial to assess the clinical effectiveness and safety of time-lapse imaging in in vitro fertilisation treatment. Trials 2020, 21, 600. [Google Scholar] [CrossRef]
- Cheredath, A.; Uppangala, S.; Asha, C.S.; Jijo, A.; Vani Lakshmi, R.; Kumar, P.; Joseph, D.; Nagana, G.A.; Kalthur, G.; Adiga, S.K. Combining Machine Learning with Metabolomic and Embryologic Data Improves Embryo Implantation Prediction. Reprod. Sci. 2022. [Google Scholar] [CrossRef]
- Heidari, M.; Darbandi, S.; Darbani, M.; Amirjanati, N.; Bozorgmehr, M.; Zeraati, H.; Akhondi, M.M.; Sadeghi, M.R. Evaluating the Potential of Three Sperm Surface Antigens as Egg-adhesion Biomarkers for Human Sperm Selection. J. Reprod. Infertil. 2018, 19, 203–210. [Google Scholar] [PubMed]
- Nikshad, A.; Aghlmandi, A.; Safaralizadeh, R.; Aghebati-Maleki, L.; Warkiani, M.E.; Khiavi, F.M.; Yousefi, M. Advances of microfluidic technology in reproductive biology. Life Sci. 2020, 265, 118767. [Google Scholar] [CrossRef] [PubMed]
- Nair, T.M. Statistical and artificial neural network-based analysis to understand complexity and heterogeneity in preeclampsia. Comput. Biol. Chem. 2018, 75, 222–230. [Google Scholar] [CrossRef]
- Gracie, S.; Pennell, C.; Ekman-Ordeberg, G.; Lye, S.; McManaman, J.; Williams, S.; Palmer, L.; Kelley, M.; Menon, R.; Gravett, M.; et al. An integrated systems biology approach to the study of preterm birth using "-omic" technology—a guideline for research. BMC Pregnancy Childbirth 2011, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Hulka, B.S.; Wilcosky, T. Biological Markers in Epidemiologic Research. Arch. Environ. Health Int. J. 1988, 43, 83–89. [Google Scholar] [CrossRef]
- Sufriyana, H.; Wu, Y.-W.; Su, E.C.-Y. Prediction of Preeclampsia and Intrauterine Growth Restriction: Development of Machine Learning Models on a Prospective Cohort. JMIR Med. Inform. 2020, 8, e15411. [Google Scholar] [CrossRef]
- Haller-Kikkatalo, K.; Salumets, A.; Uibo, R. Review on Autoimmune Reactions in Female Infertility: Antibodies to Follicle Stimulating Hormone. Clin. Dev. Immunol. 2012, 2012, 762541. [Google Scholar] [CrossRef]
- Deroux, A.; Dumestre-Perard, C.; Dunand-Faure, C.; Bouillet, L.; Hoffmann, P. Female Infertility and Serum Auto-antibodies: A Systematic Review. Clin. Rev. Allergy Immunol. 2017, 53, 78–86. [Google Scholar] [CrossRef]
- Empson, M.B.; Lassere, M.; Craig, J.C.; Scott, J.R. Prevention of recurrent miscarriage for women with antiphospholipid antibody or lupus anticoagulant. Cochrane Database Syst. Rev. 2005, 2012, CD002859. [Google Scholar] [CrossRef]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.; Krawetz, S.A.; Reproductive Medicine Network. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Yang, Q.; Hua, J.; Wang, L.; Xu, B.; Zhang, H.; Ye, N.; Zhang, Z.; Yu, D.; Cooke, H.J.; Zhang, Y.; et al. MicroRNA and piRNA Profiles in Normal Human Testis Detected by Next Generation Sequencing. PLoS ONE 2013, 8, e66809. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.-F.; Ouyang, Y.; Coyne, C.B.; Sadovsky, Y. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015, 213, S163–S172. [Google Scholar] [CrossRef]
