Abstract
Non-syndromic tooth agenesis (ns-TA) is one of the most common dental anomalies characterized by the congenital absence of at least one permanent tooth (excluding third molars). Regarding the essential role of genetic factors in ns-TA aetiology, the present study aimed to identify novel pathogenic variants underlying hypodontia and oligodontia. In a group of 65 ns-TA patients and 127 healthy individuals from the genetically homogenous Polish population, the coding sequences of 423 candidate genes were screened using targeted next-generation sequencing. Pathogenic and likely pathogenic variants were identified in 37 (56.92%) patients, including eight nucleotide alternations of genes not previously implicated in ns-TA (CHD7, CREBBP, EVC, LEF1, ROR2, TBX22 and TP63). However, since only single variants were detected, future research is required to confirm and fully understand their role in the aetiology of ns-TA. Additionally, our results support the importance of already known ns-TA candidate genes (AXIN2, EDA, EDAR, IRF6, LAMA3, LRP6, MSX1, PAX9 and WNT10A) and provide additional evidence that ns-TA might be an oligogenic condition involving the cumulative effect of rare variants in two or more distinct genes.
1. Introduction
Odontogenesis is a complex and strictly regulated process that requires a series of sequential and reciprocal epithelial-mesenchymal cell interactions mediated by conserved signal transduction pathways, including Wnt/β-catenin, Bmp, Fgf, Shh and Eda [1,2]. Since these networks of signalling molecules and transcription factors play essential and widespread roles during embryogenesis, dentition provides an easily accessible and potentially general model to study organ development and regeneration [2]. Moreover, various anomalies in teeth size, shape, number and structure may be associated with defects in other tissues and organs [2]. For example, tooth agenesis (TA), which is a component of many clinically recognizable syndromes and multisystem disorders, including hypohidrotic ectodermal dysplasia (OMIM # 305100), Kallmann syndrome (OMIM # 147950), odonto-onycho-dermal dysplasia (OMIM # 257980), Witkop syndrome (OMIM # 189500) and van der Woude syndrome (OMIM # 119300). The congenital lack of teeth is also observed as an isolated condition (non-syndromic TA, ns-TA) and is classified as hypodontia or oligodontia based on the number of missing teeth. In most studies, the prevalence of hypodontia (agenesis of one to five teeth, excluding third molars) ranges from 3% to 10%, while the incidence of oligodontia (agenesis of 6 or more teeth, excluding third molars) varies from 0.1% to 0.5% [3]. The lack of wisdom teeth is more common and occurs in up to 30% of the general population [4].
The aetiology of ns-TA is multifactorial, with genetic and environmental factors considered possible contributing agents [3]. It has been estimated that in about 80% of affected individuals, ns-TA is caused by pathogenic variants of genes, the protein products of which are involved in craniofacial and tooth development [3]. In the remaining 20% of cases, ns-TA is attributed to exogenous factors, including trauma, chemotherapy and radiotherapy treatment in early infancy, as well as maternal smoking, viral infections, and medication intake during pregnancy [5]. In addition, Wang et al. have demonstrated that sporadic ns-TA might also be related to disturbances in DNA methylation patterns [6].
To date, numerous genes and nucleotide variants have been associated with the risk of ns-TA [3]. One of the key susceptibility genes for this dental anomaly is WNT10A (OMIM * 606268), which encodes a secreted signalling protein of the Wnt/β-catenin pathway involved in multiple stages of tooth development [7,8]. It has been demonstrated that more than half of individuals with hypodontia and oligodontia are carriers of the WNT10A risk variants, comprising the most frequently observed p.Phe228Ile (rs121908120) and p.Cys107Ter (rs121908119) [9]. The well-known genes implicated in ns-TA aetiology include also AXIN2 (OMIM * 604025), EDA (OMIM * 300451), EDAR (OMIM * 604095), LRP6 (OMIM * 603507), MSX1 (OMIM * 142983) and PAX9 (OMIM * 167416) [3,10]. It is worth noting that pathogenic variants of some of these genes are associated with syndromic forms of dental agenesis, suggesting that an apparent ns-TA exists either as a distinct entity or as part of a spectrum of syndromic conditions.
Recently, progress in identifying novel ns-TA risk variants has been made through next-generation sequencing (NGS) studies. Moreover, these investigations have demonstrated that in some familial cases, ns-TA might be an oligogenic condition involving the cumulative effect of more than one causal variant [11,12]. In most cases with suggestive evidence for a multilocus inheritance, one of the rare genetic alternations required for the phenotype expression is a missense or nonsense WNT10A variant [11,12,13,14,15,16]. These observations adding a layer of complexity to the genetic characterization of ns-TA might explain the inter or intrafamilial phenotypic variability, and apparent incomplete penetrance frequently observed in this dental anomaly [17].
The present retrospective study aimed to identify rare pathogenic variants underlying ns-TA by screening the coding sequences of 423 candidate genes using an NGS-based multi-gene panel testing in a group of 65 patients and 127 controls from the genetically homogenous Polish population.
2. Materials and Methods
2.1. Study Population
Peripheral blood samples were collected from 65 individuals with ns-TA (49.23% males) who were participants of the Comprehensive Therapy Programme for Craniofacial Anomalies at Poznan University of Medical Sciences. The median age of patients was 12 years (range from 7–17 years). The inclusion criterion was congenital agenesis of at least one permanent tooth, excluding third molars and lack of other apparent structural anomalies. The diagnosis of TA was based on clinical and panoramic radiographic examinations. Microdontia was not considered a form of tooth agenesis. Additionally, peg-shaped maxillary incisors were not considered as exclusion criterion in the current study. Case eligibility and non-syndromic designation were ascertained by clinicians, including an orthodontist, paediatrician, maxillofacial surgeon and speech therapist, using detailed diagnostic information from medical records. The patient group comprised 32 (49.23%) individuals with hypodontia and 33 (50.77%) individuals with oligodontia (Table 1). The median number of missing teeth per person was six (ranging from 1 to 24). The most commonly missing teeth were maxillary lateral incisors (16.63%), followed by mandibular second premolars (15.35%) and maxillary second premolars (11.94%). Tooth agenesis was associated with minor ectodermal features like fine hair, dry skin or brittle nails in eight patients (12.31%; seven with oligodontia and one with hypodontia). They were included in the study cohort since no known syndromes were diagnosed. A positive family history of ns-TA was reported by 36 (55.39%) study participants. Peripheral blood samples were also collected from 127 healthy individuals (45.67% males) with all permanent teeth present (not considering the third molars) and no family history of TA. All study participants were unrelated Caucasians of Polish origin. Genomic DNA was extracted from peripheral blood lymphocytes by salting-out extraction procedure. DNA concentration and quality were determined using a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). The study was conducted according to the Declaration of Helsinki [18] and was approved by the Institutional Review Board of Poznan University of Medical Sciences, Poland (approval number 1115/18). Written informed consent was obtained from all study participants or their legal guardians.

