Implications of Decreased Expression of miR-125a with Respect to Its Variant Allele in the Pathogenesis of Recurrent Pregnancy Loss: A Study in a High Incidence Zone
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
2.3. DNA Extraction
2.4. miRNA Extraction from Plasma
2.5. Genotyping of Polymorphisms by PCR-RFLP
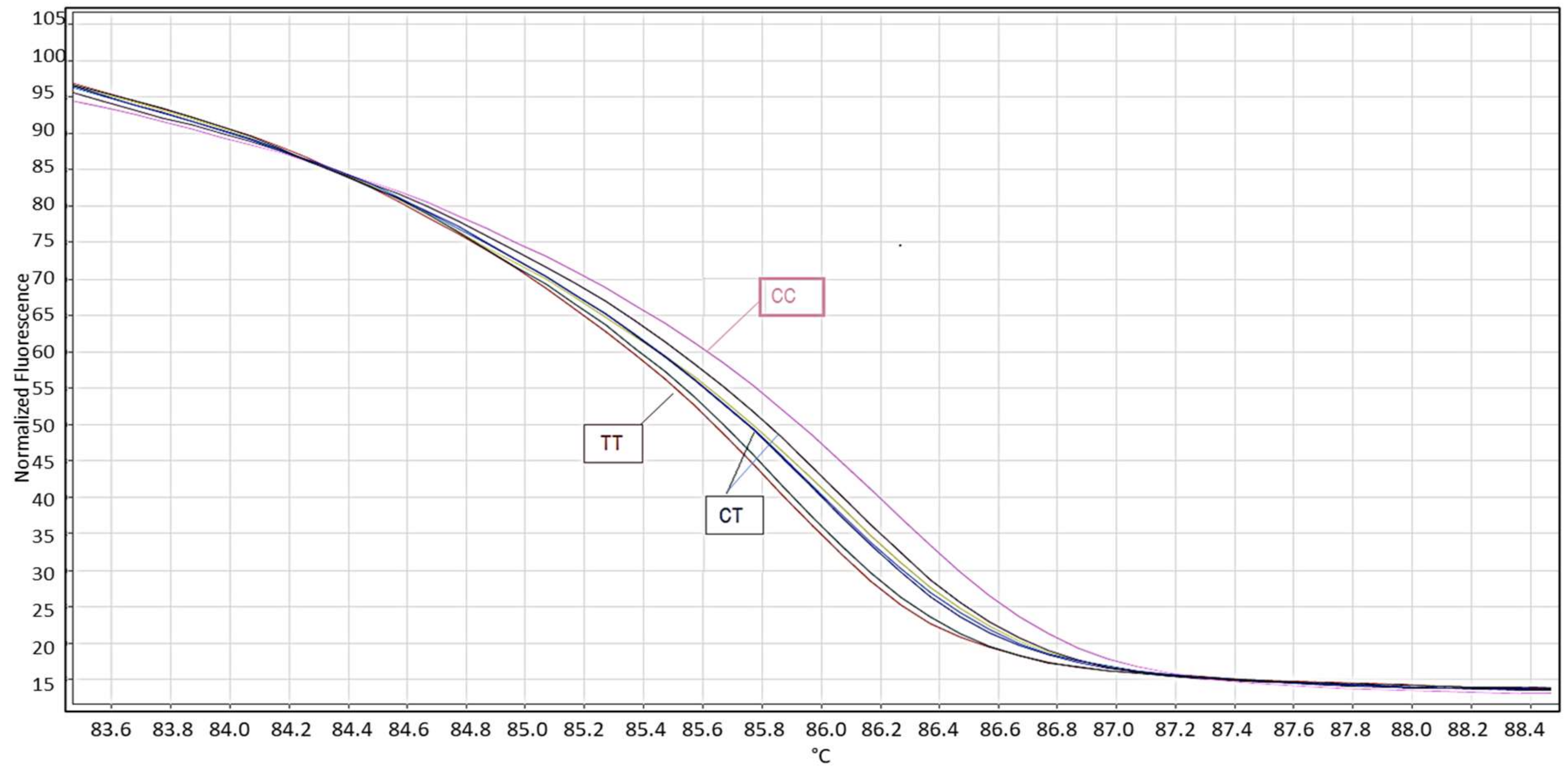
2.6. High Resolution Melting (HRM) Analysis
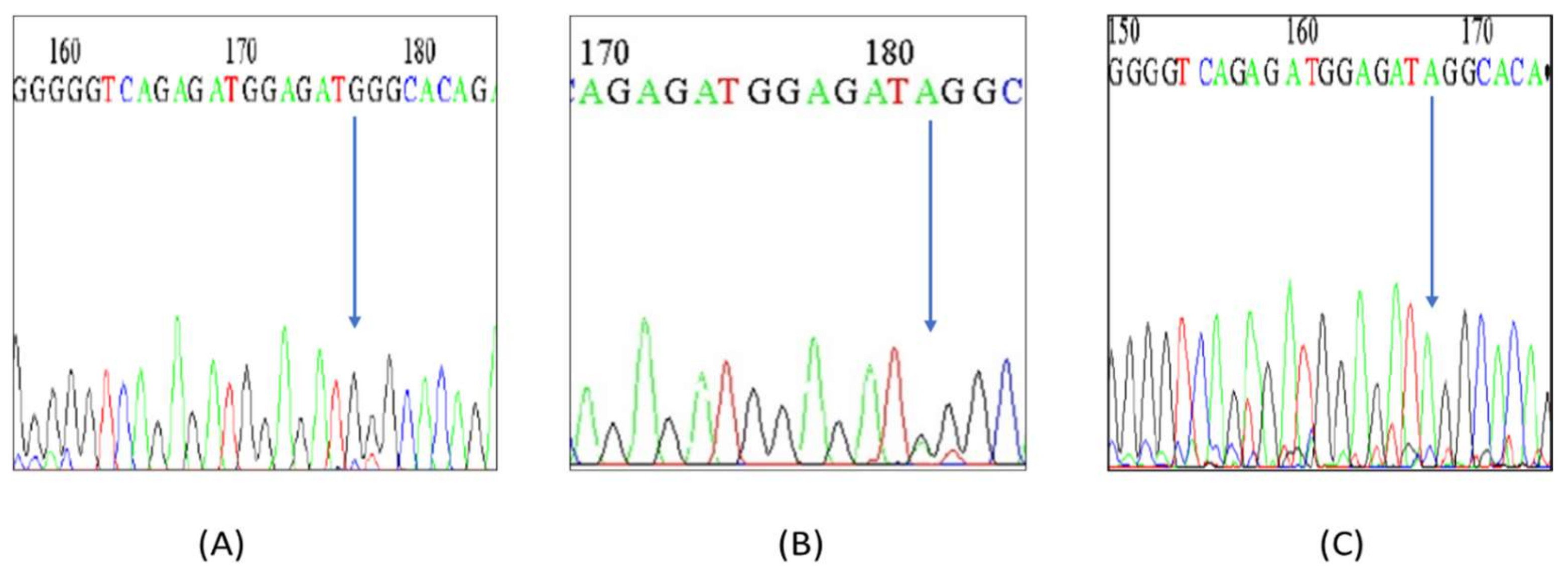
2.7. Sequencing
2.8. Expression Analysis of miR-125a by q–PCR
2.9. Ethical Compliance
2.10. Statistical Analysis
3. Results
3.1. Genotyping of miR-125a by PCR-RFLP/HRM/DNA Sequencing
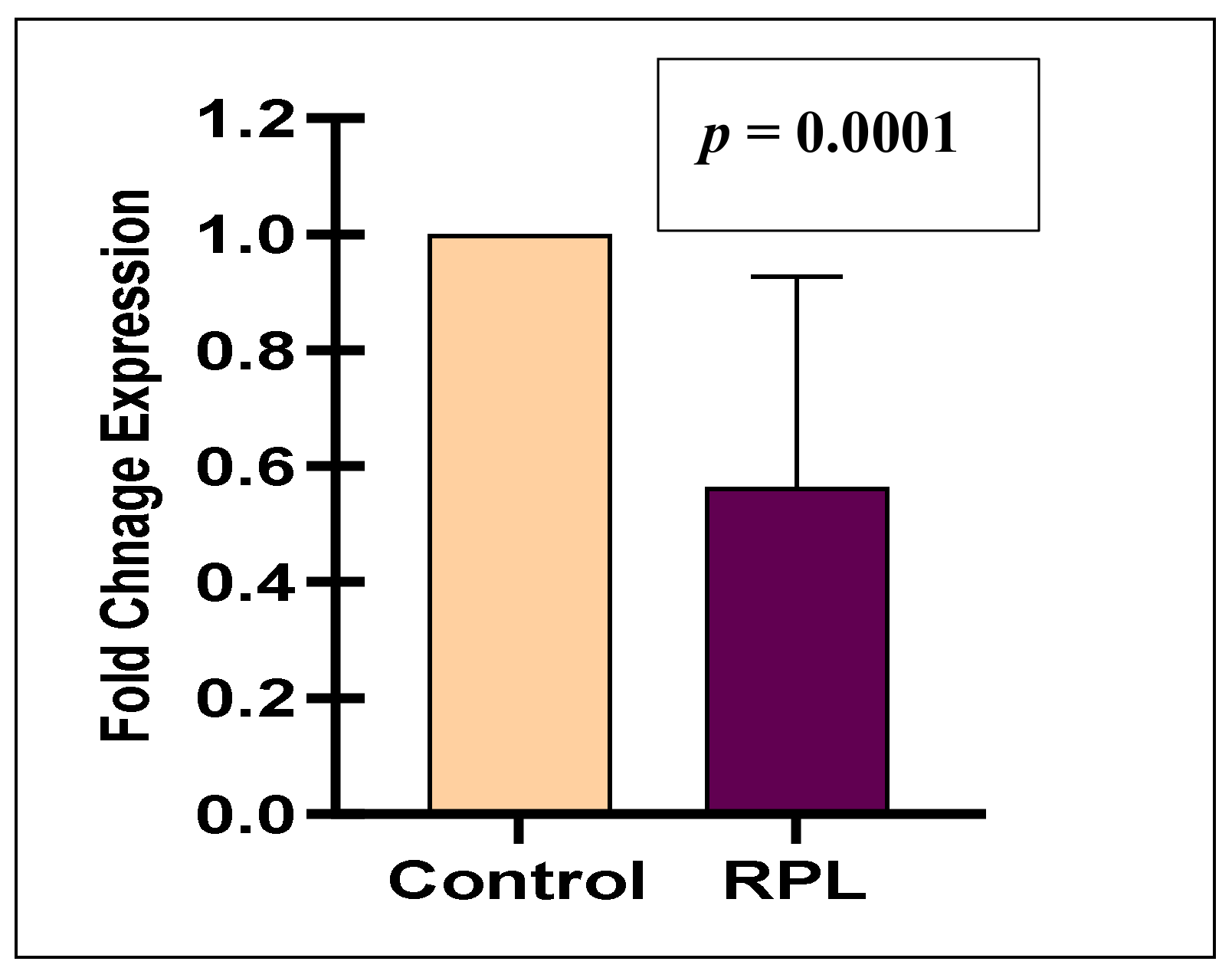
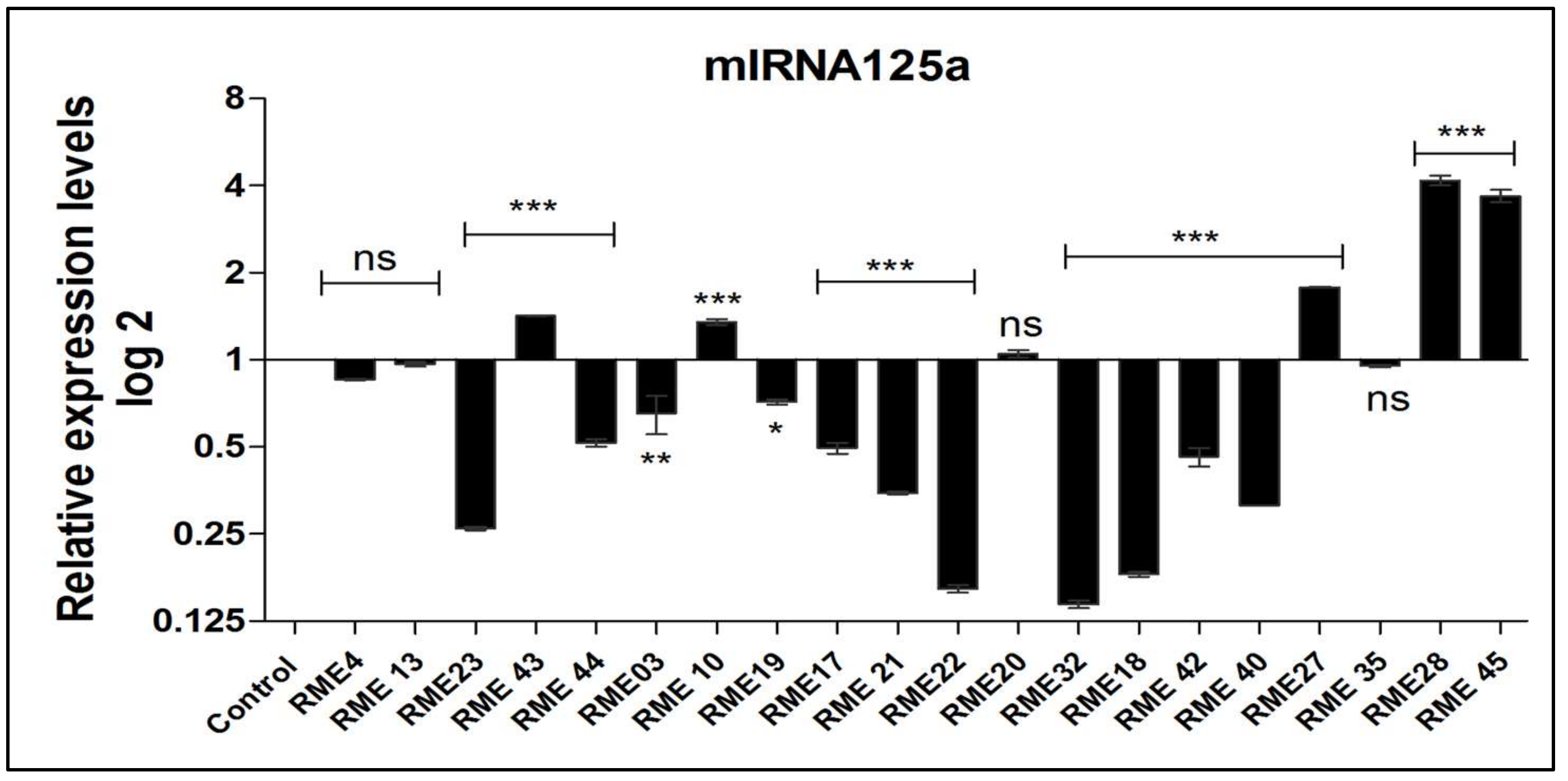
3.2. Expression Analysis of miR-125a with Respect to pri-miR-125a SNP
3.3. Multivariat Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Parazzini, F.; La Vecchia, C.; Bocciolone, L.; Franceschi, S. The epidemiology of endometrial cancer. Gynecol. Oncol. 1991, 41, 1–6. [Google Scholar] [CrossRef]
- Christiansen, O.B. Epidemiology of recurrent pregnancy loss. In Recurrent Pregnancy Loss: Causes, Controversies and Treatment; Carp, H., Ed.; Informa Healthcare: London, UK, 2007; pp. 1–13. [Google Scholar]
- Sugiura-Ogasawara, M.; Suzuki, S.; Ozaki, Y.; Katano, K.; Suzumori, N.; Kitaori, T. Frequency of recurrent spontaneous abortion and its influence on further marital relationship and illness: The Okazaki Cohort Study in Japan. J. Obstet. Gynaecol. Res. 2013, 39, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, U.; Amin, I.; Pandith, A.A.; Afroze, D.; Sanadhya, D.; Rashid, M.; Ahmad, A.; Syed, H.M.; Anwar, I.; Koul, A. Frequency of Recurrent Miscarriages in Kashmiri Population (a high incidence zone). Int. J. Health Sci. Res. 2021, 11, 272–278. [Google Scholar]