- Yoffe, L.; Gilam, A.; Yaron, O.; Polsky, A.; Farberov, L.; Syngelaki, A.; Nicolaides, K.; Hod, M.; Shomron, N. Early Detection of Preeclampsia Using Circulating Small non-coding RNA. Sci. Rep. 2018, 8, 3401. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Gimeno, P.; Horcajadas, J.A.; Martinez-Conejero, J.A.; Esteban, F.J.; Alama, P.; Pellicer, A.; Simon, C. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil. Steril. 2011, 95, 50–60e15. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Gimeno, P.; Ruiz-Alonso, M.; Blesa, D.; Bosch, N.; Martínez-Conejero, J.A.; Alamá, P.; Garrido, N.; Pellicer, A.; Simón, C. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil. Steril. 2013, 99, 508–517. [Google Scholar] [CrossRef]
- Ben Rafael, Z. Endometrial Receptivity Analysis (ERA) test: An unproven technology. Hum. Reprod. Open 2021, 2021, hoab010. [Google Scholar] [CrossRef] [PubMed]
- Raff, M.; Jacobs, E.; Van Voorhis, B.V. End of an endometrial receptivity array? Fertil. Steril. 2022, 118, P737. [Google Scholar] [CrossRef]
- Gupta, S.; Agarwal, A.; Sekhon, L.; Krajcir, N.; Cocuzza, M.; Falcone, T. Serum and peritoneal abnormalities in endometriosis: Potential use as diagnostic markers. Minerva Ginecol. 2006, 58, 527–551. [Google Scholar]
- Kitawaki, J.; Ishihara, H.; Koshiba, H.; Kiyomizu, M.; Teramoto, M.; Kitaoka, Y.; Honjo, H. Usefulness and limits of CA-125 in diagnosis of endometriosis without associated ovarian endometriomas. Hum. Reprod. 2005, 20, 1999–2003. [Google Scholar] [CrossRef]
- Hirsch, M.; Duffy, J.M.N.; Deguara, C.S.; Davis, C.J.; Khan, K.S. Diagnostic accuracy of Cancer Antigen 125 (CA125) for endometriosis in symptomatic women: A multi-center study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 210, 102–107. [Google Scholar] [CrossRef]
- Karimi-Zarchi, M.; Dehshiri-Zadeh, N.; Sekhavat, L.; Nosouhi, F. Correlation of CA-125 serum level and clinico-pathological characteristic of patients with endometriosis. Int. J. Reprod. Biomed. 2016, 14, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Knific, T.; Vouk, K.; Vogler, A.; Osredkar, J.; Gstöttner, M.; Wenzl, R.; Rižner, T.L. Models including serum CA-125, BMI, cyst pathology, dysmenorrhea or dyspareunia for diagnosis of endometriosis. Biomarkers Med. 2018, 12, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Farkas, Z.; Török, P. Role of CA 125 level in management of endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 234, e109–e110. [Google Scholar] [CrossRef]
- Reynolds, K.; Sarangi, S.; Bardia, A.; Dizon, D.S. Precision medicine and personalized breast cancer: Combination pertuzumab therapy. Pharmgenom. Pers. Med. 2014, 7, 95–105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. New Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Becker, C.M.; Laufer, M.R.; Stratton, P.; Hummelshoj, L.; Missmer, S.A.; Zondervan, K.T.; Adamson, G.D.; Allaire, C.; Anchan, R.; Bedaiwy, M.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: I. Surgical phenotype data collection in endometriosis research. Fertil. Steril. 2014, 102, 1213–1222. [Google Scholar] [CrossRef]
- A Yatsenko, S.; Rajkovic, A. Genetics of human female infertility. Biol. Reprod. 2019, 101, 549–566. [Google Scholar] [CrossRef]
- Miyamoto, T.; Minase, G.; Shin, T.; Ueda, H.; Okada, H.; Sengoku, K. Human male infertility and its genetic causes. Reprod. Med. Biol. 2017, 16, 81–88. [Google Scholar] [CrossRef]