Table 1.
Characteristics of the patient and control group.
2.2. Next-Generation Sequencing (NGS)
The coding regions, including exon-intron boundaries of 423 genes, were screened in all patients and controls using targeted NGS to identify rare pathogenic variants associated with ns-TA. The multi-gene panel was developed previously to identify rare variants underlying craniofacial anomalies, including non-syndromic cleft lip with or without cleft palate (ns-CL/P). The detailed methodology for panel design, library preparation, NGS sequencing and data analysis has been described previously [19]. Briefly, the panel was created to target the coding sequence of genes associated with non-syndromic and syndromic forms of orofacial clefts (OFC) and TA and genes encoding proteins implicated in signalling pathways critical for craniofacial and orodental development. Detailed characteristics of all panel genes are presented in Table S1. The DNA probe set complementary to target regions with a total size of 1.59 Mb was designed using the NimbleDesign online tool (Roche, Madison, WI, USA). All sequencing libraries were constructed using the KAPA HyperPlus kit (Roche Sequencing Solutions, Pleasanton, CA, USA) and sequenced with 150-bp paired-end reads on a HiSeq 4000 platform (Illumina Inc., San Diego, CA, USA) according to the manufacturer’s protocol.
2.2.1. Variant Detection and Annotation
The raw sequencing data were demultiplexed and converted to standard FASTQ format using bcl2fastq software (Illumina Inc., San Diego, CA, USA). After the quality control step, read sequences were aligned using the Burrows-Wheeler Aligner software (http://bio-bwa.sourceforge.net/, access on 1 March 2022) to the GRCh38/hg38 reference genome. Variant calling was conducted using the GATK tool (https://www.broadinstitute.org/gatk/, access on 1 March 2022) Identified variants were annotated with functional information, population frequency (http://gnomad.broadinstitute.org/, access on 1 March 2022) and association with clinically relevant phenotypes based on ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, access on 1 March 2022) and Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk, access on 1 March 2022) online resources. Variant effects were predicted using 16 in silico algorithms, including MutationTaster, FATHMM, FATHMM-MKL, FATHMM-XF, LRT, DEOGEN2, EIGEN, EIGEN PC, SIFT, SIFT4G, PROVEAN, MVP, REVEL, PrimateAI, MetaSVM and MetaLR. In addition, the combined annotation dependent depletion (CADD) and adaptive boosting (ADA) scores were extracted from the genomic variant search engine VarSome (https://varsome.com/, access on 1 March 2022) for the missense variants and variants predicted to affect splicing.
2.2.2. Variant Filtering and Prioritizing
To reduce the list of candidate variants for association with ns-TA, only exonic and splicing alternations (excluding synonymous) with a minor allele frequency (MAF) < 0.001 in the gnomAD European (non-Finnish) population were analyzed. The only exceptions were variants of the WTN10A gene, all of which were taken into evaluation. Stop-gain, stop-loss, frameshift, splicing with ADA score ≥ 0.9, and missense variants with CADD score > 20, which were designed as pathogenic/damaging by at least half of the in silico prediction algorithms, were included in likely gene disrupting variants. Variants identified within genes previously associated with the risk of non-syndromic or syndromic forms of TA, genes with a well-documented role in the odontogenesis and genes with a probability of being loss-of-function intolerant (pLi) score ≥ 0.95 were prioritized. The variants fulfilling the rigorous selection criteria were classified as pathogenic or likely pathogenic depending on whether they were annotated in the ClinVar or HGMD databases.
2.2.3. Confirmation Analyses
The identified pathogenic and likely pathogenic variants were validated using Sanger sequencing, which was carried out using BigDyeTM Terminator Version 3.1 Ready Reaction Cycle Sequencing Kit on an ABI3730 Genetic Analyzer (Applied Biosystems, Waltham, CA, USA). The sequencing template PCR conditions and sequencing primers are presented in Table S2.
2.3. Common Variant Association Analysis
An analysis of common exonic and splicing alternations (MAF ≥ 0.1 in the case group) was conducted on genes (n = 16) with identified rare pathogenic and likely pathogenic variants. Their association with the risk of ns-TA was tested with the Cochran-Armitage trend test. The odds ratio (OR, allelic model) and corresponding 95% confidence intervals (95%CIs) were used to assess the strength of the association. ptrend-values below 1.72 × 10−3 (0.05/29 tested variants) were interpreted as statistically significant.
3. Results
NGS Analysis of 423-Panel Genes
After filtering, prioritization and validation steps, a final set of 27 pathogenic and likely pathogenic variants was identified. The detailed characteristics and in silico pathogenicity scores for all these genetic alternations identified in 37 ns-TA patients (56.92%) are presented in Table 2.

Table 2.
Characteristics of variants identified in patients with ns-TA.
Eight likely pathogenic variants were detected in genes not previously correlated with the risk of ns-TA (Table 3, Table 4 and Table S3; Figures S1–S8), including one frameshift variant (EVC_p.Ala565ValfsTer23) and one nonsense variant (ROR2_p.Ser632Ter) predicted to cause nonsense-mediated mRNA decay (NMD). The other six genetic alterations were novel missense variants (CHD7_ p.Glu1856Gln, CBP_ p.Pro344Ser, CBP_ p.Glu1560Lys, LEF1_ p.Lys95Asn, TBX22_ p.Pro242Leu and TP63_ p.Pro532Ala) with a CADD score > 22 and classified as pathogenic or damaging/deleterious by most in silico prediction tools (Table 2). The only exception was the TP63_ p.Pro532Ala, which did not fulfil the project’s strict selection criteria (CADD = 18.76; verdict pathogenic/damaging by 7 out of 16 in silico tools). It was detected in a patient with hypodontia, abnormally shaped maxillary right lateral incisor and taurodontic maxillary first and second molars, who was diagnosed with complex odontoma at the age of 12 (Table 4). Three out of eight likely pathogenic variants identified in novel ns-TA candidate genes, including TP63_p.Pro532Ala, CBP_p.Glu1560Lys and EVC_p.Ala565ValfsTer23 were found in patients carrying one of the WNT10A missense alterations (Table 4).

Table 3.
Dental phenotype of patients with variants identified in genes other than WNT10A.

