Prognostic Value and Clinical Significance of FGFR Genomic Alterations (GAs) in Metastatic Urothelial Cancer Patients
Abstract
:1. Introduction
2. Materials (Patients) and Methods
2.1. Study Design and Patients
2.2. Genomic Analysis
2.3. Outcomes
2.4. Statistical Analysis
3. Results
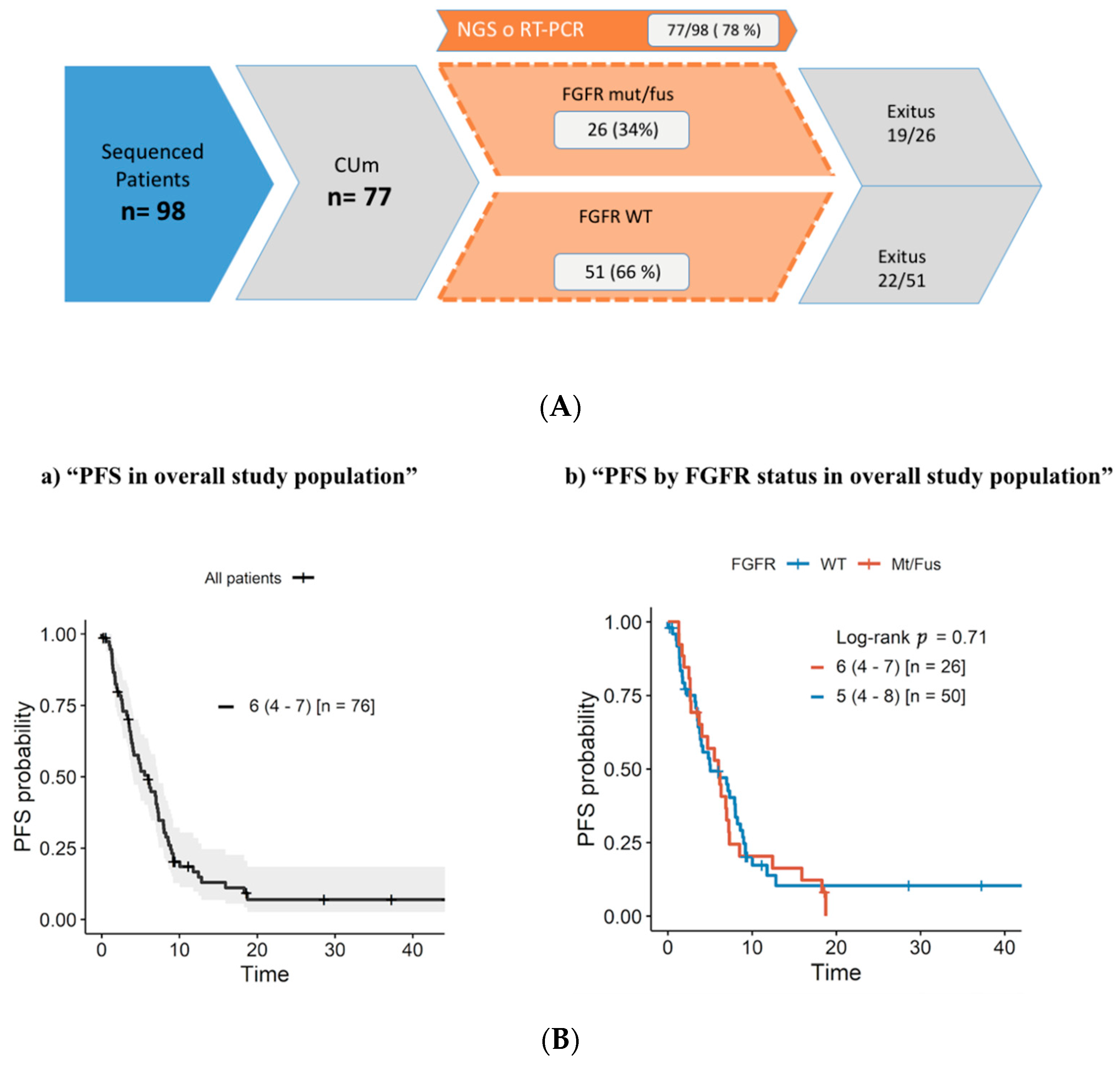
3.1. Overall Study Population
3.2. Patient Characteristics
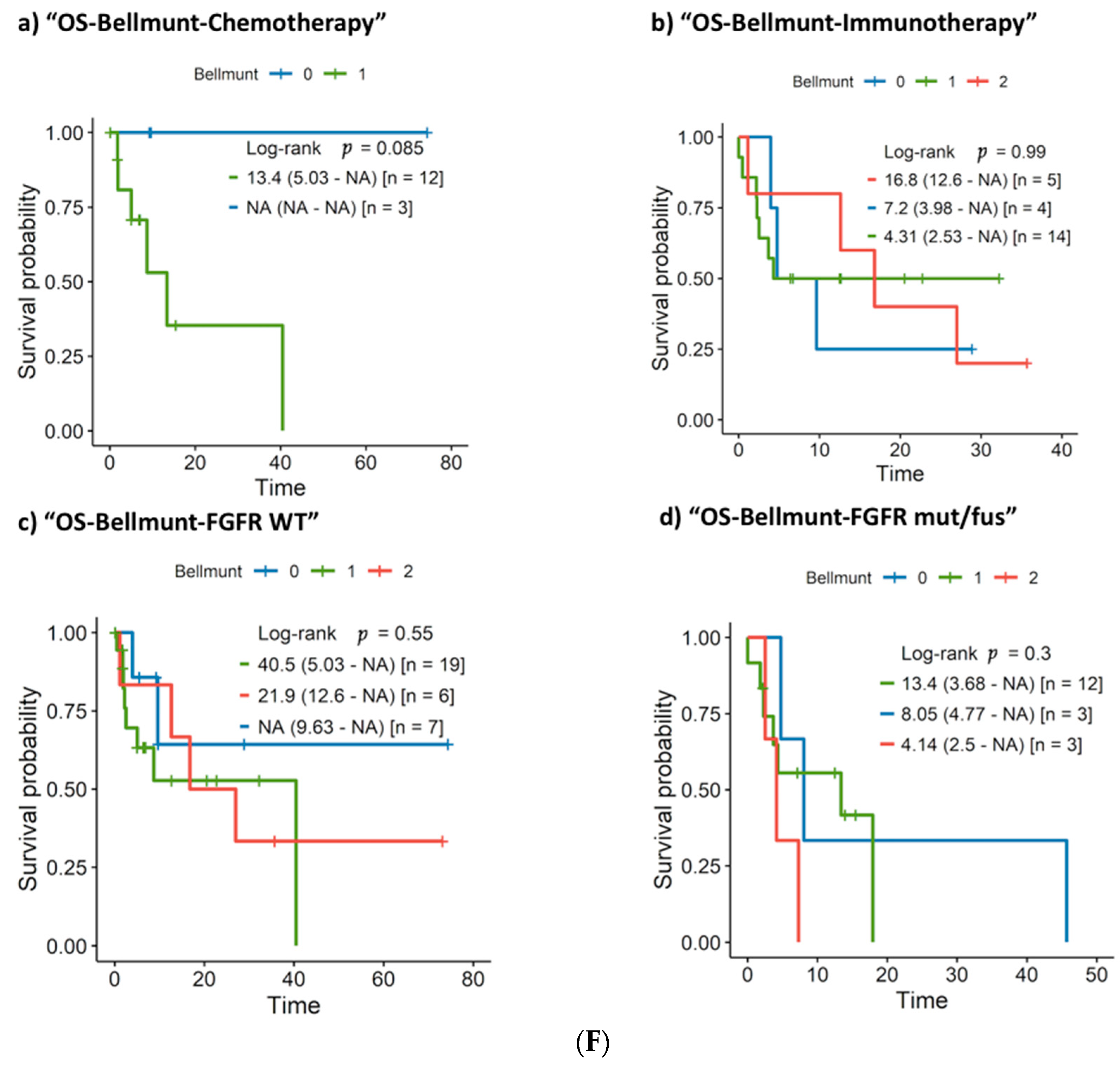
3.3. First-Line Treatment Outcome
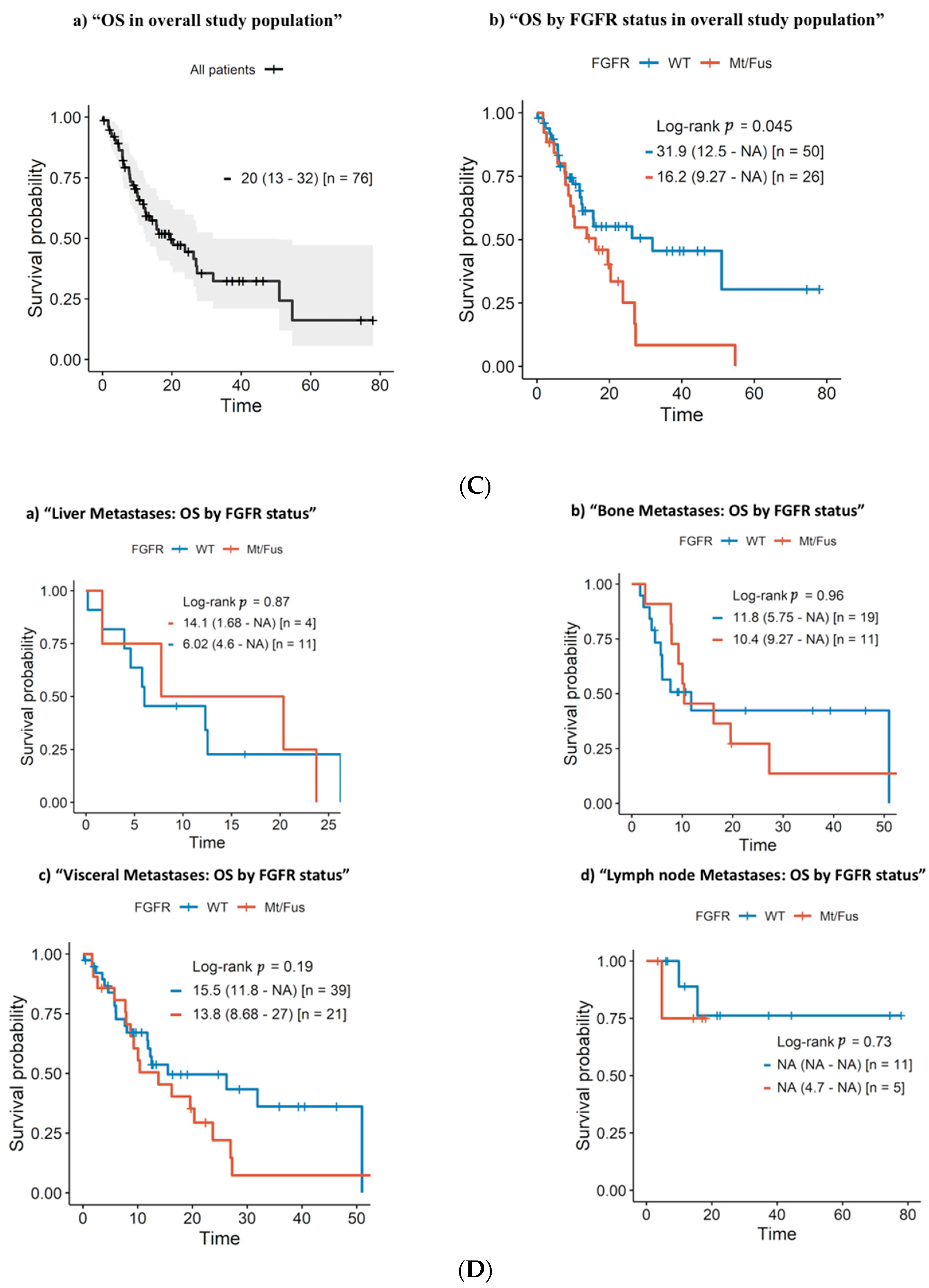
3.4. Progression-Free Survival and Overall Survival
3.5. Clinical and Molecular Prognostic Factors: Univariate and Multivariate Survival Analysis
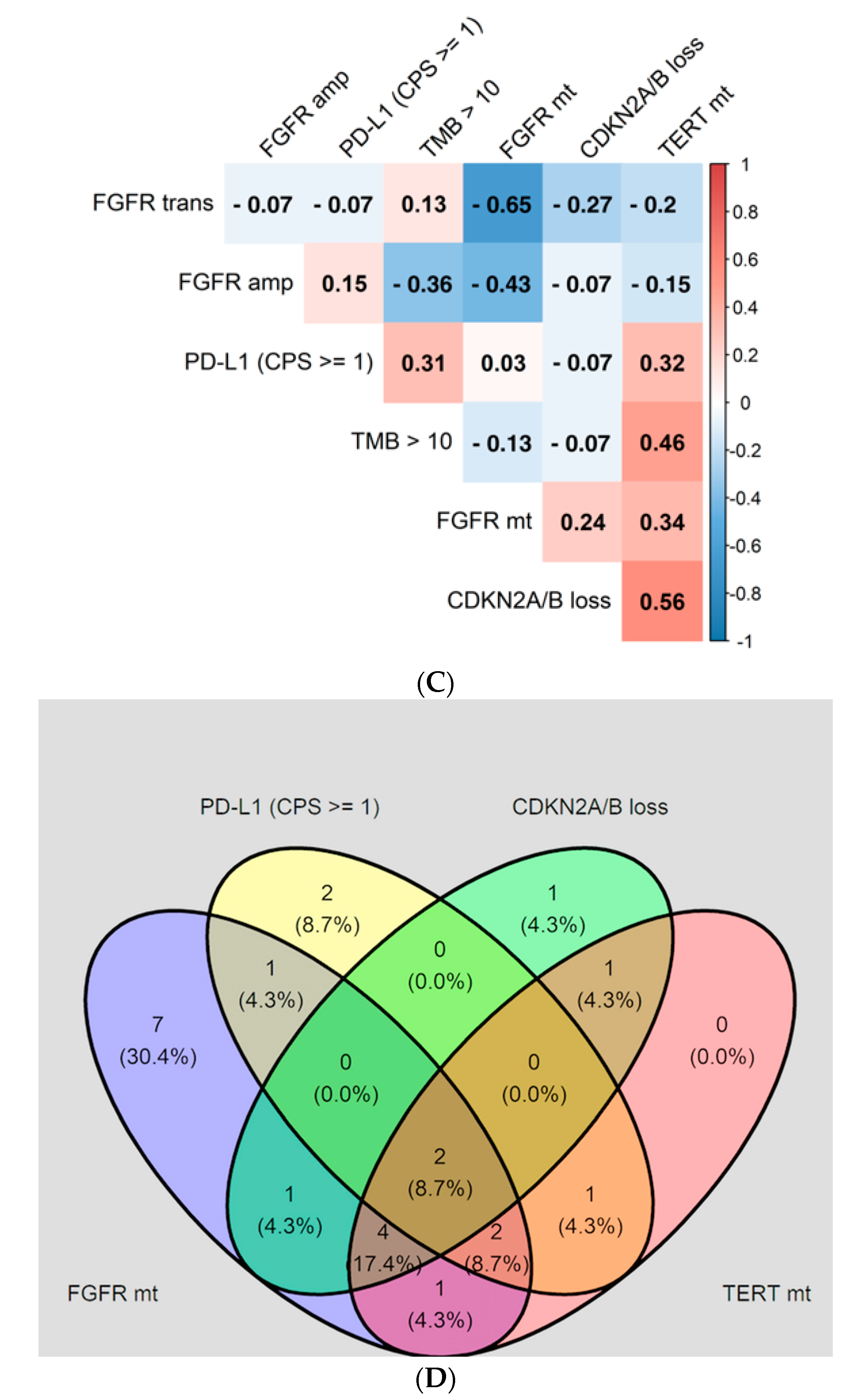
3.6. Interaction of FGFR GAs with Additional Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar]
- Teoh, J.Y.C.; Huang, J.; Ko, W.Y.K.; Lok, V.; Choi, P.; Ng, C.F.; Sengupta, S.; Mostafid, H.; Kamat, A.M.; Black, P.C.; et al. Global Trends of Bladder Cancer Incidence and Mortality, and Their Associations with Tobacco Use and Gross Domestic Product per Capita. Eur. Urol. 2020, 78, 893–906. [Google Scholar]
- Lin, J.; Spitz, M.R.; Dinney, C.P.; Etzel, C.J.; Grossman, H.B.; Wu, X. Bladder cancer risk as modified by family history and smoking. Cancer 2006, 107, 705–711. [Google Scholar]
- Bajorin, D.F.; Dodd, P.M.; Mazumdar, M.; Fazzari, M.; McCaffrey, J.A.; Scher, H.I.; Herr, H.; Higgins, G.; Boyle, M.G. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1999, 17, 3173–3181. [Google Scholar]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar]
- Powles, T.; Rosenberg, J.E.; Sonpavde, G.P.; Loriot, Y.; Durán, I.; Lee, J.L.; Matsubara, N.; Vulsteke, C.; Castellano, D.; Wu, C.; et al. Enfortumab Vedotin in Previously Treated Advanced Urothelial Carcinoma. N. Engl. J. Med. 2021, 384, 1125–1135. [Google Scholar]
- Tagawa, S.T.; Balar, A.V.; Petrylak, D.P.; Kalebasty, A.R.; Loriot, Y.; Fléchon, A.; Jain, R.K.; Agarwal, N.; Bupathi, M.; Barthelemy, P.; et al. TROPHY-U-01: A Phase II Open-Label Study of Sacituzumab Govitecan in Patients with Metastatic Urothelial Carcinoma Progressing After Platinum-Based Chemotherapy and Checkpoint Inhibitors. J. Clin. Oncol. 2021, 39, 2474–2485. [Google Scholar]
- Garje, R.; An, J.; Obeidat, M.; Kumar, K.; Yasin, H.A.; Zakharia, Y. Fibroblast Growth Factor Receptor (FGFR) Inhibitors in Urothelial Cancer: Fibroblast Growth Factor Receptor (FGFR) Inhibitors in Urothelial Cancer. Oncologist 2020, 25, e1711–e1719. [Google Scholar] [CrossRef]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar]
