Molecular Features of SLC26A4 Common Variant p.L117F
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Constructs
2.3. Functional Testing
2.4. Determination of Protein Expression Levels by Quantitative Imaging
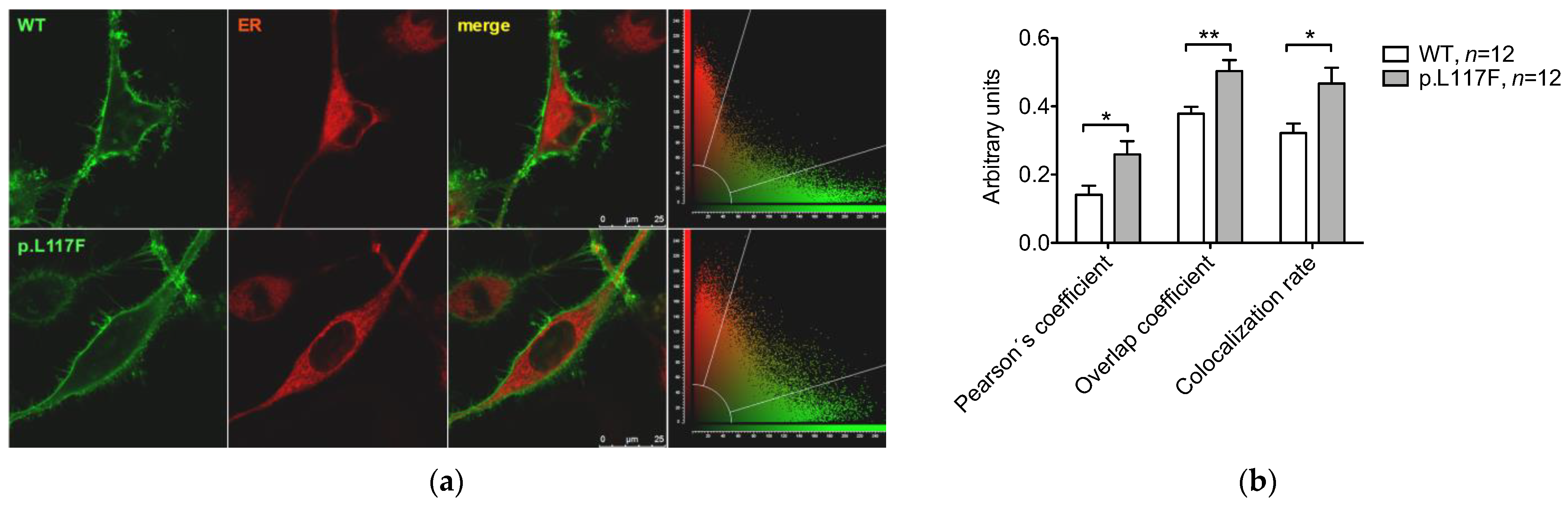
2.5. Colocalization
2.6. Western Blotting
2.7. Real-Time qPCR
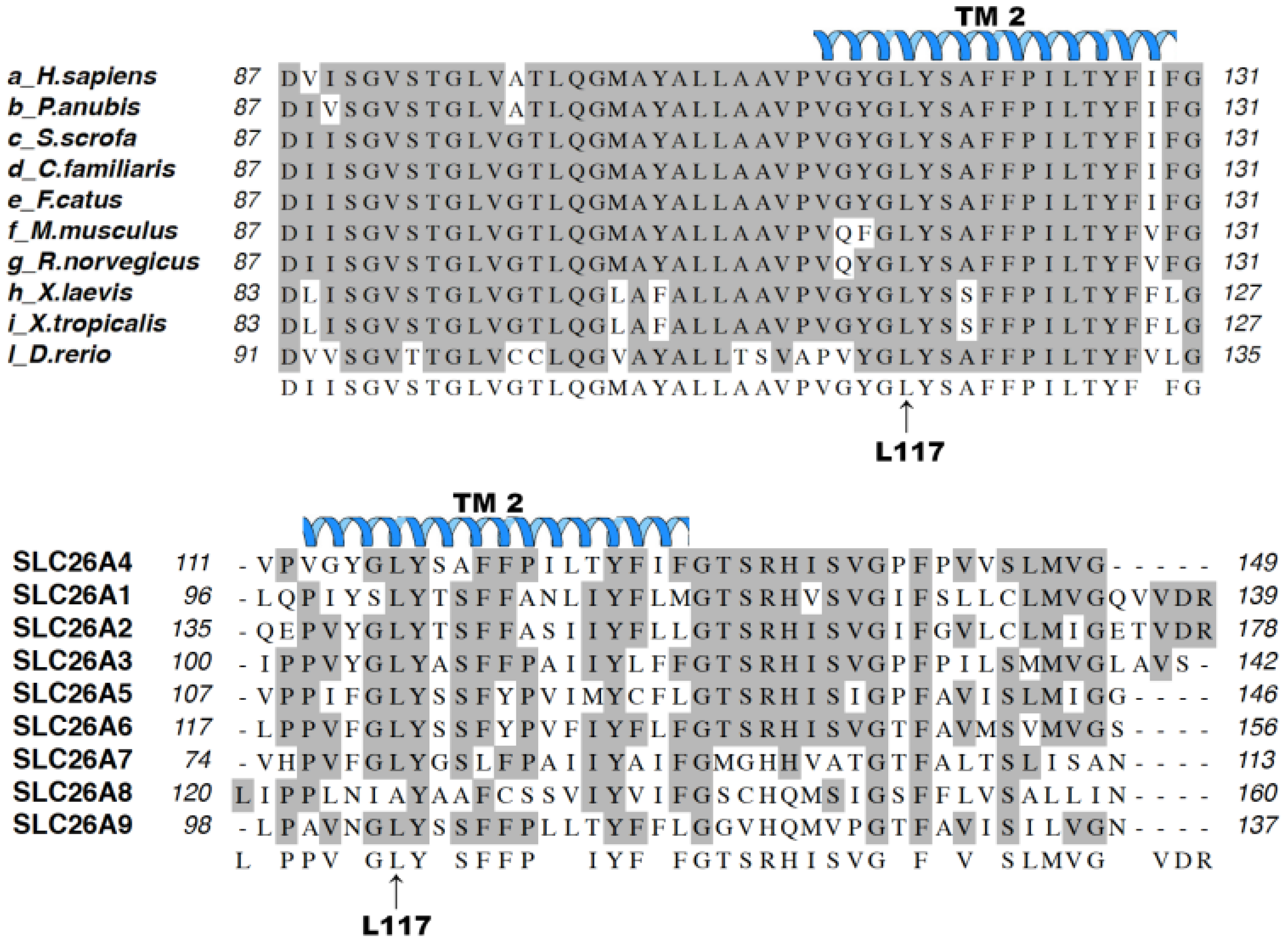
2.8. Bioinformatics
2.9. Salt and Chemicals
2.10. Statistical Analysis
3. Results
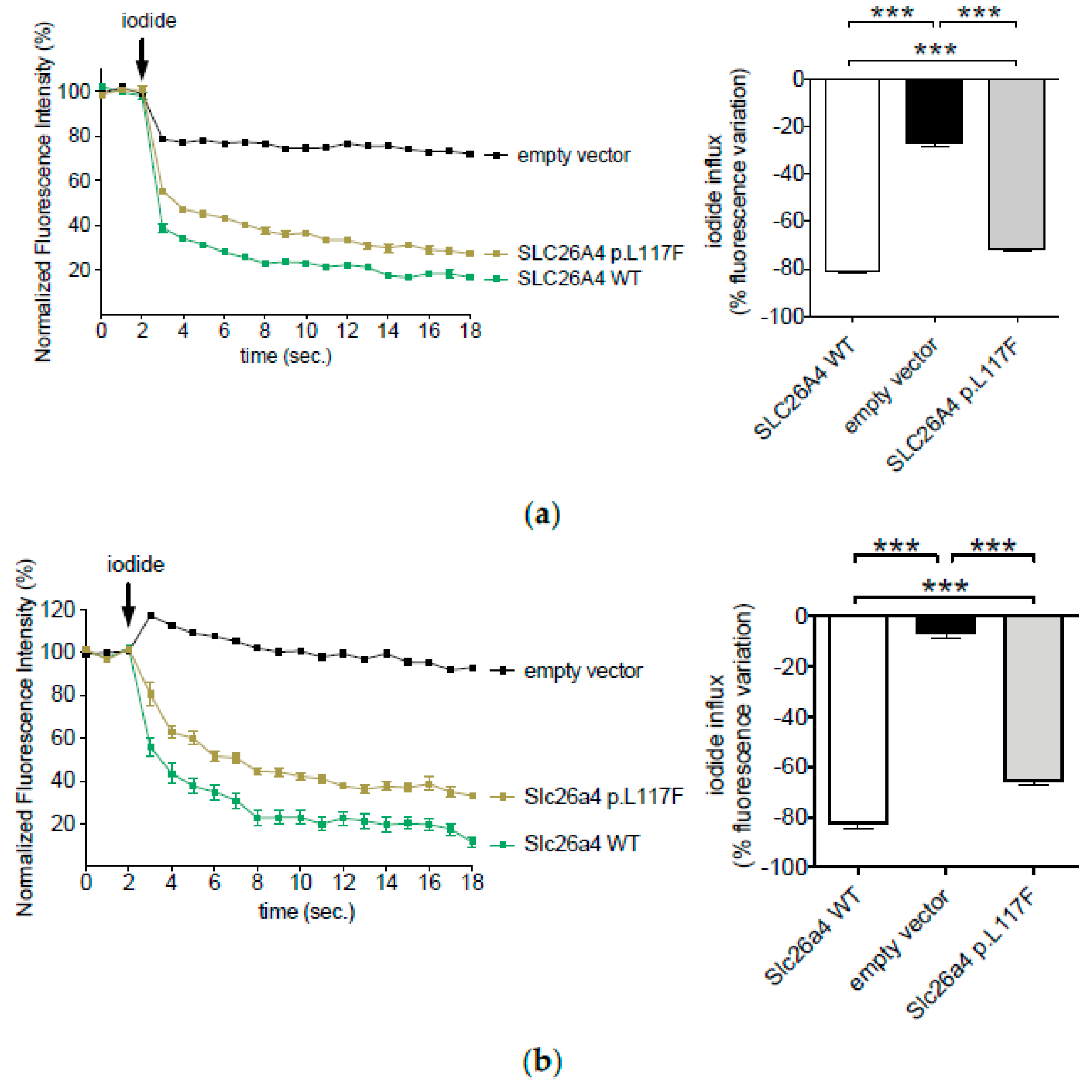
3.1. The Ion Transport Activity of Pendrin p.L117F Is Moderately but Significantly Reduced Compared to the Wild Type
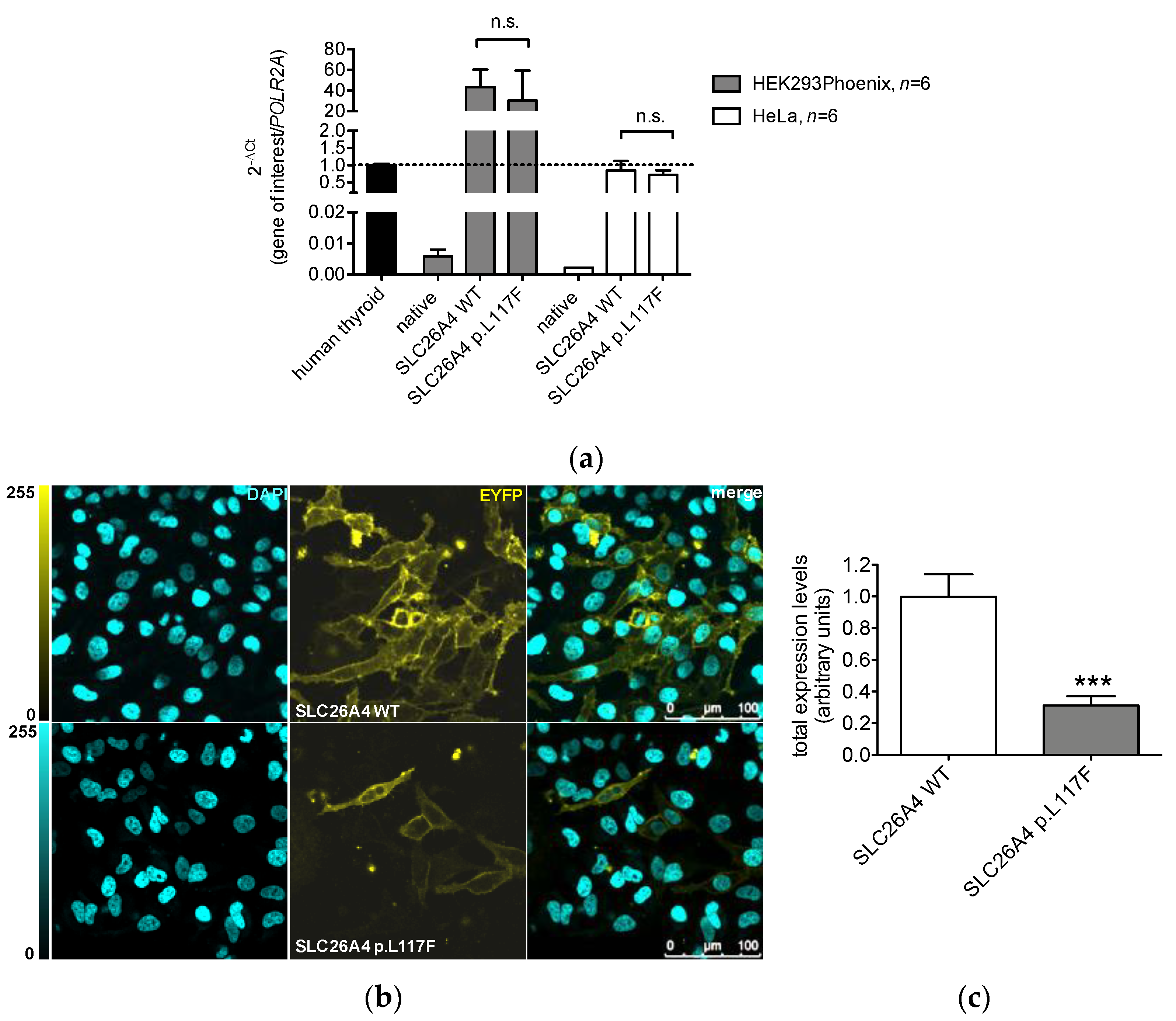
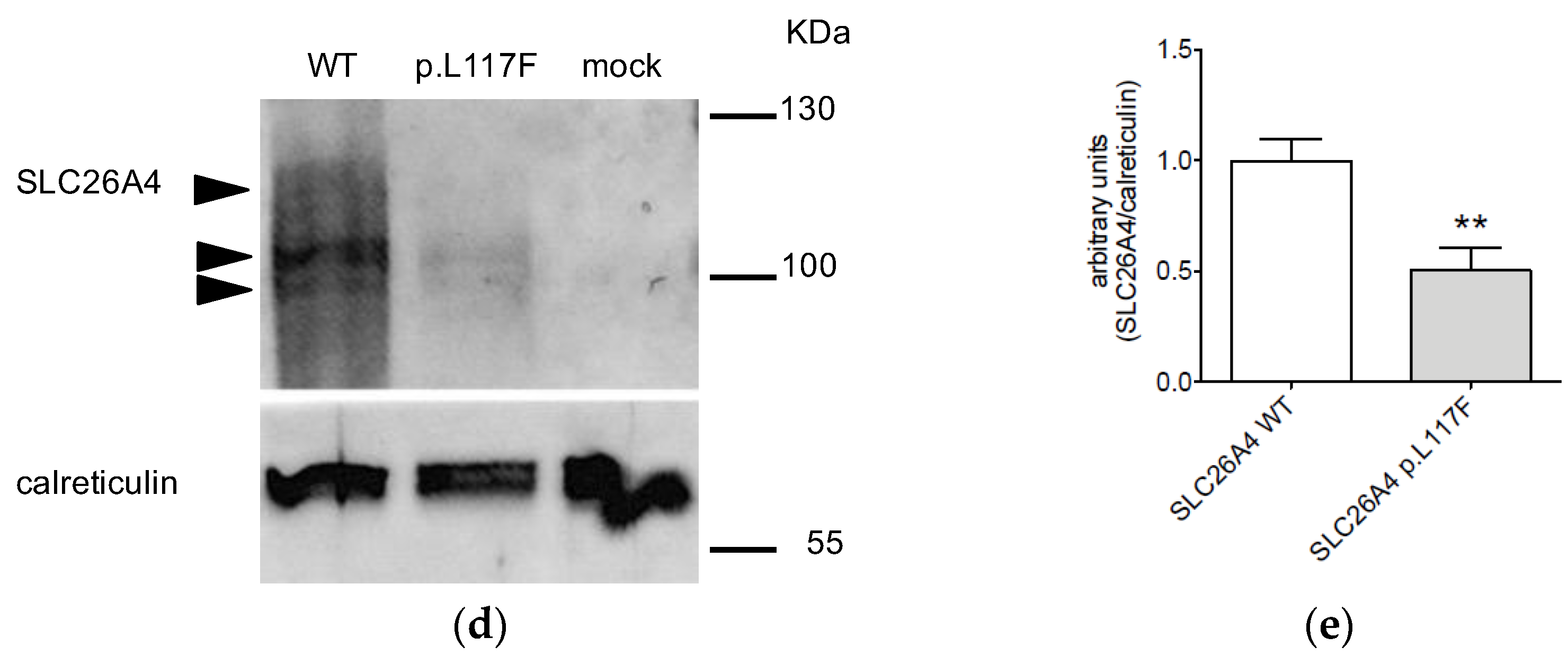
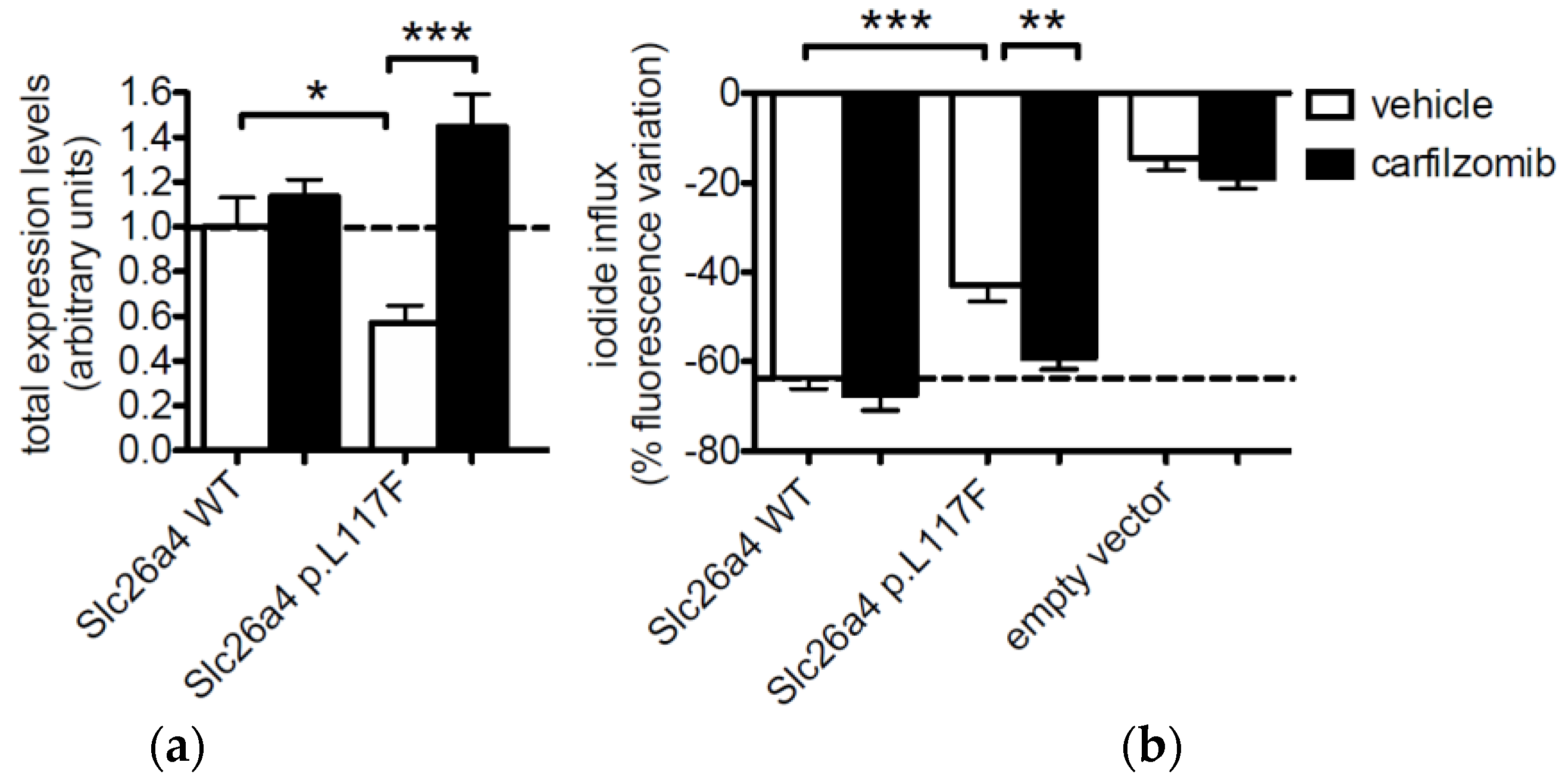
3.2. The Protein Expression of Pendrin p.L117F Is Substantially Reduced Compared to the Wild Type
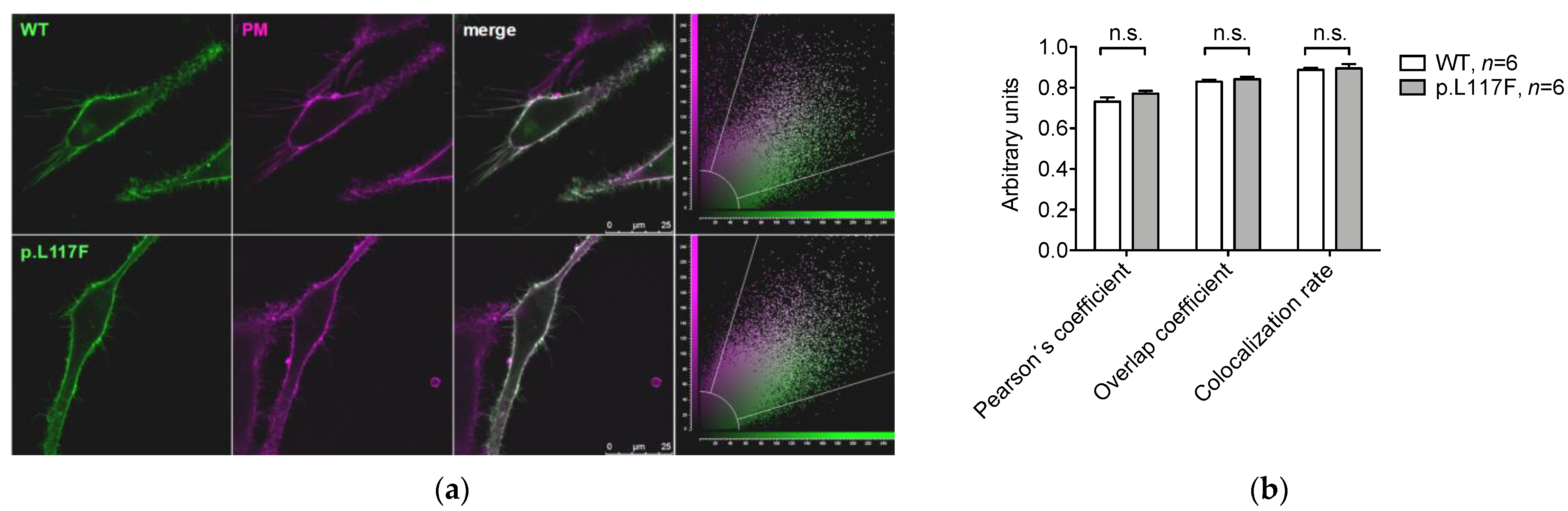
3.3. Pendrin p.L117F Is Mostly but Not Exclusively Localized at the Plasma Membrane
3.4. Pendrin p.L117F Is Degraded by the Ubiquitin Proteasome System
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Everett, L.A.; Glaser, B.; Beck, J.C.; Idol, J.R.; Buchs, A.; Heyman, M.; Adawi, F.; Hazani, E.; Nassir, E.; Baxevanis, A.D.; et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet. 1997, 17, 411–422. [Google Scholar] [CrossRef]
- Dossena, S.; Nofziger, C.; Tamma, G.; Bernardinelli, E.; Vanoni, S.; Nowak, C.; Grabmayer, E.; Kossler, S.; Stephan, S.; Patsch, W.; et al. Molecular and functional characterization of human pendrin and its allelic variants. Cell. Physiol. Biochem. 2011, 28, 451–466. [Google Scholar] [CrossRef]
- Everett, L.A.; Morsli, H.; Wu, D.K.; Green, E.D. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA 1999, 96, 9727–9732. [Google Scholar] [CrossRef] [PubMed]