Table 4.
Dental phenotype of patients with WNT10A gene variants.
Six nucleotide variants detected in 18 ns-TA patients (27.69%) were known variants of the WNT10A gene (Table 2 and Table 4). All of them, except p.Arg113Cys, were changes with a CADD score > 22 and classified as pathogenic or damaging/deleterious by most in silico algorithms (Table 2). The p.Cys107Ter (rs121908119) and p.Phe228Ile (rs121908120) variants were reported in the ClinVar database as pathogenic alternations for selective TA (OMIM # 150400), Schopf-Schulz-Passarge syndrome (OMIM # 224750) and odontoonychodermal dysplasia (OMIM # 257980). In patients with the WNT10A variants, the mean number of missing permanent teeth per person, excluding third molars, was 9.67 (range 2–20). The most commonly missing teeth were second premolars, followed by lateral incisors and first premolars (Table 4 and Figure 1). A positive family history of TA was reported by 11 (61.11%) of these affected individuals.
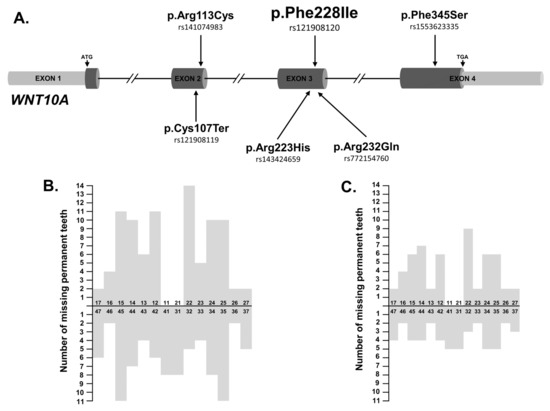
Figure 1.
(A) Distribution of WNT10A nucleotide variants identified in ns-TA patients. (B) Number of missing permanent teeth in 18 patients with identified WNT10 nucleotide variant/variants (n = 174, the mean number of missing permanent teeth per person, excluding third molars = 9.67, range 2–20). The most commonly missing teeth were second premolars, followed by lateral incisors and first premolars. (C) Number of missing permanent teeth in 9 patients exclusively harbouring the WNT10A_p.Phe228Ile variant (n = 104, the mean number of missing permanent teeth per person, excluding third molars = 11.6, range 3–20). The most commonly missing teeth were lateral incisors and first premolars followed by second premolars.
The most frequently observed WNT10A variant in the patients’ group was the nonsynonymous substitution p.Phe228Ile. It was identified in either heterozygous or homozygous form in 15 ns-TA patients (MAF = 0.16), as well as in one healthy individual. Under an assumption of an allelic inheritance model, the calculated OR for the p.Phe228Ile variant was 47.86 (95%CI: 6.36–360.45, ptrend = 9.15 × 10−7; Table 5). In four patients harbouring the p.Phe228Ile substitution, another heterozygous WNT10A variant was also detected (Table 4). In addition, two ns-TA patients carrying this missense substitution had likely pathogenic variant identified in a gene not previously correlated with the risk of ns-TA, including CREBBP (OMIM * 600140) and TP63 (OMIM * 603273) as described above (Table 4 and Figures S6 and S8).

Table 5.
Association of common exonic variants with the risk of ns-TA.
The list of variants that met the selection criteria also comprised 13 variants of well-known ns-TA risk genes, such as AXIN2, EDA, EDAR, IRF6 (OMIM * 607199), LAMA3 (OMIM * 600805), LRP6, MSX1 and PAX9 (Table 2 and Table 3). The three top likely pathogenic missense changes with the highest CADD scores were EDA_ p.Arg289His (CADD = 31.0), IRF6_ p.Arg339Gly (CADD = 28.7) and LRP6_p.Arg473Pro (CADD = 25.0). All identified stop-gain variants (AXIN2_p.Arg675ProfsTer32, AXIN2_p.Val765LeufsTer24, LAMA3_p.Glu306Ter, MSX1_p.Leu123ThrfsTer52 and PAX9_p.Gln136Ter) were predicted to cause NMD. Two out of three EDA variants detected in male patients with hypodontia were classified by HGMD and ClinVar as pathogenic for ectodermal dysplasia (p.Arg69Pro; HGMD accession number CM960503) and pathogenic/likely pathogenic for hypohidrotic X-linked ectodermal dysplasia and X-linked selective TA (p.Arg289His; ClinVar interpretation). The heterozygous nonsense variant of the LAMA3 gene identified in a patient with oligodontia (p.Glu306Ter, rs771405735) was reported in a ClinVar as a variant of uncertain significance for the autosomal recessive junctional epidermolysis bullosa gravis of Herlitz. The PAX9 transition leading to premature stop codon formation (p.Gln136Ter) was described previously in a Finnish family with non-syndromic oligodontia [13]. The remaining nine variants were novel and not previously reported in databases or the available literature. The group of patients with identified variants in known ns-TA risk genes included four (28.57%) individuals with hypodontia and ten (71.43%) individuals with oligodontia. A family history of TA was reported by nine of them (64.29%).
Except for the WNT10A_p.Phe228Ile substitution, none of the identified pathogenic and likely pathogenic variants for ns-TA were detected in the control group. In addition, no variants that fulfilled the project’s selection criteria were detected in genes harbouring these coding variants in healthy individuals. The only exception was the LAMA3 gene (pLi score of 0.00), in which a missense variant p.Asn2433Thr with a CADD score of 26 was identified in one control sample.
Association Analysis of Common Variants
Within genes harbouring pathogenic and likely pathogenic variants, 29 common exonic changes (11 missense and 18 silent) were identified (Table 5). Three of them were nominally associated with the risk of ns-TA in the tested population, including EVC_ p.Asn323Asn (ptrend = 4.90 × 10−2), IRF6_ p.Ser153Ser (ptrend = 3.29 × 10−2) and WNT10A_ p.Phe228Ile (ptrend = 9.15 × 10−7). The results for the latest variant, presented above, remained statistically significant even after adjusting for multiple comparisons.