- Al-Obaidy, K.I.; Cheng, L. Fibroblast growth factor receptor (FGFR) gene: Pathogenesis and treatment implications in urothelial carcinoma of the bladder. J. Clin. Pathol. 2021, 74, 491–495. [Google Scholar]
- Costa, R.; Carneiro, B.A.; Taxter, T.; Tavora, F.A.; Kalyan, A.; Pai, S.A.; Chae, Y.K.; Giles, F.J. FGFR3-TACC3 fusion in solid tumors: Mini review. Oncotarget 2016, 7, 55924–55938. [Google Scholar]
- Tomlinson, D.C.; L’Hôte, C.G.; Kennedy, W.; Pitt, E.; Knowles, M.A. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005, 65, 10441–10449. [Google Scholar]
- Pal, S.K.; Bajorin, D.; Dizman, N.; Hoffman-Censits, J.; Quinn, D.I.; Petrylak, D.P.; Galsky, M.D.; Vaishampayan, U.; de Giorgi, U.; Gupta, S.; et al. Infigratinib in upper tract urothelial carcinoma versus urothelial carcinoma of the bladder and its association with comprehensive genomic profiling and/or cell-free DNA results. Cancer 2020, 126, 2597–2606. [Google Scholar]
- Li, J.J.; Yan, S.; Pan, Y.; Liu, Z.; Liu, Y.; Deng, Q.; Tan, Q.; Woodward, E.R. FGFR genes mutation is an independent prognostic factor and associated with lymph node metastasis in squamous non-small cell lung cancer. Cancer Biol. Ther. 2018, 19, 1108–1116. [Google Scholar]
- Ipenburg, N.A.; Koole, K.; Liem, K.S.; van Kempen, P.M.W.; Koole, R.; van Diest, P.J.; van Es, R.J.J.; Willems, S.M. Fibroblast Growth Factor Receptor Family Members as Prognostic Biomarkers in Head and Neck Squamous Cell Carcinoma: A Systematic Review. Target Oncol. 2016, 11, 17–27. [Google Scholar]
- Amina, B.; Lynda, A.K.; Sonia, S.; Adel, B.; Jelloul, B.H.; Miloud, M.; Tewfik, S. Fibroblast growth factor receptor 1 protein (FGFR1) as potential prognostic and predictive marker in patients with luminal B breast cancers overexpressing human epidermal receptor 2 protein (HER2). Indian J. Pathol. Microbiol. 2021, 64, 254–260. [Google Scholar]
- Lee, H.W.; Seo, H.K. Fibroblast Growth Factor Inhibitors for Treating Locally Advanced/Metastatic Bladder Urothelial Carcinomas via Dual Targeting of Tumor-Specific Oncogenic Signaling and the Tumor Immune Microenvironment. Int. J. Mol. Sci. 2021, 22, 9526. [Google Scholar]