- Stephenson, M.; Kutteh, W. Evaluation and management of recurrent early pregnancy loss. Clin. Obstet. Gynecol. 2007, 50, 132–145. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.-Z. Micromanagers of gene expression: The potentially wide spread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of micro-RNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of micro RNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, C.M.; Qi, L.; He, T.Z.; Shi-Guo, L.; Hao, C.J.; Cui, Y.; Zhang, N.; Xia, H.F.; Ma, X. Two common SNPs in pri-miR- 125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 2011, 8, 861–872. [Google Scholar] [CrossRef]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef]
- Shi, X.B.; Xue, L.; Yang, J.; Ma, A.H.; Zhao, J.; Xu, M.; Tepper, C.G.; Evans, C.P.; Kung, H.J.; deVere White, R.W. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. USA 2007, 104, 19983–19988. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequentlydown-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Mitomo, S.; Maesawa, C.; Ogasawara, S.; Iwaya, T.; Shibazaki, M.; Yashima-Abo, A.; Kotani, K.; Oikawa, H.; Sakurai, E.; Izutsu, N.; et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008, 99, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; Errico, M.C.; Bottero, L.; Segnalini, P.; Stoppacciaro, A.; Biffoni, M.; Felli, N.; Mattia, G.; Petrini, M.; Colombo, M.P.; et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008, 68, 2745–2754. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J. Overexpression of the microRHAShsa-miR-200c leads toreduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007, 67, 7972–7976. [Google Scholar] [CrossRef]