- Royaux, I.E.; Wall, S.M.; Karniski, L.P.; Everett, L.A.; Suzuki, K.; Knepper, M.A.; Green, E.D. Pendrin, encoded by the Pendred syndrome gene, resides in the apical region of renal intercalated cells and mediates bicarbonate secretion. Proc. Natl. Acad. Sci. USA 2001, 98, 4221–4226. [Google Scholar] [CrossRef]
- Kim, H.M.; Wangemann, P. Epithelial cell stretching and luminal acidification lead to a retarded development of stria vascularis and deafness in mice lacking pendrin. PLoS ONE 2011, 6, e17949. [Google Scholar]
- Nakaya, K.; Harbidge, D.G.; Wangemann, P.; Schultz, B.D.; Green, E.D.; Wall, S.M.; Marcus, D.C. Lack of pendrin HCO3− transport elevates vestibular endolymphatic [Ca2+] by inhibition of acid-sensitive TRPV5 and TRPV6 channels. Am. J. Physiol. Renal Physiol. 2007, 292, F1314–F1321. [Google Scholar] [CrossRef]
- Fugazzola, L.; Cerutti, N.; Mannavola, D.; Vannucchi, G.; Beck-Peccoz, P. The role of pendrin in iodide regulation. Exp. Clin. Endocrinol. Diabetes 2001, 109, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, M. The multiple roles of pendrin in the kidney. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc. -Eur. Ren. Assoc. 2015, 30, 1257–1266. [Google Scholar] [CrossRef]
- Smith, R.J.H. Pendred Syndrome/Nonsyndromic Enlarged Vestibular Aqueduct. In GeneReviews((R)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Griffith, A.J.; Wangemann, P. Hearing loss associated with enlargement of the vestibular aqueduct: Mechanistic insights from clinical phenotypes, genotypes, and mouse models. Hear. Res. 2011, 281, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Roesch, S.; Rasp, G.; Sarikas, A.; Dossena, S. Genetic Determinants of Non-Syndromic Enlarged Vestibular Aqueduct: A Review. Audiol. Res. 2021, 11, 423–442. [Google Scholar] [CrossRef]