- Kardoust Parizi, M.; Margulis, V.; Lotan, Y.; Mori, K.; Shariat, S.F. Fibroblast growth factor receptor: A systematic review and meta-analysis of prognostic value and therapeutic options in patients with urothelial bladder carcinoma. Urol. Oncol. Semin. Orig. Investig. 2021, 39, 409–421. [Google Scholar]
- Santiago-Walker, A.E.; Chen, F.; Loriot, Y.; Siefker-Radtke, A.O.; Sun, L.; Sundaram, R.; de Porre, P.; Patel, K.; Wan, Y. Predictive value of fibroblast growth factor receptor (FGFR) mutations and gene fusions on anti-PD-(L)1 treatment outcomes in patients (pts) with advanced urothelial cancer (UC). J. Clin. Oncol. 2019, 37 (Suppl. S7), 419. [Google Scholar]
- Rose, T.L.; Hayward, M.C.; Salazar, A.H.; Eulitt, P.; McGinty, K.; Drier, A.; Wobker, S.E.; Whang, Y.E.; Brower, B.Y.; Dunn, M.; et al. Fibroblast growth factor receptor status and response to immune checkpoint inhibition in metastatic urothelial cancer. J. Clin. Oncol. 2019, 37 (Suppl. S7), 458. [Google Scholar]
- Wang, L.; Gong, Y.; Saci, A.; Szabo, P.M.; Martini, A.; Necchi, A.; Pal, S.; Plimack, E.R.; Sfakianos, J.P.; Bhardwaj, N.; et al. Fibroblast Growth Factor Receptor 3 Alterations and Response to PD-1/PD-L1 Blockade in Patients with Metastatic Urothelial Cancer. Eur. Urol. 2019, 76, 599–603. [Google Scholar]
- Teo, M.Y.; Mota, J.M.; Whiting, K.A.; Li, H.A.; Funt, S.A.; Lee, C.H.; Solit, D.B.; Al-Ahmadie, H.; Milowsky, M.I.; Balar, A.V.; et al. Fibroblast Growth Factor Receptor 3 Alteration Status is Associated with Differential Sensitivity to Platinum-based Chemotherapy in Locally Advanced and Metastatic Urothelial Carcinoma. Eur. Urol. 2020, 78, 907–915. [Google Scholar]
- Bellmunt, J.; Choueiri, T.K.; Fougeray, R.; Schutz, F.A.B.; Salhi, Y.; Winquist, E.; Culine, S.; von der Maase, H.; Vaughn, D.J.; Rosenberg, J.E. Prognostic Factors in Patients with Advanced Transitional Cell Carcinoma of the Urothelial Tract Experiencing Treatment Failure With Platinum-Containing Regimens. J. Clin. Oncol. 2010, 28, 1850–1855. [Google Scholar]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar]
- Audenet, F.; Isharwal, S.; Cha, E.K.; Donoghue, M.T.A.; Drill, E.N.; Ostrovnaya, I.; Pietzak, E.J.; Sfakianos, J.P.; Bagrodia, A.; Murugan, P.; et al. Clonal Relatedness and Mutational Differences between Upper Tract and Bladder Urothelial Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 967–976. [Google Scholar]