- Gajbhiye, R.; Fung, J.N.; Montgomery, G.W. Complex genetics of female fertility. NPJ Genom. Med. 2018, 3, 29. [Google Scholar] [CrossRef]
- Laven, J.S.E. Follicle Stimulating Hormone Receptor (FSHR) Polymorphisms and Polycystic Ovary Syndrome (PCOS). Front. Endocrinol. 2019, 10, 23. [Google Scholar] [CrossRef]
- Zhang, P.-Y.; Yu, Y. Precise Personalized Medicine in Gynecology Cancer and Infertility. Front. Cell Dev. Biol. 2019, 7, 382. [Google Scholar]
- Legro, R.S.; Barnhart, H.X.; Schlaff, W.D.; Carr, B.R.; Diamond, M.; Carson, S.A.; Steinkampf, M.P.; Coutifaris, C.; McGovern, P.G.; Cataldo, N.A.; et al. Ovulatory Response to Treatment of Polycystic Ovary Syndrome Is Associated with a Polymorphism in the STK11 Gene. J. Clin. Endocrinol. Metab. 2008, 93, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Fishel, S.; Baker, D.; Elson, J.; Ragunath, M.; Atkinson, G.; Shaker, A.; Omar, A.; Kazem, R.; Beccles, A.; Greer, I.A. Precision Medicine in Assisted Conception: A Multicenter Observational Treatment Cohort Study of the Annexin A5 M2 Haplotype as a Biomarker for Antithrombotic Treatment to Improve Pregnancy Outcome. eBioMedicine 2016, 10, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef]
- Chen, L.-W.; Wu, Y.; Neelakantan, N.; Chong, M.F.-F.; Pan, A.; van Dam, R.M. Maternal caffeine intake during pregnancy and risk of pregnancy loss: A categorical and dose–response meta-analysis of prospective studies. Public Health Nutr. 2016, 19, 1233–1244. [Google Scholar] [CrossRef]
- Gaskins, A.J.; Rich-Edwards, J.W.; Williams, P.L.; Toth, T.L.; A Missmer, S.; E Chavarro, J. Prepregnancy Low to Moderate Alcohol Intake Is Not Associated with Risk of Spontaneous Abortion or Stillbirth. J. Nutr. 2015, 146, 799–805. [Google Scholar] [CrossRef]
- Carré, J.; Gatimel, N.; Moreau, J.; Parinaud, J.; Léandri, R. Does air pollution play a role in infertility?: A systematic review. Environ. Health 2017, 16, 82. [Google Scholar] [CrossRef]
- Rattan, S.; Zhou, C.; Chiang, C.; Mahalingam, S.; Brehm, E.; Flaws, J.A. Exposure to endocrine disruptors during adulthood: Consequences for female fertility. J. Endocrinol. 2017, 233, R109–R129. [Google Scholar] [CrossRef]
- Krzastek, S.C.; Farhi, J.; Gray, M.; Smith, R.P. Impact of environmental toxin exposure on male fertility potential. Transl. Androl. Urol. 2020, 9, 2797–2813. [Google Scholar] [CrossRef]
- Magiakou, M.A.; Mastorakos, G.; Webster, E.; Chrousos, G.P. The hypothalamic-pituitary-adrenal axis and the female reproductive system. Ann. N. Y. Acad. Sci. 1997, 816, 42–56. [Google Scholar] [CrossRef]
- Lathi, R.B.; Liebert, C.A.; Brookfield, K.F.; Taylor, J.A.; Saal, F.S.V.; Fujimoto, V.Y.; Baker, V.L. Conjugated bisphenol A in maternal serum in relation to miscarriage risk. Fertil. Steril. 2014, 102, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Stephens, V.R.; Rumph, J.T.; Ameli, S.; Bruner-Tran, K.L.; Osteen, K.G. The Potential Relationship Between Environmental Endocrine Disruptor Exposure and the Development of Endometriosis and Adenomyosis. Front. Physiol. 2022, 12, 807685. [Google Scholar] [CrossRef]