- Azadegan-Dehkordi, F.; Ahmadi, R.; Bahrami, T.; Yazdanpanahi, N.; Farrokhi, E.; Tabatabaiefar, M.A.; Hashemzadeh-Chaleshtori, M. A novel variant of SLC26A4 and first report of the c.716T>A variant in Iranian pedigrees with non-syndromic sensorineural hearing loss. Am. J. Otolaryngol. 2018, 39, 719–725. [Google Scholar] [CrossRef]
- Bassot, C.; Minervini, G.; Leonardi, E.; Tosatto, S.C. Mapping pathogenic mutations suggests an innovative structural model for the pendrin (SLC26A4) transmembrane domain. Biochimie 2017, 132, 109–120. [Google Scholar] [CrossRef]
- Fraser, G.R. Association of Congenital Deafness with Goitre (Pendred’s Syndrome) a Study of 207 Families. Ann. Hum. Genet. 1965, 28, 201–249. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Oza, A.M.; DiStefano, M.T.; Hemphill, S.E.; Cushman, B.J.; Grant, A.R.; Siegert, R.K.; Shen, J.; Chapin, A.; Boczek, N.J.; Schimmenti, L.A.; et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018, 39, 1593–1613. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.A.; Wang, R.; Kreman, T.M.; Sheffield, V.C.; Karniski, L.P. The Pendred syndrome gene encodes a chloride-iodide transport protein. Nat. Genet. 1999, 21, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Metcalfe, R.A.; Watson, P.F.; Weetman, A.P.; Trembath, R.C. Mutations of the PDS gene, encoding pendrin, are associated with protein mislocalization and loss of iodide efflux: Implications for thyroid dysfunction in Pendred syndrome. J. Clin. Endocrinol. Metab. 2002, 87, 1778–1784. [Google Scholar] [CrossRef] [PubMed]
- Gillam, M.P.; Sidhaye, A.R.; Lee, E.J.; Rutishauser, J.; Stephan, C.W.; Kopp, P. Functional characterization of pendrin in a polarized cell system. Evidence for pendrin-mediated apical iodide efflux. J. Biol. Chem. 2004, 279, 13004–13010. [Google Scholar] [CrossRef]