- Galsky, M.D.; Arija, J.Á.A.; Bamias, A.; Davis, I.D.; De Santis, M.; Kikuchi, E.; Garcia-Del-Muro, X.; de Giorgi, U.; Mencinger, M.; Izumi, K.; et al. Atezolizumab with or without chemotherapy in metastatic urothelial cancer (IMvigor130): A multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2020, 395, 1547–1557. [Google Scholar]
- Jing, W.; Wang, G.; Cui, Z.; Xiong, G.; Jiang, X.; Li, Y.; Li, W.; Han, B.; Chen, S.; Shi, B.; et al. FGFR3 Destabilizes PD-L1 via NEDD4 to Control T-cell–Mediated Bladder Cancer Immune Surveillance. Cancer Res. 2022, 82, 114–129. [Google Scholar]
- Robinson, B.D.; Vlachostergios, P.J.; Bhinder, B.; Liu, W.; Li, K.; Moss, T.J.; Bareja, R.; Park, K.; Tavassoli, P.; Cyrta, J.; et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat. Commun. 2019, 10, 2977. [Google Scholar]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018, 174, 1033. [Google Scholar]
- Palakurthi, S.; Kuraguchi, M.; Zacharek, S.J.; Zudaire, E.; Huang, W.; Bonal, D.M.; Liu, J.; Dhaneshwar, A.; DePeaux, K.; Gowaski, M.R.; et al. The Combined Effect of FGFR Inhibition and PD-1 Blockade Promotes Tumor-Intrinsic Induction of Antitumor Immunity. Cancer Immunol. Res. 2019, 7, 1457–1471. [Google Scholar]
- Halbert, B.; Einstein, D.J. Hot or Not: Tumor Mutational Burden (TMB) as a Biomarker of Immunotherapy Response in Genitourinary Cancers. Urology 2021, 147, 119–126. [Google Scholar]
- Pal, S.K.; Agarwal, N.; Choueiri, T.K.; Stephens, P.J.; Ross, J.S.; Miller, V.A.; Ali, S.M.; Chung, J.; Grivas, P. Comparison of tumor mutational burden (TMB) in relevant molecular subsets of metastatic urothelial cancer (MUC). Ann. Oncol. 2017, 28, v297. [Google Scholar]
- Hosen, I.; Rachakonda, P.S.; Heidenreich, B.; de Verdier, P.J.; Ryk, C.; Steineck, G.; Hemminki, K.; Kumar, R. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer: TERT, FGFR3, Telomere Length in Bladder Cancer. Int. J. Cancer 2015, 137, 1621–1629. [Google Scholar]
- Rachakonda, P.S.; Hosen, I.; de Verdier, P.J.; Fallah, M.; Heidenreich, B.; Ryk, C.; Wiklund, N.P.; Steineck, G.; Schadendorf, D.; Hemminki, K.; et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. USA 2013, 110, 17426–17431. [Google Scholar]