- Barbarotto, E.; Schmittgen, T.D.; Calin, G.A. MicroRNAs and cancer: Profile, profile, profile. Int. J. Cancer 2008, 122, 969–977. [Google Scholar] [CrossRef]
- Yang, Y.; Li, H.; Hou, S.; Hu, B.; Liu, J.; Wang, J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS ONE 2013, 8, e65309. [Google Scholar] [CrossRef]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Agodi, A. The role of miRNAs as biomarkers for pregnancy outcomes: A comprehensive review. Int. J. Genom. 2017, 2017, 8067972. [Google Scholar] [CrossRef]
- Chakrabarty, A.; Tranguch, S.; Daikoku, T.; Jensen, K.; Furneaux, H.; Dey, S.K. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. USA 2007, 104, 15144–15149. [Google Scholar] [CrossRef]
- El-Shorafa, H.M.; Sharif, F.A. Dysregulation of micro-RNA contributes to the risk of unexplained recurrent pregnancy loss. Int. J. Reprod. Contracept. Obstet. Gynecol. 2013, 2, 350. [Google Scholar] [CrossRef]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Lu, J.; Schlanger, R.; Zhang, H.; Wang, J.Y.; Fox, M.C.; Purton, L.E.; Fleming, H.H.; Cobb, B.; Merkenschlage, M.; et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA 2010, 107, 14229–14234. [Google Scholar] [CrossRef] [PubMed]
- Leotta, M.; Biamonte, L.; Raimondi, L.; Ronchetti, D.; Di Martino, M.T.; Botta, C.; Leone, E.; Pitari, M.R.; Neri, A.; Giordano, A.; et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J. Cell Physiol. 2014, 229, 2106–2116. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Pak, C.; Jin, P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007, 16, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Watanabe, M.; Inoue, N.; Kagawa, T.; Shibutani, S.; Otsu, H.; Saeki, M.; Takuse, Y.; Hidaka, Y.; Iwatani, Y.; et al. Associations of single nucleotide polymorphisms in precursor-microRNA (miR)-125a and the expression of mature miR-125a with the development and prognosis of autoimmune thyroid diseases. Clin. Exp. Immunol. 2014, 178, 229–235. [Google Scholar] [CrossRef]