- Dossena, S.; Vezzoli, V.; Cerutti, N.; Bazzini, C.; Tosco, M.; Sironi, C.; Rodighiero, S.; Meyer, G.; Fascio, U.; Furst, J.; et al. Functional characterization of wild-type and a mutated form of SLC26A4 identified in a patient with Pendred syndrome. Cell. Physiol. Biochem. 2006, 17, 245–256. [Google Scholar] [CrossRef]
- Choi, B.Y.; Stewart, A.K.; Madeo, A.C.; Pryor, S.P.; Lenhard, S.; Kittles, R.; Eisenman, D.; Kim, H.J.; Niparko, J.; Thomsen, J.; et al. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: Genotype-phenotype correlation or coincidental polymorphisms? Hum. Mutat. 2009, 30, 599–608. [Google Scholar] [CrossRef]
- Dai, P.; Stewart, A.K.; Chebib, F.; Hsu, A.; Rozenfeld, J.; Huang, D.; Kang, D.; Lip, V.; Fang, H.; Shao, H.; et al. Distinct and novel SLC26A4/Pendrin mutations in Chinese and U.S. patients with nonsyndromic hearing loss. Physiol. Genom. 2009, 38, 281–290. [Google Scholar] [CrossRef]
- Palos, F.; Garcia-Rendueles, M.E.; Araujo-Vilar, D.; Obregon, M.J.; Calvo, R.M.; Cameselle-Teijeiro, J.; Bravo, S.B.; Perez-Guerra, O.; Loidi, L.; Czarnocka, B.; et al. Pendred syndrome in two Galician families: Insights into clinical phenotypes through cellular, genetic, and molecular studies. J. Clin. Endocrinol. Metab. 2008, 93, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Dossena, S.; Rodighiero, S.; Vezzoli, V.; Bazzini, C.; Sironi, C.; Meyer, G.; Furst, J.; Ritter, M.; Garavaglia, M.L.; Fugazzola, L.; et al. Fast fluorometric method for measuring pendrin (SLC26A4) Cl-/I- transport activity. Cell. Physiol. Biochem. 2006, 18, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Park, H.J.; Yoo, S.Y.; Namkung, W.; Jo, M.J.; Koo, S.K.; Park, H.Y.; Lee, W.S.; Kim, K.H.; Lee, M.G. Heterogeneity in the processing defect of SLC26A4 mutants. J. Med. Genet. 2008, 45, 411–419. [Google Scholar] [CrossRef]