- Li, H.; Li, J.; Zhang, C.; Zhang, C.; Wang, H. TERT mutations correlate with higher TMB value and unique tumor microenvironment and may be a potential biomarker for anti-CTLA4 treatment. Cancer Med. 2020, 9, 7151–7160. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- Rebouissou, S.; Hérault, A.; Letouzé, E.; Neuzillet, Y.; Laplanche, A.; Ofualuka, K.; Maillé, P.; Leroy, K.; Riou, A.; Lepage, M.-L.; et al. CDKN2A homozygous deletion is associated with muscle invasion in FGFR3-mutated urothelial bladder carcinoma. J. Pathol. 2012, 227, 315–324. [Google Scholar]
- Nassar, A.; Adib, E.; Akl, E.W.; Abou Alaiwi, S.; Nuzzo, P.V.; Mouhieddine, T.H.; Sonpavde, G.; Haddad, R.I.; Giannakis, M.; Hodi, F.S.; et al. CDKN2A alterations as markers of immune checkpoint blockade (ICB) resistance in urothelial carcinoma (UC). J. Clin. Oncol. 2021, 39 (Suppl. S6), 475. [Google Scholar]
- Banchereau, R.; Leng, N.; Zill, O.; Sokol, E.; Liu, G.; Pavlick, D.; Maund, S.; Liu, L.-F.; Kadel, E., III; Balswin, N.; et al. Molecular determinants of response to PD-L1 blockade across tumor types. Nat. Commun. 2021, 12, 3969. [Google Scholar]
- Stangl-Kremser, J.; Muto, G.; Grosso, A.A.; Briganti, A.; Comperat, E.; Di Maida, F. The impact of lymphovascular invasion in patients treated with radical nephroureterectomy for upper tract urothelial carcinoma: An extensive updated systematic review and meta-analysis. Urol. Oncol. 2022, 40, 243–261. [Google Scholar]
- Di Maida, F.; Scalici Gesolfo, C.; Tellini, R.; Mari, A.; Sanfilippo, C.; Lambertini, L. Fibronectin urothelial gene expression as a new reliable biomarker for early detection of local toxicity secondary to adjuvant intravesical therapy for non-muscle invasive bladder cancer. Ther. Adv. Urol. 2021, 13, 1756287221995683. [Google Scholar]


| Variable | Modality | Metastatic | Mt/Fus (n = 26) | WT (n = 51) | p-Value |
|---|---|---|---|---|---|
| Age | Median (IQR) | 69 (62–76) | 69 (63–77) | 69 (61–75) | 0.45 |
| Sex | Male | 55 (71.4%) | 17 (65.4%) | 38 (74.5%) | 0.57 |
| Female | 22 (28.6%) | 9 (34.6%) | 13 (25.5%) | ||
| ECOG PS | 0 | 26 (33.8%) | 9 (34.6%) | 17 (33.3%) | 0.15 |
| 1 | 25 (32.4%) | 14 (53.8%) | 11 (21.6%) | ||
| 2 | 2 (2.6%) | 0 (0%) | 2 (3.9%) | ||
| Not available | 24 (31.2%) | 3 (11.5%) | 21 (41.2%) | ||
| Smoking | Never smoked | 16 (25.4%) | 7 (26.9%) | 9 (17.6%) | 0.55 |
| Current smoker | 9 (14.3%) | 3 (11.5%) | 6 (11.8%) | ||
| Former smoker | 38 (60.3%) | 11 (42.3%) | 27 (52.9%) | ||
| Not available | 14 (18.2%) | 5 (19.2%) | 9 (17.6%) | ||
| Tumor location | Bladder | 62 (80.5%) | 17 (65.4%) | 45 (88.2%) | 0.037 |
| Nonbladder | 15 (19.5%) | 9 (34.6%) | 6 (11.8%) | ||