- Hu, Y.; Huo, Z.H.; Liu, C.M.; Liu, S.G.; Zhang, N.; Yin, K.L.; Qi, L.; Ma, X.; Xia, H.F. Functional study of one nucleotide mutation in pri-miR-125a coding region which related to recurrent pregnancy loss. PLoS ONE 2014, 9, e114781. [Google Scholar]
- Kuntla, S.; Goli, S.; Sekher, T.V.; Doshi, R. Consanguineous marriages and their effects on pregnancy outcomes in India. Int. J. Sociol. Soc. Policy 2013, 33, 437–452. [Google Scholar] [CrossRef]
- De Leeneer, K.; Coene, I.; Poppe, B.; De Paepe, A.; Claes, K. Rapid and sensitive detection of BRCA1/2 mutations in a diagnostic setting: Comparison of two high-resolution melting platforms. Clin. Chem. 2008, 54, 982–989. [Google Scholar] [CrossRef]
- Takano, E.A.; Mitchell, G.; Fox, S.B.; Dobrovic, A. Rapid detection of carriers with BRCA1 and BRCA2 mutations using high resolution melting analysis. BMC Cancer 2008, 8, 59. [Google Scholar] [CrossRef]
- Reed, G.H.; Wittwer, C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis 14. Clin. Chem. 2004, 50, 1748–1754. [Google Scholar] [CrossRef] [PubMed]
- Gerhardus, A.; Schleberger, H.; Schlegelberger, B.; Gadzicki, D. Diagnostic accuracy of methods for the detection of BRCA1 and BRCA2 mutations: A systematic review 2. Eur. J. Hum. Genet. 2007, 15, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Jazdzewski, K.; Murray, E.L.; Franssila, K.; Jarzab, B.; Schoenberg, D.R.; de la Chapelle, A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2008, 105, 7269–7274. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.A.; Liang, H.; Li, W.H. Human Polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, J.; Tian, T.; Zhou, X.; Gu, H.; Xu, L.; Zeng, Y.; Miao, R.; Jin, G.; Ma, H.; et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Investig. 2008, 118, 2600–2608. [Google Scholar] [CrossRef]
- Jazdzewski, K.; Liyanarachchi, S.; Swierniak, M.; Pachucki, J.; Ringel, M.D.; Jarzab, B.; de la Chapelle, A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 1502–1505. [Google Scholar] [CrossRef]
- Lehmann, T.P.; Korski, K.; Ibbs, M.; Zawierucha, P.; Grodecka-Gazdecka, S.; Jagodzinski, P.P. rs12976445 variant in thepri-miR-125a correlates with a lower level hashsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol. Lett. 2013, 5, 569–573. [Google Scholar] [CrossRef][Green Version]
- Li, G.; Ao, S.; Hou, J.; Lyu, G. Low expression of miR-125a-5p is associated with poor prognosis in patients with gastric cancer. Oncol. Lett. 2019, 18, 1483–1490. [Google Scholar] [CrossRef]
- Hosseini, M.K.; Gunel, T.; Gumusoglu, E.; Benian, A.; Aydinli, K. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol. Med. Rep. 2018, 17, 4941–4952. [Google Scholar] [CrossRef]
- Li, D.; Li, J. Association of miR-34a-3p/5p, miR-141-3p/5p, and miR-24 in decidual natural killer cells with unexplained recurrent spontaneous abortion. Med. Sci. Monit. 2016, 22, 922–929. [Google Scholar] [CrossRef]
- Kozinn, S.I.; Harty, N.J.; Delong, J.M.; Deliyiannis, C.; Logvinenko, T.; Summerhayes, I.C.; Libertino, J.A.; Holway, A.H.; Rieger-Christ, K.M. MicroRNA profile to predict gemcitabine resistance in bladder carcinoma cell lines. Genes Cancer 2013, 4, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, C.; Chen, X.; Yang, C.; Li, K.; Wang, J.; Dai, J.; Hu, Z.; Zhou, X.; Chen, L.; et