- Wasano, K.; Takahashi, S.; Rosenberg, S.K.; Kojima, T.; Mutai, H.; Matsunaga, T.; Ogawa, K.; Homma, K. Systematic quantification of the anion transport function of pendrin (SLC26A4) and its disease-associated variants. Hum. Mutat. 2020, 41, 316–331. [Google Scholar] [CrossRef]
- Rotman-Pikielny, P.; Hirschberg, K.; Maruvada, P.; Suzuki, K.; Royaux, I.E.; Green, E.D.; Kohn, L.D.; Lippincott-Schwartz, J.; Yen, P.M. Retention of pendrin in the endoplasmic reticulum is a major mechanism for Pendred syndrome. Hum. Mol. Genet. 2002, 11, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, V.C.S.; Bernardinelli, E.; Zocal, N.; Fernandez, J.A.; Nofziger, C.; Castilho, A.M.; Sartorato, E.L.; Paulmichl, M.; Dossena, S. Reduction of Cellular Expression Levels Is a Common Feature of Functionally Affected Pendrin (SLC26A4) Protein Variants. Mol. Med. 2016, 22, 41–53. [Google Scholar] [CrossRef]
- Roesch, S.; Bernardinelli, E.; Nofziger, C.; Toth, M.; Patsch, W.; Rasp, G.; Paulmichl, M.; Dossena, S. Functional Testing of SLC26A4 Variants-Clinical and Molecular Analysis of a Cohort with Enlarged Vestibular Aqueduct from Austria. Int. J. Mol. Sci. 2018, 19, 209. [Google Scholar] [CrossRef]
- Brownstein, Z.; Gulsuner, S.; Walsh, T.; Martins, F.T.A.; Taiber, S.; Isakov, O.; Lee, M.K.; Bordeynik-Cohen, M.; Birkan, M.; Chang, W.; et al. Spectrum of genes for inherited hearing loss in the Israeli Jewish population, including the novel human deafness gene ATOH1. Clin. Genet. 2020, 98, 353–364. [Google Scholar] [CrossRef]
- DiCiommo, D.P.; Duckett, A.; Burcescu, I.; Bremner, R.; Gallie, B.L. Retinoblastoma protein purification and transduction of retina and retinoblastoma cells using improved alphavirus vectors. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3320–3329. [Google Scholar] [CrossRef]