| Surgery | No | 4 (5.2%) | 1 (3.8%) | 3 (5.9%) | 1 |
| Yes | 73 (94.8%) | 25 (96.2%) | 48 (94.1%) | ||
| Surgery extension | Cystectomy (RC) | 38 (49.3%) | 7 (28.0%) | 31 (64.6%) | 0.0076 |
| Nephroureterectomy/nephrectomy(NU) | 12 (15.6%) | 8 (32.0%) | 4 (8.3%) | ||
| NU + RC | 3 (3.9%) | 1 (4.0%) | 2 (4.2%) | ||
| TURBT | 20 (26%) | 9 (36.0%) | 11 (22.9%) | ||
| Lymphadenectomy | No | 36 (46.8%) | 15 (57.7%) | 21 (41.2%) | 0.37 |
| Yes | 38 (49.3%) | 11 (42.3%) | 27 (52.9%) | ||
| Not available | 3 (3.9%) | 0 (0.0%) | 3 (5.9%) | ||
| Bladder preservation | Radiotherapy | 4 (5.2%) | 2 (7.7%) | 2 (3.9%) | 0.86 |
| Chemo-radiotherapy | 6 (7.8%) | 2 (7.7%) | 4 (7.8%) | ||
| No | 67 (87.0%) | 22 (84.6%) | 45 (88.2%) | ||
| pT | 1 | 7 (9.1%) | 3 (11.5%) | 4 (7.8%) | 0.022 |
| 2 | 28 (36.4%) | 8 (30.8%) | 20 (39.2%) | ||
| 3 | 28 (36.4%) | 6 (23.1%) | 22 (43.1%) | ||
| 4 | 13 (16.8%) | 9 (34.6%) | 4 (7.8%) | ||
| Not available | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| pN | 0 | 14 (36.8%) | 3 (27.3%) | 11 (40.7%) | 0.81 |
| 1 | 11 (28.9%) | 4 (36.4%) | 7 (25.9%) | ||
| 2 | 12 (31.6%) | 4 (36.4%) | 8 (29.6%) | ||
| 3 | 1 (2.6%) | 0 (0%) | 1 (3.7%) | ||
| Grade | 2 | 2 (2.6%) | 1 (3.8%) | 1 (2%) | 1 |
| 3 | 73 (94.8%) | 25 (96.2%) | 48 (94.1%) | ||
| NA, n (%) | 2 (2.6%) | 0 (0.0%) | 2 (3.9%) | ||
| Variant histologies | Transitional cells | 70 (90.9%) | 23 (88.5%) | 47 (92.2%) | 0.71 |
| Squamous | 5 (6.5%) | 2 (7.7%) | 3 (5.9%) | ||
| Anaplastic | 1 (1.3%) | 1 (3.8%) | 0 (0%) | ||
| Neuroendocrine | 1 (1.3%) | 0 (0%) | 1 (2%) | ||
| Perioperative chemotherapy | No | 52 (67.5%) | 16 (61.5%) | 36 (70.6%) | 0.67 |
| Neoadjuvant | 12 (15.6%) | 5 (19.2%) | 7 (13.7%) | ||
| Adjuvant | 13 (16.9%) | 5 (19.2%) | 8 (15.7%) | ||
| Liver metastases | No | 61 (79.2%) | 22 (84.6%) | 39 (76.5%) | 0.7 |
| Yes | 15 (19.5%) | 4 (15.4%) | 11 (21.6%) | ||
| Not available | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| Bone metastases | No | 46 (59.7%) | 15 (57.7%) | 31 (60.8%) | 0.91 |
| Yes | 30 (39.0%) | 11 (42.3%) | 19 (37.3%) | ||
| Not available | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| Lymph node only metastases | Yes | 15 (19.5%) | 5 (19.2%) | 10 (19.6%) | 1 |
| Visceral metastases | Yes | 60 (77.9%) | 21 (80.8%) | 39 (76.5%) | 1 |
| Not available | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| First-line treatment for mUC | Cisplatin-based | 33 (42.8%) | 11 (42.3%) | 22 (44.0%) | |