4. Discussion
To investigate the role of rare nucleotide variants in the aetiology of ns-TA, a panel of functionally and clinically relevant candidate genes was analyzed using NGS in a group of 65 Polish patients with hypodontia and oligodontia. After applying a stringent variant filtering strategy, pathogenic and likely pathogenic nucleotide alternations were detected in 57% of affected individuals. Eight of these variants were identified in genes not previously associated with the ns-TA risk, including CHD7 (OMIM * 608892), CREBBP, EVC (OMIM * 604831), LEF1 (OMIM + 153245), ROR2 (OMIM * 602337), TBX22 (OMIM * 300307) and TP63. Expression and functional studies have demonstrated that protein products of all these novel candidate genes play crucial roles during odontogenesis and craniofacial development [20,21,22,23,24,25,26]. Mice deficient in Lef1, an essential epithelial survival factor in tooth morphogenesis, show arrested tooth development at the late bud stage before forming a mesenchymal dental papilla [20,27]. Impaired tooth development and morphology were also observed in Crebbp, Evc, Ror2 and Trp63 (orthologue of human TP63) knockout mice [28,29,30,31]. Moreover, pathogenic variants of novel ns-TA candidate genes identified in this study have been reported in human clinically distinguishable syndromes and multisystem disorders associated with various craniofacial abnormalities, including anomalies in the number, size and shape, structure, position and eruption of teeth. Recently, de novo microdeletions encompassing the LEF1 gene have also been identified in two unrelated patients with severe oligodontia and other features compatible with hypohidrotic ectodermal dysplasia [32]. In addition, monoallelic and biallelic LEF1 variants have been proposed as causative factors for a putatively novel syndrome characterized by limb malformations and ectodermal dysplasia [33]. It is worth noting that the heterozygous LEF1 missense variant (p.Lys95Asn) identified in the current study was found in an individual with a mild form of sporadic TA. This patient had congenital agenesis of a maxillary right lateral incisor and an abnormally shaped maxillary left lateral incisor without any other ectodermal features and structural anomalies.
Among the nucleotide alternations of these newly identified ns-TA candidate genes, three were found within sequences encoding highly conserved protein domains. One of them was the nonsense variant predicted to truncate the ROR2 receptor by 311 amino acids at the cytoplasmic kinase domain, which, together with the distal C-terminal regions, is required for receptor stability and downstream signalling [34,35]. In addition, this ROR2_p.Ser632Ter variant identified in a patient with non-syndromic hypodontia was also predicted to trigger NMD, a highly conserved RNA quality control pathway targeting mRNAs harbouring premature termination codons for degradation [36]. To date, pathogenic variants of the ROR2 receptor have been associated with Brachydactyly B (OMIM # 113000) and Robinow syndrome (OMIM # 268310), in which some individuals show dental problems such as delayed eruption and delayed root formation of permanent teeth [37]. Recent findings indicate that heterozygous recurrent nucleotide alternations, including the p.Gly559Ser missense variant located within the ROR2 kinase domain, might also be associated with isolated short stature [38].
In a male patient with the congenital lack of seven permanent teeth, a novel missense variant (p.Pro242Leu) was identified within the T-box domain of the transcription factor TBX22, acting as an essential transcriptional repressor during facial and palatal development [21]. Mutations in TBX22 are associated with X-linked cleft palate and ankyloglossia (OMIM # 303400), which is a disorder characterized by phenotypic heterogeneity ranging from ankyloglossia only to submucous cleft palate, bifid uvula or cleft of the soft and hard palate, all with or without tongue-tie [39]. Pathogenic variants of this gene were also detected in patients with non-syndromic CLP and patients with CLP and micro/hypodontia [40,41,42]. The current study expands the phenotypic spectrum associated with TBX22 pathogenic variants to include the ns-TA and highlights the essential role of the transcription factor TBX22 during odontogenesis. Interestingly, analyses in both humans and mice have demonstrated that during embryogenesis, TBX22 is specifically expressed in craniofacial tissues that are affected in TBX22 mutation carriers, including the mesenchyme of the palatal shelves, the first branchial arch, developing nose and base of the tongue [43]. High expression level in both species is also observed in the developing tooth buds [43].
Furthermore, a likely pathogenic variant for ns-TA was identified within the histone acetyltransferase (HAT) domain of CREB-binding protein (CBP), which acts as a transcriptional coactivator and epigenetic factor involved in the regulation of gene expression by modifying the chromatin structure [44]. It has been noted that the CBP HAT contains about 52% of missense mutations reported to cause Rubinstein-Taybi syndrome (OMIM # 180849), characterized by short stature, typical facial features, broad thumbs and halluces, and intellectual disability [45,46]. Dental anomalies such as talon cusps, hypodontia, supernumerary teeth and natal teeth are also frequently observed in patients with this rare autosomal dominant syndrome [44]. Animal studies have suggested that during odontogenesis, CBP provides positional information for tooth crown morphogenesis and regulates the growth of tooth cusps [22]. It is noteworthy that in our hypodontia patient with the CPB missense variant located within the HAT domain (p.Glu1560Lys), a recurrent pathogenic variant of WNT10A was also detected. In contrast, the second CREBB variant identified in the current study was the only variant fulfilling the strict project selection criteria detected in a patient with oligodontia. This novel likely pathogenic missense alteration (p.Pro344Ser) was situated outside the CBP HAT domain.
Our study provides additional evidence that ns-TA may have an oligogenic inheritance. Besides the patient harbouring variants in WNT10A and CREBBP, two other affected individuals were carriers of two pathogenic or likely pathogenic nucleotide alternations. In both cases, one variant was located within WNT10A while the other was within a gene not previously associated with ns-TA risk, including EVC and TP63. The heterozygous EVC_p.Ala565ValfsTer23 sequence change detected in a patient with the congenital lack of maxillary lateral incisors was categorized in the ClinVar database as pathogenic for Ellis-van Creveld syndrome (OMIM # 225500). All patients with this autosomal recessive chondrodysplasia present a wide range of dental anomalies, including hypoplastic enamel, abnormally shaped teeth, TA and premature tooth eruption [25]. Interestingly, there is evidence that heterozygous EVC mutations might also be associated with Weyers acrodental dysostosis (OMIM # 193530) [47]. Based on mice studies, it has been proposed that dental defects observed in patients with these congenital disorders result from disruption of the Sonic hedgehog signalling pathway combined with displaced Wnt signalling [48].