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Toppari, J.; Zoeller, R.T. EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr. Rev. 2015, 36, E1–E150. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, S.M.; Barupal, D.K.; Wishart, D.S.; Vineis, P.; Scalbert, A. The Blood Exposome and Its Role in Discovering Causes of Disease. Environ. Health Perspect. 2014, 122, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.; Eklund, M.; Madlensky, L.; Sawyer, S.D.; Thompson, C.K.; Fiscalini, A.S.; Ziv, E.; an’t Veer, L.J.; Esserman, L.J.; Tice, J.A. Breast Cancer Screening in the Precision Medicine Era: Risk-Based Screening in a Population-Based Trial. J. Natl. Cancer Inst. 2017, 109, djw290. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J.; Investigators WSaA. The WISDOM Study: Breaking the deadlock in the breast cancer screening debate. NPJ Breast Cancer 2017, 3, 34. [Google Scholar] [CrossRef]
- Ghobadi, C.; Gregory, A.; Crewe, H.K.; Rostami-Hodjegan, A.; Lennard, M.S. CYP2D6 is Primarily Responsible for the Metabolism of Clomiphene. Drug Metab. Pharmacokinet. 2008, 23, 101–105. [Google Scholar] [CrossRef]
- Ji, M.; Kim, K.-R.; Lee, W.; Choe, W.; Chun, S.; Min, W.-K. Genetic Polymorphism of CYP2D6and Clomiphene Concentrations in Infertile Patients with Ovulatory Dysfunction Treated with Clomiphene Citrate. J. Korean Med. Sci. 2016, 31, 310–314. [Google Scholar] [CrossRef]
- Mürdter, T.E.; Kerb, R.; Turpeinen, M.; Schroth, W.; Ganchev, B.; Böhmer, G.M.; Igel, S.; Schaeffeler, E.; Zanger, U.; Brauch, H.; et al. Genetic polymorphism of cytochrome P450 2D6 determines oestrogen receptor activity of the major infertility drug clomiphene via its active metabolites. Hum. Mol. Genet. 2012, 21, 1145–1154. [Google Scholar] [CrossRef]
- Simoni, M.; Gromoll, J.; Nieschlag, E. The Follicle-Stimulating Hormone Receptor: Biochemistry, Molecular Biology, Physiology, and Pathophysiology. Endocr. Rev. 1997, 18, 739–773. [Google Scholar] [CrossRef]
- Themmen, A.P.N.; Huhtaniemi, I.T. Mutations of Gonadotropins and Gonadotropin Receptors: Elucidating the Physiology and Pathophysiology of Pituitary-Gonadal Function. Endocr. Rev. 2000, 21, 551–583. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Kuroi, N.; Sano, M.; Tabara, Y.; Katsuya, T.; Ogihara, T.; Makita, Y.; Hata, A.; Yamada, M.; Takahashi, N.; et al. Mutation of the Follicle-Stimulating Hormone Receptor Gene 5′-Untranslated Region Associated With Female Hypertension. Hypertension 2006, 48, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Achrekar, S.K.; Modi, D.N.; Desai, S.K.; Mangoli, V.S.; Mangoli, R.V.; Mahale, S.D. Poor ovarian response to gonadotrophin stimulation is associated with FSH receptor polymorphism. Reprod. Biomed. Online 2009, 18, 509–515. [Google Scholar] [CrossRef]
- Mayorga, M.P.; Gromoll, J.; Behre, H.M.; Gassner, C.; Nieschlag, E.; Simoni, M. Ovarian Response to Follicle-Stimulating Hormone (FSH) Stimulation Depends on the FSH Receptor Genotype. J. Clin. Endocrinol. Metab. 2000, 85, 3365–3369. [Google Scholar] [CrossRef]
- Behre, H.M.; Greb, R.R.; Mempel, A.; Sonntag, B.; Kiesel, L.; Kaltwaer, P.; Seliger, E.; Röpke, F.; Gromoll, J.; Nieschlag, E.; et al. Significance of a common single nucleotide polymorphism in exon 10 of the follicle-stimulating hormone (FSH) receptor gene for the ovarian response to FSH: A pharmacogenetic approach to controlled ovarian hyperstimulation. Pharm. Genom. 2005, 15, 451–456. [Google Scholar] [CrossRef]