al. Expression profile of microRNAs in serum: A fingerprint for esophageal squamous cell carcinoma. Clin. Chem. 2010, 56, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Bi, Q.; Tang, S.; Xia, L.; Du, R.; Fan, R.; Gao, L.; Jin, J.; Liang, S.; Chen, Z.; Xu, G.; et al. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS ONE 2012, 7, e40169. [Google Scholar]
- Nishida, N.; Yokobori, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Kuwano, H.; Mori, M. MicroRNA miR-125b is a prognostic marker in human colorectal cancer. Int. J. Oncol. 2011, 38, 1437–1443. [Google Scholar] [PubMed]
- Fassan, M.; Pizzi, M.; Realdon, S.; Balistreri, M.; Guzzardo, V.; Zagonel, V.; Castoro, C.; Mastracci, L.; Farinati, F.; Nitti, D.; et al. The HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesis. Hum. Pathol. 2013, 44, 1804–1810. [Google Scholar] [CrossRef]
- Nishida, N.; Mimori, K.; Fabbri, M.; Yokobori, T.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA- 125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin. Cancer Res. 2011, 17, 2725–2733. [Google Scholar] [CrossRef]
- Li, M.; Sendtner, M.; Smith, A. Essential function of LIF receptor in motor neurons. Nature 1995, 378, 724–727. [Google Scholar] [CrossRef]
- Ware, C.B.; Horowitz, M.C.; Renshaw, B.R.; Hunt, J.S.; Liggitt, D.; Koblar, S.A. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 1995, 121, 1283–1299. [Google Scholar] [CrossRef]
| SNP Single Nucleotide Polymorphism | Genotype | Case n = 150 (%) | Control n = 180 (%) | OR (95CI%) | p Value |
|---|---|---|---|---|---|
| rs12976445 C/T Overall Genotyping | TT CT CC | 51 (34) 70 (46.6) 29 (19.3) | 74 (41.1) 87 (48.3) 19(10.5) | Ref 1.1 (0.7–1.9) 2.2 (1.2–4.3) | Ref 0.5 0.02 |