The novel TP63_p.Pro532Ala substitution together with the pathogenic WNT10A_p.Phe228Ile variant was identified in a patient with hypodontia and taurodontism of maxillary molars. In addition, one of her impacted maxillary lateral incisors was removed with complex odontoma. There is strong evidence from human and mouse studies for the TP63 transcription factor involvement in tooth development. Mutations of this gene have been associated with six autosomal dominant syndromes characterized mainly by ectodermal dysplasia, limb malformation and OFC [49]. Dental anomalies in these disorders are common and include TA, conoid teeth, taurodontism, enamel hypoplasia and dentinal dysplasia [50]. Classical animal studies have shown that p63 is an essential factor in embryonic epidermal development and epidermal keratinocyte proliferation and differentiation [51,52]. Moreover, p63 establishes enhancers at craniofacial development genes modifying their expression levels [53] and P63-deficient embryos fail to form ectodermal placodes that mark early tooth and hair follicle morphogenesis [30]. It should be noted that TP63 germline mutations associated with ectodermal-related syndromes are found almost exclusively within sequences encoding functional domains of the transcription factor [54]. In contrast, the TP63 missense variant identified in our patient is located outside these domains and does not match the rigorous project criteria used to select ns-TA risk variants. Therefore, future research will be required to approve TP63 as a novel candidate gene for the congenital lack of teeth.
The current study also confirms the status of WNT10A as a key susceptibility gene for ns-TA. Its known coding variants were identified in nearly 30% of patients with a mean number of missing teeth of 9.7 (range 2 to 20). Interestingly, in our previous research conducted in an independent group of patients with ns-TA, nucleotide alternations of WNT10A were detected in 62% of individuals with this common dental anomaly [9]. Therefore, we can conclude, similarly to van den Boogaard et al. [55], that WNT10A variants may account for about 50% of all cases of hypodontia and oligodontia in the Polish population. In accordance with other reports [9,13,55,56], the most frequently observed WNT10A variation was p.Phe228Ile (rs121908120), which was detected in heterozygous or homozygous form in 15 individuals with variable dental phenotypes. The most severe TA was observed in homozygous carriers of this recurrent variant; however, even among them, the number of missing permanent teeth varied and ranged from 6 to 20. The presence of phenotypic variability among carriers of the same DNA lesion might be explained not only by the co-occurrence of secondary genetic events but also by epigenetic modifications or environmental factors. It is worth noting that other likely pathogenic variants have been found in six heterozygous Phe228Ile carriers, including variants of WNT10A, CREBBP and TP63, as described above. In ns-TA patients, the WNT10A alternations have been previously identified in combination with nucleotide changes of BCOR (OMIM * 300485), EDA, EDAR, EDARADD (OMIM * 606603), LAMA3 or LRP6 [11,12,13,14,15,16].
Thirteen pathogenic and likely pathogenic nucleotide alternations were also detected in other known susceptibility genes for ns-TA, including AXIN2, EDA, EDAR, IRF6, LAMA3, LRP6, MSX1 and PAX9. The most severe dental phenotype with 15 or more missing permanent teeth was observed in female patients carrying newly discovered variants in AXIN2, EDA and MSX1 (p.Arg675ProfsTer32, p.Asn280Lys and p.Leu123ThrfsTer52, respectively). Interestingly, the oligodontia phenotype was also present in an individual with the IRF6 variant. The patient inherited this novel p.Arg339Gly missense substitution from her father with cleft palate only (results not shown). This result may support the hypothesis of a common genetic link between the congenital lack of teeth and orofacial clefts, which are frequently co-occurring craniofacial defects [19,57,58,59]. In addition, it may confirm the assumption that dental anomalies are subclinical phenotypes of the OFC spectrum. It should be noted that the increased incidence of dental abnormalities among unaffected family members of patients with overt clefts has already been noted [60,61].
The IRF6 gene is one of the the key susceptibility genes susceptibility genes for OFC, of which common and rare alleles have been found etiologic in both non-syndromic and syndromic forms of this structural anomaly [62,63]. The IRF6 deleterious variants have also been detected in patients with CLP associated with TA [19,64]. Moreover, common variants of this gene are associated with an increased risk of ns-TA [65,66]. Research in mouse models has confirmed the essential role of Irf6 during tooth development, with a null mutation of this gene resulting in evagination of the incisor epithelium and upregulation of the canonical Wnt signalling pathway [67]. Dental epithelium-specific Irf6 conditional knockout mice display hypodontia, occasional supernumerary incisors and molars, and alternations in the crown and root morphology [68]. In addition, their ameloblasts exhibit disturbances in adhesion, polarity and enamel formation [68].
The present study has several limitations, including the lack of segregation analysis of identified pathogenic and likely pathogenic variants in families of affected individuals. Another limitation was searching for variants underlying ns-TA only in the coding regions of a limited number of candidate genes. Since odontogenesis is a complex and not fully understood process, our criteria for selecting panel genes could cause the omission of potentially novel risk genes for ns-TA. Furthermore, the applied research approach was not intended to detect copy number variants, large deletions, duplications and insertions, and gene fusions. In addition, the in silico evaluation of the impact of the identified nucleotide changes by selected prediction tools or our variant filtering and prioritization strategy might be inappropriate, leading to the omission of clinically relevant genetic alternations. Despite its limitations, the study certainly adds to our understanding of the molecular basis of both hypodontia and oligodontia.
5. Conclusions
In conclusion, we have identified novel genes implicated in ns-TA, however, since mostly single nucleotide variants were detected, future research is required to confirm and fully understand their role in the aetiology of the congenital lack of permanent teeth. Additionally, our results support the importance of already known ns-TA candidate genes and provide additional evidence that ns-TA might be an oligogenic condition involving the combined effects of rare or private variants in two or more distinct genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm11206089/s1, Table S1: NGS targeted multigene panel for ns-TA; Table S2: Sequencing template PCR conditions and sequencing primers; Table S3: Characteristics of genes harbouring rare variants identified in patients with ns-TA (gene-phenotype relationships); Figure S1: Detection of a novel CHD7 missense variant (p.Glu1856Gln); Figure S2: Detection of a novel CREBBP missense variant (p.Pro344Ser); Figure S3: Detection of a novel LEF1 missense variant (p.Lys95As); Figure S4: Detection of a novel ROR2 nonsense variant (p.Ser632Ter); Figure S5: Detection of a novel TBX22 missense variant (p.Pro242Leu); Figure S6: Detection of a novel CREBBP missense variant (p.Glu1560Lys); Figure S7: Detection of a known EVC frameshift variant (Ala565ValfsTer23); Figure S8: Detection of a novel TP63 missense variant (p.Pro532Ala).
Author Contributions
Conceptualization, B.B., E.F., J.D., A.B. and A.M.; Formal analysis, B.B., E.F., J.D., A.B., M.Z. and A.M.; Funding acquisition, B.B. and A.M.; Investigation, J.D. and A.M.; Methodology, J.D., A.B. and A.M., Supervision, A.M.; Writing—original draft, A.M.; Writing—review and editing, B.B., E.F., J.D., A.B., M.Z. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
The Comprehensive Therapy Program for Craniofacial Anomalies (RPWP.07.02.02-30-0037/16) was supported by European Social Fund and Wielkopolska Voivodeship (Regional Operational Programme for Wielkopolskie Voivodeship 2014–2020).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki [18] and approved by the Institutional Review Board of Poznan University of Medical Sciences, Poland (approval number 1115/18).