| Checkpoint inhibitors | 23 (29.9%) | 5 (19.2%) | 18 (36.0%) | ||
| Carboplatin-based | 8 (10.4%) | 2 (7.7%) | 6 (12.0%) | ||
| FGFR inhibitor | 5 (6.5%) | 5 (19.2%) | 0 (0.0%) | ||
| Vinflunine | 3 (3.9%) | 1 (3.8%) | 2 (4.0%) | ||
| Best supportive care | 2 (2.6%) | 2 (7.7%) | 0 (0.0%) | ||
| Paclitaxel | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| Surgery | 1 (1.3%) | 0 (0.0%) | 1 (2.0%) | ||
| Not available | 1 (1.3%) | (0%) | 1 (2.0%) |
| TYPE OF FGFR | N (26) | % |
|---|---|---|
| GENOMIC ALTERATION (Number of Cases) | ||
| MUTATION | 15 | 57.7% |
| -FGFR3 S249C (13) | ||
| -FGFR3 S249C // S783 frameshift mutation (1) | ||
| -FGFR3 S249C // H349D (1) | ||
| FUSION | 6 | 23.1% |
| -FGFR1-FGFR1 (1) | ||
| -FGFR3-TACC3 (3) | ||
| -FGFR2-OFD1 (1) -FGFR2-AFF3 (1) | ||
| MUTATION + AMPLIFICATION | 2 | 7.7% |
| -FGFR3 S249C + FGFR1 amplification (1) | ||
| -FGFR3 S249C + FGFR1 amplification (1) | ||
| MUTATION + FUSION | 1 | 3.8% |
| -FGFR3 R248C // S249C + FGFR3-TACC3 (1) | ||
| FUSION + AMPLIFICATION | 2 | 7.7% |
| -FGFR2-RTKN2 + FGFR2 amplification (1) -FGFR3-TACC3 + FGFR1 amplification (1) |
| Population | Treatment (n) | Type of Response | ||||||
|---|---|---|---|---|---|---|---|---|
| CR | PR | SD | PD | ORR | p-Value * | p-Value ORR | ||
| Overall | Any (70) | 5 (7.1%) | 30 (42.9%) | 8 (11.4%) | 27 (38.6%) | 35 (50.0%) | ||
| FGFR WT | 3 (6.5%) | 18 (39.1%) | 5 (10.9%) | 20 (43.5%) | 21 (45.7%) | 0.71 | 0.57 | |
| FGFR mut/fus | 2 (8.3%) | 12 (50.0%) | 3 (12.5%) | 7 (29.2%) | 14 (58.3%) | |||
| Overall | Cisplatinum-based (32) | 2 (6.3%) | 17 (53,1%) | 5 (15.6%) | 8 (25.0%) | 19 (59.4%) | ||
| FGFR WT | 1 (4.8%) | 10 (47.6%) | 3 (14.3%) | 7 (33.3%) | 11 (52.4%) | 0.43 | 0.45 | |
| FGFR mut/fus | 1 (9.1%) | 7 (63.6%) | 2 (18.2%) | 1 (9.1%) | 8 (72.7%) | |||
| Overall | Immunotherapy (21) | 2 (9.5%) | 6 (28.6%) | 2 (9.5%) | 11 (52.4%) | 8 (38.1%) | ||
| FGFR WT | 1 (6.3%) | 4 (25.0%) | 2 (12.5%) | 9 (56.3%) | 5 (31.3%) | 0.59 | 0.33 | |
| FGFR mut/fus | 1 (20.0%) | 2 (40.0%) | 0 (0.0%) | 2 (40.0%) | 3 (60.0%) | |||
| FGFR mut/fus | FGFR inhibitors (5) Other (19) | 0 (0.0%) | 1 (20.0%) | 0 (0.0%) | 4 (80.0%) | 1 (20.0%) | 0.065 | 0.12 |
| 2 (10.5%) | 11 (57.9%) | 3 (15.8%) | 3 (15.8%) | 13 (68.4%) | ||||
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Variable | Modality | HR | 95% CI | HR | 95% CI |