| Dominant Model | TT CT + CC | 51 (34) 99 (66) | 74 (41.1) 106 (58.8) | Ref 1.35 (0.8–2.1) | Ref 0.21 |
| Recessive Model | CC CT + TT | 29 (18.6) 121 (81.3) | 19 (10.5) 106 (89.4) | Ref 0.7 (0.4–1.4) | Ref 0.4 |
| Additive Model | TT CC | 51 (64.5) 29 (35.4) | 74 (79.5) 19 (20.5) | Ref 2.2(1.2–4.3) | Ref 0.02 |
| Allele Frequency | T C | 172 (57.6) 128 (42.3) | 235 (65.2) 125 (34.7) | Ref 1.4 (1.0–1.9) | Ref 0.04 |
| rs10404453 A/G Overall Genotyping | GG AG AA | 150 (100) 0 0 | 180 (100) 0 0 | Ref 0.70 0.70 |
| SNP | Genotype | POC Product of Conception n = 50(%) | Control n = 180 (%) | OR (95 CI%) | p Value |
|---|---|---|---|---|---|
| rs12976445 C/T Overall Genotyping | TT CT CC | 11 (22) 31 (62) 8 (16) | 74 (41.1) 87 (48.3) 19 (10.5) | Ref 2.4 (1.1–5.09) 2.8 (1.0–8.0) | Ref 0.02 0.07 |
| Recessive model | CC CT + TT | 08 (16) 42 (84) | 19 (10.5) 161 (89.4) | Ref 0.9 (0.3–2.7) | Ref 0.9 |
| Allele Frequency | T C | 53 (53) 47 (47) | 235 (65.2) 25 (34.7) | Ref 1.7 (1.06–2.6) | Ref 0.02 |
| SNP | Case (%) | Control (%) | OR (95 CI %) | p Value | ||
|---|---|---|---|---|---|---|
| TT | CT + CC | TT | CT + CC | |||
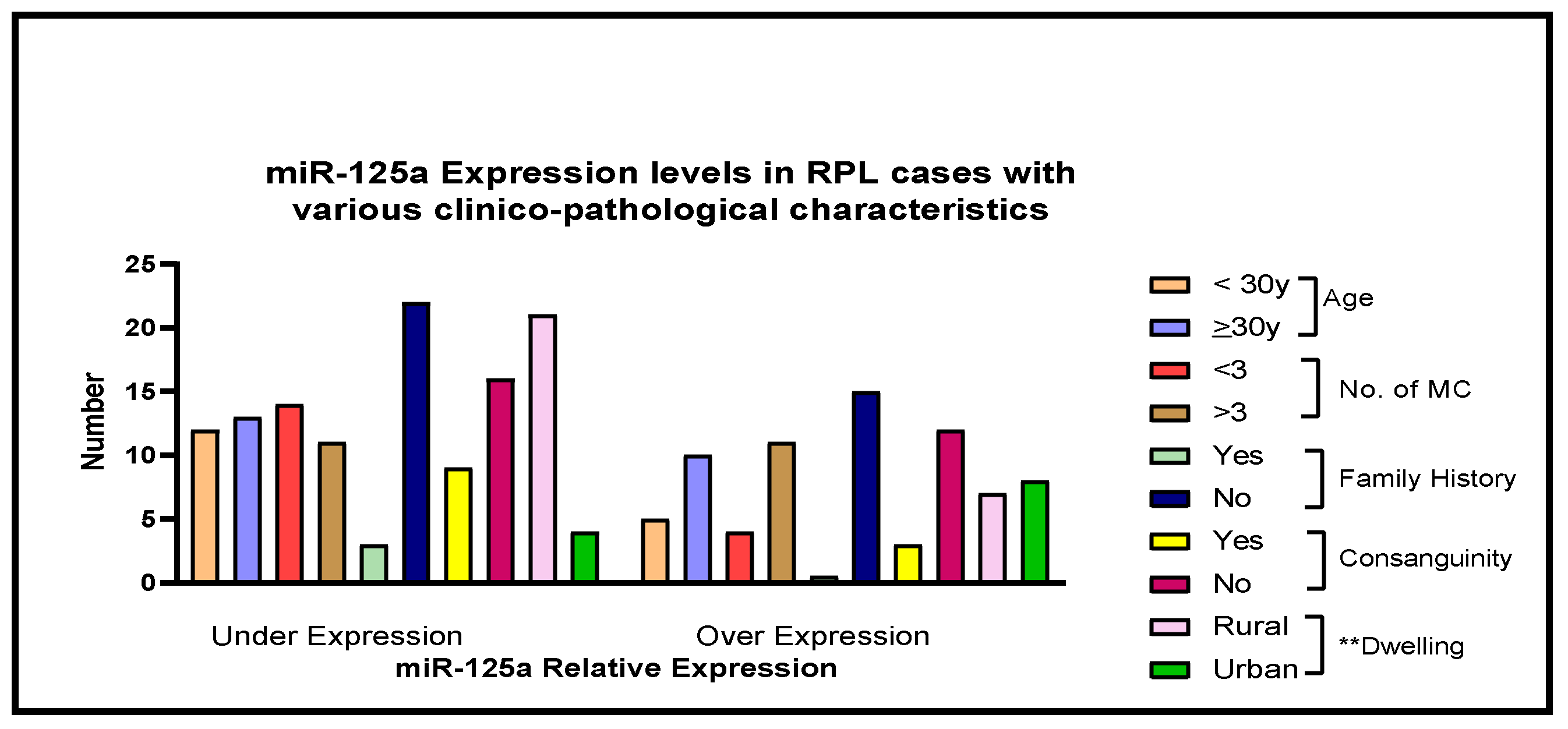
| Age <30 ≥30 | 21 (27) 30 (41) | 56 (73) 43 (59) | 32 (40) 42 (42) | 48 (60) 58 (58) | 1.79 (0.9–3.4) 1.03 (0.56–1.91) | 0.1 0.9 |
| Miscarriages <3 ≥3 | 19 (27) 32 (41) | 52 (73) 47 (59) | 0.04 * 0.09 | |||
| Family History Yes No | 10 (43) 41 (32) | 13 (57) 86 (68) | 4 (40) 70 (41) | 6 (60) 100 (59) | 0.8 (0.1–3.9) 1.4 (0.9–2.3) | 0.9 0.1 |
| Consanguinity Yes No | 09 (28) 42 (36) | 23 (72) 76 (64) | 8 (29) 66 (43) | 20 (71) 86 (57) | 1.0 (0.3–3.1) 1.3 (0.8–2.2) | 0.9 0.2 |
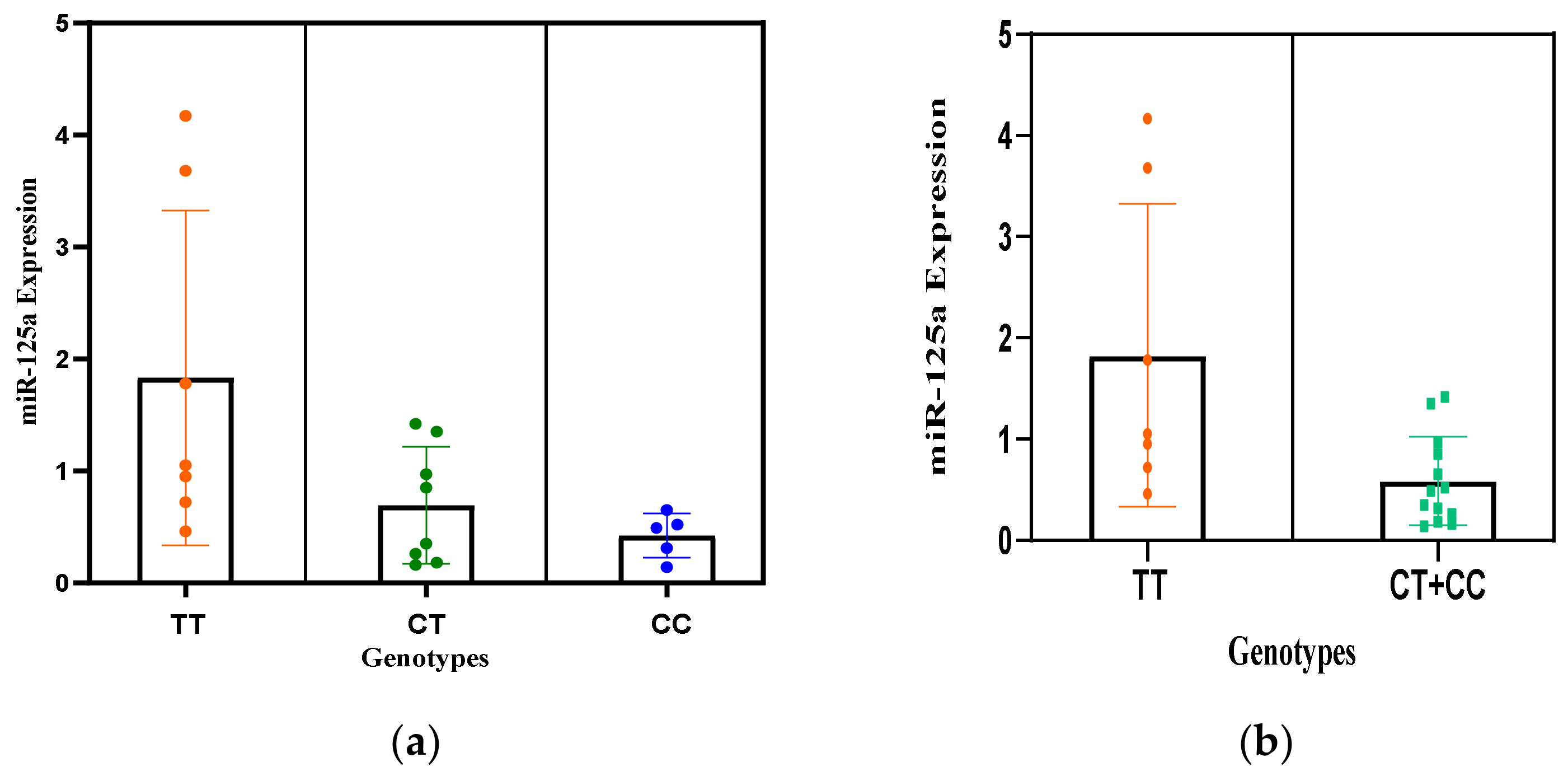
| Expression Analysis of miR-125a in Plasma Samples of RPL Cases | ||||
|---|---|---|---|---|