Informed Consent Statement
Written informed consent was obtained from all study participants or their legal guardians.
Data Availability Statement
The de-identified datasets generated through this study can be provided by the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lan, Y.; Jia, S.; Jiang, R. Molecular patterning of the mammalian dentition. Semin. Cell Dev. Biol. 2014, 25–26, 61–70. [Google Scholar] [CrossRef]
- Balic, A.; Thesleff, I. Tissue Interactions Regulating Tooth Development and Renewal. Curr. Top. Dev. Biol. 2015, 115, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Letra, A.; Chiquet, B.; Hansen-Kiss, E.; Menezes, S.; Hunter, E. Nonsyndromic Tooth Agenesis Overview. In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK572295/ (accessed on 1 March 2022).
- Polder, B.J.; Van’t Hof, M.A.; Van der Linden, F.P.; Kuijpers-Jagtman, A.M. A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent. Oral Epidemiol. 2004, 32, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Al-Ani, A.H.; Antoun, J.S.; Thomson, W.M.; Merriman, T.R.; Farella, M. Maternal Smoking during Pregnancy Is Associated with Offspring Hypodontia. J. Dent. Res. 2017, 96, 1014–1019. [Google Scholar] [CrossRef]
- Wang, J.; Sun, K.; Shen, Y.; Xu, Y.; Xie, J.; Huang, R.; Zhang, Y.; Xu, C.; Zhang, X.; Wang, R.; et al. DNA methylation is critical for tooth agenesis: Implications for sporadic non-syndromic anodontia and hypodontia. Sci. Rep. 2016, 6, 19162. [Google Scholar] [CrossRef]
- Liu, F.; Millar, S.E. Wnt/beta-catenin signaling in oral tissue development and disease. J. Dent. Res. 2010, 89, 318–330. [Google Scholar] [CrossRef]
- Tamura, M.; Nemoto, E.; Sato, M.M.; Nakashima, A.; Shimauchi, H. Role of the Wnt signaling pathway in bone and tooth. Front. Biosci. 2010, 2, 1405–1413. [Google Scholar] [CrossRef]
- Mostowska, A.; Biedziak, B.; Zadurska, M.; Dunin-Wilczynska, I.; Lianeri, M.; Jagodzinski, P.P. Nucleotide variants of genes encoding components of the Wnt signalling pathway and the risk of non-syndromic tooth agenesis. Clin. Genet. 2013, 84, 429–440. [Google Scholar] [CrossRef]
- Gaczkowska, A.D.; Jagodziński, P.P.; Mostowska, A. The molecular basis of non-syndromic orofacial clefts and tooth agenesis. JMS 2017, 86, 321–324. [Google Scholar] [CrossRef][Green Version]
- Dinckan, N.; Du, R.; Petty, L.E.; Coban-Akdemir, Z.; Jhangiani, S.N.; Paine, I.; Baugh, E.H.; Erdem, A.P.; Kayserili, H.; Doddapaneni, H.; et al. Whole-Exome Sequencing Identifies Novel Variants for Tooth Agenesis. J. Dent Res. 2018, 97, 49–59. [Google Scholar] [CrossRef]
- Du, R.; Dinckan, N.; Song, X.; Coban-Akdemir, Z.; Jhangiani, S.N.; Guven, Y.; Aktoren, O.; Kayserili, H.; Petty, L.E.; Muzny, D.M.; et al. Identification of likely pathogenic and known variants in TSPEAR, LAMB3, BCOR, and WNT10A in four Turkish families with tooth agenesis. Hum. Genet. 2018, 137, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Arte, S.; Parmanen, S.; Pirinen, S.; Alaluusua, S.; Nieminen, P. Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS ONE 2013, 8, e73705. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Han, D.; Feng, H.; Qu, H.; Song, S.; Bai, B.; Zhang, Z. Involvement of and interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS ONE 2013, 8, e80393. [Google Scholar] [CrossRef]
- Salvi, A.; Giacopuzzi, E.; Bardellini, E.; Amadori, F.; Ferrari, L.; De Petro, G.; Borsani, G.; Majorana, A. Mutation analysis by direct and whole exome sequencing in familial and sporadic tooth agenesis. Int. J. Mol. Med. 2016, 38, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.Y.; Wang, Y.L.; Chou, Y.R.; Chen, J.T.; Wang, Y.P.; Simmer, J.P.; Hu, J.C.; Wang, S.K. Synergistic Mutations of LRP6 and WNT10A in Familial Tooth Agenesis. J. Pers. Med. 2021, 11, 1217. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.A.; Letra, A. The Changing Landscape in the Genetic Etiology of Human Tooth Agenesis. Genes 2018, 9, 255. [Google Scholar] [CrossRef] [PubMed]
- Sawicka-Gutaj, N.; Gruszczyński, D.; Guzik, P.; Mostowska, A.; Walkowiak, J. Publication ethics of human studies in the light of the Declaration of Helsinki—A mini-review. JMS 2022, 91, e700. [Google Scholar] [CrossRef]
- Dąbrowska, J.; Biedziak, B.; Szponar-Żurowska, A.; Budner, M.; Jagodziński, P.P.; Płoski, R.; Mostowska, A. Identification of novel susceptibility genes for non-syndromic cleft lip with or without cleft palate using NGS-based multigene panel testing. Mol. Genet. Genomics. 2022, 297, 1315–1327. [Google Scholar] [CrossRef]
- Sasaki, T.; Ito, Y.; Xu, X.; Han, J.; Bringas, P.; Maeda, T., Jr.; Slavkin, H.C.; Grosschedl, R.; Chai, Y. LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev. Biol. 2005, 278, 130–143. [Google Scholar] [CrossRef]