- Sudo, S.; Kudo, M.; Wada, S.-I.; Sato, O.; Hsueh, A.J.; Fujimoto, S. Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol. Hum. Reprod. 2002, 8, 893–899. [Google Scholar] [CrossRef]
- Laven, J.S.; Mulders, A.G.; A Suryandari, D.; Gromoll, J.; Nieschlag, E.; Fauser, B.C.; Simoni, M. Follicle-stimulating hormone receptor polymorphisms in women with normogonadotropic anovulatory infertility. Fertil. Steril. 2003, 80, 986–992. [Google Scholar] [CrossRef]
- Achrekar, S.K.; Modi, D.N.; Desai, S.K.; Mangoli, V.S.; Mangoli, R.V.; Mahale, S.D. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyperstimulation syndrome in Indian women. Fertil. Steril. 2009, 91, 432–439. [Google Scholar] [CrossRef]
- Moreno, I.; Simon, C. Deciphering the effect of reproductive tract microbiota on human reproduction. Reprod. Med. Biol. 2018, 18, 40–50. [Google Scholar] [CrossRef]
- Chen, C.; Song, X.; Chunwei, Z.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef]
- Hou, D.; Zhou, X.; Zhong, X.; Settles, M.L.; Herring, J.; Wang, L.; Abdo, Z.; Forney, L.J.; Xu, C. Microbiota of the seminal fluid from healthy and infertile men. Fertil. Steril. 2013, 100, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.-L.; Chiu, C.-M.; Lin, F.-M.; Huang, W.-C.; Liang, C.; Yang, T.; Yang, T.-L.; Liu, C.-Y.; Wu, W.-Y.; Chang, Y.-A.; et al. Bacterial Communities in Semen from Men of Infertile Couples: Metagenomic Sequencing Reveals Relationships of Seminal Microbiota to Semen Quality. PLoS ONE 2014, 9, e110152. [Google Scholar] [CrossRef] [PubMed]
- Kyono, K.; Hashimoto, T.; Nagai, Y.; Sakuraba, Y. Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: A single-center pilot study. Reprod. Med. Biol. 2018, 17, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kyono, K.; Hashimoto, T.; Kikuchi, S.; Nagai, Y.; Sakuraba, Y. A pilot study and case reports on endometrial microbiota and pregnancy outcome: An analysis using 16S rRNA gene sequencing among IVF patients, and trial therapeutic intervention for dysbiotic endometrium. Reprod. Med. Biol. 2018, 18, 72–82. [Google Scholar] [CrossRef]
- Vitagliano, A.; Saccardi, C.; Noventa, M.; Sardo, A.D.S.; Saccone, G.; Cicinelli, E.; Pizzi, S.; Andrisani, A.; Litta, P.S. Effects of chronic endometritis therapy on in vitro fertilization outcome in women with repeated implantation failure: A systematic review and meta-analysis. Fertil. Steril. 2018, 110, 103–112.e1. [Google Scholar] [CrossRef]
- Cicinelli, E.; Matteo, M.; Trojano, G.; Mitola, P.C.; Tinelli, R.; Vitagliano, A.; Crupano, F.M.; Lepera, A.; Miragliotta, G.; Resta, L. Chronic endometritis in patients with unexplained infertility: Prevalence and effects of antibiotic treatment on spontaneous conception. Am. J. Reprod. Immunol. 2018, 79, e12782. [Google Scholar] [CrossRef]
- UK CR. We’re Saving Lives Through Research: Annual Report & Accounts 2018/19. Available online: https://www.cancerresearchuk.org/sites/default/files/ec1060588_cruk_ar_2019_interactive.pdf (accessed on 2 July 2020).
- NIHR. Reproductive Health. Available online: https://www.nihr.ac.uk/explore-nihr/specialties/reproductive-health.htm (accessed on 2 July 2020).