| Age | (continuous) | 1.02 | 0.992–1.05 | 1.03 | 1.00–1.07 |
| Location | Nonbladder | 1 (ref) * | - | 1 (ref) | - |
| Bladder | 1.07 | 0.47–2.42 | 1.39 | 0.56–3.48 | |
| Treatment | Cisplatin | 1 (ref) | - | 1 (ref) | - |
| Immunotherapy | 1.32 | 0.60–2.90 | 2.40 | 0.97–5.90 | |
| Other | 1.71 | 0.84–3.48 | 3.17 | 1.38–7.24 | |
| Visceral metastases | No | 1 (ref) | - | 1 (ref) | - |
| Yes | 4.87 | 1.48–16.0 | 11.4 | 2.56–50.9 | |
| ECOG > 1 | No | 1 (ref) | - | 1 (ref) | - |
| Yes | 2.79 | 1.29–6.00 | 6.40 | 2.43–16.9 | |
| FGFR | Wild-type | 1 (ref) | - | 1 (ref) | - |
| Mutated | 1.87 | 1.01–3.48 | 2.59 | 1.21–5.55 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sevillano Fernández, E.; Madurga de Lacalle, R.; Rodriguez Moreno, J.F.; Barquín García, A.; Yagüe Fernández, M.; Navarro Alcaraz, P.; Barba Llacer, M.; Quiralte Pulido, M.; García-Donás Jiménez, J. Prognostic Value and Clinical Significance of FGFR Genomic Alterations (GAs) in Metastatic Urothelial Cancer Patients. J. Clin. Med. 2022, 11, 4483. https://doi.org/10.3390/jcm11154483
Sevillano Fernández E, Madurga de Lacalle R, Rodriguez Moreno JF, Barquín García A, Yagüe Fernández M, Navarro Alcaraz P, Barba Llacer M, Quiralte Pulido M, García-Donás Jiménez J. Prognostic Value and Clinical Significance of FGFR Genomic Alterations (GAs) in Metastatic Urothelial Cancer Patients. Journal of Clinical Medicine. 2022; 11(15):4483. https://doi.org/10.3390/jcm11154483
Chicago/Turabian StyleSevillano Fernández, Elena, Rodrigo Madurga de Lacalle, Juan Francisco Rodriguez Moreno, Arantzazu Barquín García, Mónica Yagüe Fernández, Paloma Navarro Alcaraz, María Barba Llacer, Miguel Quiralte Pulido, and Jesús García-Donás Jiménez. 2022. "Prognostic Value and Clinical Significance of FGFR Genomic Alterations (GAs) in Metastatic Urothelial Cancer Patients" Journal of Clinical Medicine 11, no. 15: 4483. https://doi.org/10.3390/jcm11154483
APA StyleSevillano Fernández, E., Madurga de Lacalle, R., Rodriguez Moreno, J. F., Barquín García, A., Yagüe Fernández, M., Navarro Alcaraz, P., Barba Llacer, M., Quiralte Pulido, M., & García-Donás Jiménez, J. (2022). Prognostic Value and Clinical Significance of FGFR Genomic Alterations (GAs) in Metastatic Urothelial Cancer Patients. Journal of Clinical Medicine, 11(15), 4483. https://doi.org/10.3390/jcm11154483