- Fuchs, A.; Inthal, A.; Herrmann, D.; Cheng, S.; Nakatomi, M.; Peters, H.; Neubüser, A. Regulation of Tbx22 during facial and palatal development. Dev. Dyn. 2010, 239, 2860–2874. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, B.; Zhou, X. Expression patterns of histone acetyltransferases p300 and CBP during murine tooth development. In Vitro Cell. Dev. Biol. Anim. 2012, 48, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Rostampour, N.; Appelt, C.M.; Abid, A.; Boughner, J.C. Expression of new genes in vertebrate tooth development and p63 signaling. Dev. Dyn. 2019, 248, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Q.; Xiao, Q.; Gong, P.; Kang, N. CHD7 Regulates Osteogenic Differentiation of Human Dental Follicle Cells via PTH1R Signaling. Stem Cells Int. 2020, 2020, 8882857. [Google Scholar] [CrossRef] [PubMed]
- Louie, K.W.; Mishina, Y.; Zhang, H. Molecular and Cellular Pathogenesis of Ellis-van Creveld Syndrome: Lessons from Targeted and Natural Mutations in Animal Models. J. Dev. Biol. 2020, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Tokavanich, N.; Wein, M.N.; English, J.D.; Ono, N.; Ono, W. The Role of Wnt Signaling in Postnatal Tooth Root Development. Front. Dent. Med. 2021, 2, 769134. [Google Scholar] [CrossRef] [PubMed]
- van Genderen, C.; Okamura, R.M.; Fariñas, I.; Quo, R.G.; Parslow, T.G.; Bruhn, L.; Grosschedl, R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994, 8, 2691–2703. [Google Scholar] [CrossRef]
- Ma, Y.; Jing, J.; Feng, J.; Yuan, Y.; Wen, Q.; Han, X.; He, J.; Chen, S.; Ho, T.V.; Chai, Y. Ror2-mediated non-canonical Wnt signaling regulates Cdc42 and cell proliferation during tooth root development. Development 2021, 148, dev196360. [Google Scholar] [CrossRef]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef]
- Laurikkala, J.; Mikkola, M.L.; James, M.; Tummers, M.; Mills, A.A.; Thesleff, I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 2006, 133, 1553–1563. [Google Scholar] [CrossRef]
- Ruiz-Perez, V.L.; Blair, H.J.; Rodriguez-Andres, M.E.; Blanco, M.J.; Wilson, A.; Liu, Y.N.; Miles, C.; Peters, H.; Goodship, J.A. Evc is a positive mediator of Ihh-regulated bone growth that localizes at the base of chondrocyte cilia. Development 2007, 134, 2903–2912. [Google Scholar] [CrossRef]
- Lévy, J.; Capri, Y.; Rachid, M.; Dupont, C.; Vermeesch, J.R.; Devriendt, K.; Verloes, A.; Tabet, A.C.; Bailleul-Forestier, I. LEF1 haploinsufficiency causes ectodermal dysplasia. Clin. Genet. 2020, 97, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Dufour, W.; Alawbathani, S.; Jourdain, A.S.; Asif, M.; Baujat, G.; Becker, C.; Budde, B.; Gallacher, L.; Georgomanolis, T.; Ghoumid, J.; et al. Monoallelic and biallelic variants in LEF1 are associated with a new syndrome combining ectodermal dysplasia and limb malformations caused by altered WNT signaling. Genet. Med. 2022, 24, 1708–1721. [Google Scholar] [CrossRef] [PubMed]
- Mikels, A.; Minami, Y.; Nusse, R. Ror2 receptor requires tyrosine kinase activity to mediate Wnt5A signaling. J. Biol. Chem. 2009, 284, 30167–30176. [Google Scholar] [CrossRef] [PubMed]
- Raivola, J.; Dini, A.; Salokas, K.; Karvonen, H.; Niininen, W.; Piki, E.; Varjosalo, M.; Ungureanu, D. New insights into the molecular mechanisms of ROR1, ROR2, and PTK7 signaling from the proteomics and pharmacological modulation of ROR1 interactome. Cell. Mol. Life Sci. 2022, 79, 276. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Jain, P.S.; Gupte, T.S.; Jetpurwala, A.M.; Dedhia, S.P. Robinow Syndrome and Fusion of Primary Teeth. Contemp. Clin. Dent. 2017, 8, 479–481. [Google Scholar] [CrossRef]
- Gui, B.; Yu, C.; Li, X.; Zhao, S.; Zhao, H.; Yan, Z.; Cheng, X.; Lin, J.; Zheng, H.; Shao, J.; et al. Heterozygous Recurrent Mutations Inducing Dysfunction of ROR2 Gene in Patients With Short Stature. Front. Cell Dev. Biol. 2021, 9, 661747. [Google Scholar] [CrossRef]
- Pauws, E.; Hoshino, A.; Bentley, L.; Prajapati, S.; Keller, C.; Hammond, P.; Martinez-Barbera, J.P.; Moore, G.E.; Stanier, P. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum. Mol. Genet. 2009, 18, 4171–4179. [Google Scholar] [CrossRef]
- Kantaputra, P.N.; Paramee, M.; Kaewkhampa, A.; Hoshino, A.; Lees, M.; McEntagart, M.; Masrour, N.; Moore, G.E.; Pauws, E.; Stanier, P. Cleft lip with cleft palate, ankyloglossia, and hypodontia are associated with TBX22 mutations. J. Dent. Res. 2011, 90, 450–455. [Google Scholar] [CrossRef]
- Kaewkhampa, A.; Jotikasthira, D.; Malaivijitnond, S.; Kantaputra, P. TBX22 mutation associated with cleft lip/palate, hypodontia, and limb anomaly. Cleft Palate Craniofac. J. 2012, 49, 240–244. [Google Scholar] [CrossRef]
- Dai, J.; Xu, C.; Wang, G.; Liang, Y.; Wan, T.; Zhang, Y.; Xu, X.; Yu, L.; Che, Z.; Han, Q.; et al. Novel TBX22 mutations in Chinese nonsyndromic cleft lip/palate families. J. Genet. 2018, 97, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Braybrook, C.; Lisgo, S.; Doudney, K.; Henderson, D.; Marçano, A.C.; Strachan, T.; Patton, M.A.; Villard, L.; Moore, G.E.; Stanier, P.; et al. Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum. Mol. Genet. 2002, 11, 2793–2804. [Google Scholar] [CrossRef] [PubMed]
- Van Gils, J.; Magdinier, F.; Fergelot, P.; Lacombe, D. Rubinstein-Taybi Syndrome: A Model of Epigenetic Disorder. Genes 2021, 12, 968. [Google Scholar] [CrossRef] [PubMed]
- Stevens, C.A. Rubinstein-Taybi Syndrome. In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1526/ (accessed on 1 March 2022).