- Fugazzola, L.; Cirello, V.; Dossena, S.; Rodighiero, S.; Muzza, M.; Castorina, P.; Lalatta, F.; Ambrosetti, U.; Beck-Peccoz, P.; Botta, G.; et al. High phenotypic intrafamilial variability in patients with Pendred syndrome and a novel duplication in the SLC26A4 gene: Clinical characterization and functional studies of the mutated SLC26A4 protein. Eur. J. Endocrinol. 2007, 157, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Pera, A.; Dossena, S.; Rodighiero, S.; Gandia, M.; Botta, G.; Meyer, G.; Moreno, F.; Nofziger, C.; Hernandez-Chico, C.; Paulmichl, M. Functional assessment of allelic variants in the SLC26A4 gene involved in Pendred syndrome and nonsyndromic EVA. Proc. Natl. Acad. Sci. USA 2008, 105, 18608–18613. [Google Scholar] [CrossRef] [PubMed]
- Dror, A.A.; Politi, Y.; Shahin, H.; Lenz, D.R.; Dossena, S.; Nofziger, C.; Fuchs, H.; Hrabe de Angelis, M.; Paulmichl, M.; Weiner, S.; et al. Calcium oxalate stone formation in the inner ear as a result of an Slc26a4 mutation. J. Biol. Chem. 2010, 285, 21724–21735. [Google Scholar] [CrossRef] [PubMed]
- Dossena, S.; Bizhanova, A.; Nofziger, C.; Bernardinelli, E.; Ramsauer, J.; Kopp, P.; Paulmichl, M. Identification of allelic variants of pendrin (SLC26A4) with loss and gain of function. Cell. Physiol. Biochem. 2011, 28, 467–476. [Google Scholar] [CrossRef]
- Dossena, S.; Nofziger, C.; Brownstein, Z.; Kanaan, M.; Avraham, K.B.; Paulmichl, M. Functional characterization of pendrin mutations found in the Israeli and Palestinian populations. Cell. Physiol. Biochem. 2011, 28, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Procino, G.; Milano, S.; Tamma, G.; Dossena, S.; Barbieri, C.; Nicoletti, M.C.; Ranieri, M.; Di Mise, A.; Nofziger, C.; Svelto, M.; et al. Co-regulated pendrin and aquaporin 5 expression and trafficking in Type-B intercalated cells under potassium depletion. Cell. Physiol. Biochem. 2013, 32, 184–199. [Google Scholar] [CrossRef]