- Gleicher, N.; Kushnir, V.A.; Barad, D.H. Worldwide decline of IVF birth rates and its probable causes. Hum. Reprod. Open 2019, 2019, hoz017. [Google Scholar] [CrossRef]
- Theobald, R.; Sengupta, S.; Harper, J. The status of preimplantation genetic testing in the UK and USA. Hum. Reprod. 2020, 35, 986–998. [Google Scholar] [CrossRef]
- Pirtea, P.; De Ziegler, D.; Tao, X.; Sun, L.; Zhan, Y.; Ayoubi, J.M.; Seli, E.; Franasiak, J.M.; Scott, R.T. Rate of true recurrent implantation failure is low: Results of three successive frozen euploid single embryo transfers. Fertil. Steril. 2021, 115, 45–53. [Google Scholar] [CrossRef]
- Xu, J.; Fang, R.; Chen, L.; Chen, D.; Xiao, J.-P.; Yang, W.; Wang, H.; Song, X.; Ma, T.; Bo, S.; et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, 11907–11912. [Google Scholar] [CrossRef]
- Ruiz-Alonso, M.; Blesa, D.; Diaz-Gimeno, P.; Garrido-Gómez, T.; Vilella, F.; Simón, C. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil. Steril. 2013, 100, 818–824. [Google Scholar] [CrossRef]
- Zaninovic, N.; Rosenwaks, Z. Artificial intelligence in human in vitro fertilization and embryology. Fertil. Steril. 2020, 114, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, I.; Zaninovic, N.; Badiola, A.C.; Bormann, C.L. Artificial intelligence in the embryology laboratory: A review. Reprod. Biomed. Online 2021, 44, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Yovich, J.; Alsbjerg, B.; Conceicao, J.; Hinchliffe, P.; Keane, K.N. PIVET rFSH dosing algorithms for individualized controlled ovarian stimulation enables optimized pregnancy productivity rates and avoidance of ovarian hyperstimulation syndrome. Drug Des. Dev. Ther. 2016, 10, 2561–2573. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, O.; Arce, J.-C. Individualized follitropin delta dosing reduces OHSS risk in Japanese IVF/ICSI patients: A randomized controlled trial. Reprod. Biomed. Online 2021, 42, 909–918. [Google Scholar] [CrossRef] [PubMed]
- You, J.B.; McCallum, C.; Wang, Y.; Riordon, J.; Nosrati, R.; Sinton, D. Machine learning for sperm selection. Nat. Rev. Urol. 2021, 18, 387–403. [Google Scholar] [CrossRef]
- Itoi, F.; Miyamoto, T.; Himaki, T.; Honnma, H.; Sano, M.; Ueda, J. Importance of real-time measurement of sperm head morphology in intracytoplasmic sperm injection. Zygote 2021, 30, 9–16. [Google Scholar] [CrossRef]
- Peschansky, C.; Patel, S.; Amir, J.; Dynia, S.; Usmani, S.; Lynn, R.; Vitale, K.; Grimm, L.; Arabi, A.; Jeelani, R.; et al. Picture perfect?: Determining the clinical utilization of artificial intelligence in oocyte cryopreservation. Fertil. Steril. 2021, 116, e157. [Google Scholar] [CrossRef]
- VerMilyea, M.; Hall, J.M.M.; Diakiw, S.M.; Johnston, A.; Nguyen, T.; Perugini, D.; Miller, A.; Picou, A.; Murphy, A.P.; Perugini, M. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum. Reprod. 2020, 35, 770–784. [Google Scholar] [CrossRef]
- Vermilyea, M.D.; Hall, J.M.; Perugini, D.; Murphy, A.P.; Ngyuen, T.; Rios, C.; Picou, A.; Miller, A.; Dinsmore, A.W.; Silverberg, K.; et al. Artificial intelligence: Non-invasive detection of morphological features associated with abnormalities in chromosomes 21 and 16. Fertil. Steril. 2019, 112, e237–e238. [Google Scholar] [CrossRef]
- Diagnosis and Management of Ectopic Pregnancy. BJOG Int. J. Obstet. Gynaecol. 2016, 123, e15–e55. [CrossRef] [PubMed]