- Cross, E.; Duncan-Flavell, P.J.; Howarth, R.J.; Hobbs, J.I.; Thomas, N.S.; Bunyan, D.J. Screening of a large Rubinstein-Taybi cohort identified many novel variants and emphasizes the importance of the CREBBP histone acetyltransferase domain. Am. J. Med. Genet. A. 2020, 182, 2508–2520. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, V.L.; Ide, S.E.; Strom, T.M.; Lorenz, B.; Wilson, D.; Woods, K.; King, L.; Francomano, C.; Freisinger, P.; Spranger, S.; et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat. Genet. 2000, 24, 283–286. [Google Scholar] [CrossRef]
- Nakatomi, M.; Hovorakova, M.; Gritli-Linde, A.; Blair, H.J.; MacArthur, K.; Peterka, M.; Lesot, H.; Peterkova, R.; Ruiz-Perez, V.L.; Goodship, J.A.; et al. Evc regulates a symmetrical response to Shh signaling in molar development. J. Dent. Res. 2013, 92, 222–228. [Google Scholar] [CrossRef]
- Harazono, Y.; Morita, K.; Tonouchi, E.; Anzai, E.; Takahara, N.; Kohmoto, T.; Imoto, I.; Yoda, T. TP63 mutation mapping information in TP63 mutation-associated syndromes. Adv. Oral Maxillofac. Surg. 2022, 5, 100253. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, H.; Zhan, Y.; Liu, Y.; Wong, S.W.; Cai, T.; Feng, H.; Han, D. Tooth defects of EEC and AEC syndrome caused by heterozygous TP63 mutations in three Chinese families and genotype-phenotype correlation analyses of TP63-related disorders. Mol. Genet. Genomic Med. 2019, 7, e704. [Google Scholar] [CrossRef]
- Carroll, D.K.; Carroll, J.S.; Leong, C.O.; Cheng, F.; Brown, M.; Mills, A.A.; Brugge, J.S.; Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell. Biol. 2006, 8, 551–561. [Google Scholar] [CrossRef]
- Truong, A.B.; Kretz, M.; Ridky, T.W.; Kimmel, R.; Khavari, P.A. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006, 20, 3185–3197. [Google Scholar] [CrossRef]
- Lin-Shiao, E.; Lan, Y.; Welzenbach, J.; Alexander, K.A.; Zhang, Z.; Knapp, M.; Mangold, E.; Sammons, M.; Ludwig, K.U.; Berger, S.L. p63 establishes epithelial enhancers at critical craniofacial development genes. Sci. Adv. 2019, 5, eaaw0946. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.; Zhou, H. Master regulatory role of p63 in epidermal development and disease. Cell. Mol. Life Sci. 2018, 75, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- van den Boogaard, M.J.; Créton, M.; Bronkhorst, Y.; van der Hout, A.; Hennekam, E.; Lindhout, D.; Cune, M.; Ploos van Amstel, H.K. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J. Med. Genet. 2012, 49, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Magruder, S.; Carter, E.; Williams, M.A.; English, J.; Akyalcin, S.; Letra, A. Further evidence for the role of WNT10A, WNT10B and GREM2 as candidate genes for isolated tooth agenesis. Orthod. Craniofac. Res. 2018, 21, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.; Conte, F.; Khandelwal, K.D.; Ockeloen, C.W.; Bartzela, T.; Kleefstra, T.; van Bokhoven, H.; Rubini, M.; Zhou, H.; Carels, C.E. Tooth agenesis and orofacial clefting: Genetic brothers in arms? Hum. Genet. 2016, 135, 1299–1327. [Google Scholar] [CrossRef]
- Möller, L.H.; Pradel, W.; Gedrange, T.; Botzenhart, U.U. Prevalence of hypodontia and supernumerary teeth in a German cleft lip with/without palate population. BMC Oral. Health 2021, 21, 60. [Google Scholar] [CrossRef]
- Konstantonis, D.; Nassika, M.; Athanasiou, M.; Vastardis, H. Subphenotypes in Non-Syndromic Orofacial Cleft Patients Based on the Tooth Agenesis Code (TAC). Children 2022, 9, 437. [Google Scholar] [CrossRef]
- Eerens, K.; Vlietinck, R.; Heidbüchel, K.; Van Olmen, A.; Derom, C.; Willems, G.; Carels, C. Hypodontia and tooth formation in groups of children with cleft, siblings without cleft, and nonrelated controls. Cleft Palate Craniofac. J. 2001, 38, 374–378. [Google Scholar] [CrossRef]
- Aspinall, A.; Raj, S.; Jugessur, A.; Marazita, M.; Savarirayan, R.; Kilpatrick, N. Expanding the cleft phenotype: The dental characteristics of unaffected parents of Australian children with non-syndromic cleft lip and palate. Int. J. Paediatr. Dent. 2014, 24, 286–292. [Google Scholar] [CrossRef]
- Reynolds, K.; Zhang, S.; Sun, B.; Garland, M.A.; Ji, Y.; Zhou, C.J. Genetics and signaling mechanisms of orofacial clefts. Birth Defects Res. 2020, 112, 1588–1634. [Google Scholar] [CrossRef]
- Nasreddine, G.; El Hajj, J.; Ghassibe-Sabbagh, M. Orofacial clefts embryology, classification, epidemiology, and genetics. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108373. [Google Scholar] [CrossRef] [PubMed]
- Neves, L.T.; Dionísio, T.J.; Garbieri, T.F.; Parisi, V.A.; Oliveira, F.V.; Oliveira, T.M.; Santos, C.F. Novel rare variations in IRF6 in subjects with non-syndromic cleft lip and palate and dental agenesis. Oral Dis. 2019, 25, 223–233. [Google Scholar] [CrossRef]
- Vieira, A.R.; Modesto, A.; Meira, R.; Barbosa, A.R.; Lidral, A.C.; Murray, J.C. Interferon regulatory factor 6 (IRF6) and fibroblast growth factor receptor 1 (FGFR1) contribute to human tooth agenesis. Am. J. Med. Genet. A. 2007, 143A, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.R.; Seymen, F.; Patir, A.; Menezes, R. Evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and isolate tooth agenesis, in a Turkish population. Arch. Oral. Biol. 2008, 53, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, J.; Ohazama, A.; Kawasaki, K.; Otsuka-Tanaka, Y.; Liu, B.; Honda, K.; Rountree, R.B.; Hu, Y.; Kawasaki, M.; Birchmeier, W.; et al. The role of Irf6 in tooth epithelial invagination. Dev. Biol. 2012, 365, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Chu, E.Y.; Tamasas, B.; Fong, H.; Foster, B.L.; LaCourse, M.R.; Tran, A.B.; Martin, J.F.; Schutte, B.C.; Somerman, M.J.; Cox, T.C. Full Spectrum of Postnatal Tooth Phenotypes in a Novel Irf6 Cleft Lip Model. J. Dent. Res. 2016, 95, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).