- Bernardinelli, E.; Costa, R.; Nofziger, C.; Paulmichl, M.; Dossena, S. Effect of Known Inhibitors of Ion Transport on Pendrin (SLC26A4) Activity in a Human Kidney Cell Line. Cell. Physiol. Biochem. 2016, 38, 1984–1998. [Google Scholar] [CrossRef]
- Galietta, L.J.; Haggie, P.M.; Verkman, A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001, 499, 220–224. [Google Scholar] [CrossRef]
- Adler, J.; Parmryd, I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytom. Part A J. Int. Soc. Anal. Cytol. 2010, 77, 733–742. [Google Scholar] [CrossRef]
- Reardon, W.; Mahoney, C.F.O.; Trembath, R.; Jan, H.; Phelps, P.D. Enlarged vestibular aqueduct: A radiological marker of pendred syndrome, and mutation of the PDS gene. QJM Mon. J. Assoc. Physicians 2000, 93, 99–104. [Google Scholar] [CrossRef]
- Albert, S.; Blons, H.; Jonard, L.; Feldmann, D.; Chauvin, P.; Loundon, N.; Sergent-Allaoui, A.; Houang, M.; Joannard, A.; Schmerber, S.; et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur. J. Hum. Genet. EJHG 2006, 14, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Shearer, A.E.; Eppsteiner, R.W.; Booth, K.T.; Ephraim, S.S.; Gurrola, J., 2nd; Simpson, A.; Black-Ziegelbein, E.A.; Joshi, S.; Ravi, H.; Giuffre, A.C.; et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am. J. Hum. Genet. 2014, 95, 445–453. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Dossena, S.; Bernardinelli, E.; Sharma, A.K.; Alper, S.L.; Paulmichl, M. The pendrin polypeptide. In The Role of Pendrin in Health and Disease; Dossena, S., Paulmichl, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Lu, Y.C.; Wu, C.C.; Yang, T.H.; Lin, Y.H.; Yu, I.S.; Lin, S.W.; Chang, Q.; Lin, X.; Wong, J.M.; Hsu, C.J. Differences in the pathogenicity of the p.H723R mutation of the common deafness-associated SLC26A4 gene in humans and mice. PLoS ONE 2014, 8, e64906. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.J.; Lu, Y.C.; Tsai, C.Y.; Chan, Y.H.; Lin, P.H.; Lee, Y.S.; Yu, I.S.; Lin, S.W.; Liu, T.C.; Hsu, C.J.; et al. Insights into phenotypic differences between humans and mice with p.T721M and other C-terminal variants of the SLC26A4 gene. Sci. Rep. 2021, 11, 20983. [Google Scholar] [CrossRef]
- Hu, C.J.; Lu, Y.C.; Yang, T.H.; Chan, Y.H.; Tsai, C.Y.; Yu, I.S.; Lin, S.W.; Liu, T.C.; Cheng, Y.F.; Wu, C.C.; et al. Toward the Pathogenicity of the SLC26A4 p.C565Y Variant Using a Genetically Driven Mouse Model. Int. J. Mol. Sci. 2021, 22, 2789. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matulevičius, A.; Bernardinelli, E.; Brownstein, Z.; Roesch, S.; Avraham, K.B.; Dossena, S. Molecular Features of SLC26A4 Common Variant p.L117F. J. Clin. Med. 2022, 11, 5549. https://doi.org/10.3390/jcm11195549
Matulevičius A, Bernardinelli E, Brownstein Z, Roesch S, Avraham KB, Dossena S. Molecular Features of SLC26A4 Common Variant p.L117F. Journal of Clinical Medicine. 2022; 11(19):5549. https://doi.org/10.3390/jcm11195549
Chicago/Turabian StyleMatulevičius, Arnoldas, Emanuele Bernardinelli, Zippora Brownstein, Sebastian Roesch, Karen B. Avraham, and Silvia Dossena. 2022. "Molecular Features of SLC26A4 Common Variant p.L117F" Journal of Clinical Medicine 11, no. 19: 5549. https://doi.org/10.3390/jcm11195549
APA StyleMatulevičius, A., Bernardinelli, E., Brownstein, Z., Roesch, S., Avraham, K. B., & Dossena, S. (2022). Molecular Features of SLC26A4 Common Variant p.L117F. Journal of Clinical Medicine, 11(19), 5549. https://doi.org/10.3390/jcm11195549