- Liu, L.; Jiao, Y.; Li, X.; Ouyang, Y.; Shi, D. Machine learning algorithms to predict early pregnancy loss after in vitro fertilization-embryo transfer with fetal heart rate as a strong predictor. Comput. Methods Programs Biomed. 2020, 196, 105624. [Google Scholar] [CrossRef] [PubMed]
- Wald, N.J.; Hackshaw, A.K. Combining ultrasound and biochemistry in first-trimester screening for Down’s syndrome. Prenat. Diagn. 1997, 17, 821–829. [Google Scholar] [CrossRef]
- Health Quality Ontario. Noninvasive Prenatal Testing for Trisomies 21, 18, and 13, Sex Chromosome Aneuploidies, and Microdeletions: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2019, 19, 1–166. [Google Scholar]
- Gynaecologists RCoOa. Amniocentesis and Chorionic Villus Sampling; Green-Top Guideline No.8; RCOG Press: London, UK, 2010. [Google Scholar]
- Kuhrt, K.; Smout, E.; Hezelgrave, N.; Seed, P.T.; Carter, J.; Shennan, A.H. Development and validation of a tool incorporating cervical length and quantitative fetal fibronectin to predict spontaneous preterm birth in asymptomatic high-risk women. Ultrasound Obstet. Gynecol. 2016, 47, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Watson, H.A.; Seed, P.T.; Carter, J.; Hezelgrave, N.L.; Kuhrt, K.; Tribe, R.M.; Shennan, A.H. Development and validation of predictive models for QUiPP App v.2: Tool for predicting preterm birth in asymptomatic high-risk women. Ultrasound Obstet. Gynecol. 2019, 55, 348–356. [Google Scholar] [CrossRef]
- Frydman, G. A patient-centric definition of participatory medicine. J. Particip. Med. 2010, 12, 2016. [Google Scholar]
- Stuppia, L.; Gatta, V. Genetic testing in infertile couples. EuroBiotech J. 2017, 1, 151. [Google Scholar] [CrossRef]
- Beim, P.Y.; Parfitt, D.-E.; Tan, L.; Sugarman, E.A.; Hu-Seliger, T.; Clementi, C.; Levy, B. At the dawn of personalized reproductive medicine: Opportunities and challenges with incorporating multigene panel testing into fertility care. J. Assist. Reprod. Genet. 2017, 34, 1573–1576. [Google Scholar] [CrossRef][Green Version]
- Petrucelli, N.; Daly, M.B.; Pal, T. BRCA1- and BRCA2-Associated hereditary breast and ovarian cancer. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Milne-Ives, M.; van Velthoven, M.H.; Meinert, E. Mobile apps for real-world evidence in health care. J. Am. Med. Inform. Assoc. 2020, 27, 976–980. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Lynam, A.L.; Dennis, J.M.; Owen, K.R.; Oram, R.A.; Jones, A.G.; Shields, B.M.; Ferrat, L.A. Logistic regression has similar performance to optimised machine learning algorithms in a clinical setting: Application to the discrimination between type 1 and type 2 diabetes in young adults. Diagn. Progn. Res. 2020, 4, 6. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Velde, E.T.; Nieboer, D.; Lintsen, A.; Braat, D.; Eijkemans, M.; Habbema, J.; Vergouwe, Y. Comparison of two models predicting IVF success; the effect of time trends on model performance. Hum. Reprod. 2014, 29, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Mulherin, S.A.; Miller, W.C. Spectrum Bias or Spectrum Effect? Subgroup Variation in Diagnostic Test Evaluation. Ann. Intern. Med. 2002, 137, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, D.F.; Feinstein, A.R. Problems of Spectrum and Bias in Evaluating the Efficacy of Diagnostic Tests. N. Engl. J. Med. 1978, 299, 926–930. [Google Scholar] [CrossRef]
- HFEA. Ehnic Diversity in Fertility Treatment. Available online: https://www.hfea.gov.uk/about-us/publications/research-and-data/ethnic-diversity-in-fertility-treatment-2018/ (accessed on 28 March 2021).
- Dyer, S.; Chambers, G.; de Mouzon, J.; Nygren, K.; Zegers-Hochschild, F.; Mansour, R.; Ishihara, O.; Banker, M.; Adamson, G. International Committee for Monitoring Assisted Reproductive Technologies world report: Assisted Reproductive Technology 2008, 2009 and 2010. Hum. Reprod. 2016, 31, 1588–1609. [Google Scholar] [CrossRef]
- Sauerbrei, W. Prognostic Factors. Confusion caused by bad quality design, analysis and reporting of many studies. Adv. Otorhinolaryngol. 2005, 62, 184–200. [Google Scholar] [CrossRef]
- Knottnerus, J.A. Between iatrotropic stimulus and interiatric referral: The domain of primary care research. J. Clin. Epidemiol. 2002, 55, 1201–1206. [Google Scholar] [CrossRef]
- HFEA. Multiple Births Minimisation Strategy. Available online: https://ifqlive.blob.core.windows.net/umbraco-website/1314/2009-12-09_authority_papers_-_527_multiple_births.pdf (accessed on 10 December 2021).
- Braude, P. One Child at a Time: Reducing Multiple Births after IVF. In Report of the Expert Group on Multiple Births after IVF; HFEA: London, UK, 2006; Available online: https://ifqlive.blob.core.windows.net/umbraco-website/1311/one-child-at-a-time-report.pdf (accessed on 2 April 2022).
- HFEA. Fertility Treatment 2017: Trends and Figures. Available online: https://www.hfea.gov.uk/media/2894/fertility-treatment-2017-trends-and-figures-may-2019.pdf (accessed on 25 August 2021).
- Janssen, K.J.; Moons, K.G.; Kalkman, C.J.; Grobbee, D.E.; Vergouwe, Y. Updating methods improved the performance of a clinical prediction model in new patients. J. Clin. Epidemiol. 2008, 61, 76–86. [Google Scholar] [CrossRef]
- Riley, R.D.; Ensor, J.; Snell, K.; Debray, T.; Altman, D.G.; Moons, K.G.M.; Collins, G. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: Opportunities and challenges. BMJ 2016, 353, i3140. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, S.; A Mateen, B.; Bohner, G.; Király, F.J.; Ghani, R.; Jonsson, P.; Cumbers, S.; Jonas, A.; McAllister, K.S.L.; Myles, P.; et al. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ 2020, 368, l6927. [Google Scholar] [CrossRef] [PubMed]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.M.; Kerlikowske, K.; Tosteson, A.N.A.; Miglioretti, D.L. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Saleiro, P.; Kuester, B.; Hinkson, L.; London, J.; Stevens, A.; Anisfeld, A.; Rodolfa, K.T.; Ghani, R. Aequitas: A bias and fairness audit toolkit. arXiv 2018, arXiv:181105577. [Google Scholar]
- Chen, I.Y.; Szolovits, P.; Ghassemi, M. Can AI Help Reduce Disparities in General Medical and Mental Health Care? AMA J. Ethics 2019, 21, E167–E179. [Google Scholar] [CrossRef]
- Abdullah, K.A.L.; Atazhanova, T.; Chavez-Badiola, A.; Shivhare, S.B. Automation in ART: Paving the Way for the Future of Infertility Treatment. Reprod. Sci. 2022. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Santiago, I.; Polanski, L. Data-Driven Medicine in the Diagnosis and Treatment of Infertility. J. Clin. Med. 2022, 11, 6426. https://doi.org/10.3390/jcm11216426
de Santiago I, Polanski L. Data-Driven Medicine in the Diagnosis and Treatment of Infertility. Journal of Clinical Medicine. 2022; 11(21):6426. https://doi.org/10.3390/jcm11216426
Chicago/Turabian Stylede Santiago, Ines, and Lukasz Polanski. 2022. "Data-Driven Medicine in the Diagnosis and Treatment of Infertility" Journal of Clinical Medicine 11, no. 21: 6426. https://doi.org/10.3390/jcm11216426
APA Stylede Santiago, I., & Polanski, L. (2022). Data-Driven Medicine in the Diagnosis and Treatment of Infertility. Journal of Clinical Medicine, 11(21), 6426. https://doi.org/10.3390/jcm11216426