| Genotypes | n = 40(%) | Mean ± SD Fold Change | p Value | |
| Under | Over | |||
| rs12976445 C/T | ||||
| TT | 13(32.5) | 4(31)0.71 ± 0.24 | 9(69)2.67 ± 1.49 | Ref |
| CT | 22(55) | 16(73)0.46 ± 0.35 | 6(27)1.38 ± 0.04 | 0.03 |
| CC | 5(12.5) | 5(100)0.42 ± 0.19 | 0 | 0.02 |
| CC + CT | 27(67.5) | 21(77.7)0.44 ± 0.28 | 6(22.2)1.38 ± 0.04 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manzoor, U.; Pandith, A.A.; Amin, I.; Wani, S.; Sanadhya, D.; Lone, T.A.; Mir, H.; Paray, B.A.; Gulnaz, A.; Anwar, I.; et al. Implications of Decreased Expression of miR-125a with Respect to Its Variant Allele in the Pathogenesis of Recurrent Pregnancy Loss: A Study in a High Incidence Zone. J. Clin. Med. 2022, 11, 3834. https://doi.org/10.3390/jcm11133834
Manzoor U, Pandith AA, Amin I, Wani S, Sanadhya D, Lone TA, Mir H, Paray BA, Gulnaz A, Anwar I, et al. Implications of Decreased Expression of miR-125a with Respect to Its Variant Allele in the Pathogenesis of Recurrent Pregnancy Loss: A Study in a High Incidence Zone. Journal of Clinical Medicine. 2022; 11(13):3834. https://doi.org/10.3390/jcm11133834
Chicago/Turabian StyleManzoor, Usma, Arshad A. Pandith, Ina Amin, Saima Wani, Dheera Sanadhya, Tawseef A. Lone, Hyder Mir, Bilal Ahamad Paray, Aneela Gulnaz, Iqra Anwar, and et al. 2022. "Implications of Decreased Expression of miR-125a with Respect to Its Variant Allele in the Pathogenesis of Recurrent Pregnancy Loss: A Study in a High Incidence Zone" Journal of Clinical Medicine 11, no. 13: 3834. https://doi.org/10.3390/jcm11133834
APA StyleManzoor, U., Pandith, A. A., Amin, I., Wani, S., Sanadhya, D., Lone, T. A., Mir, H., Paray, B. A., Gulnaz, A., Anwar, I., Ahmad, A., & Aein, Q. U. (2022). Implications of Decreased Expression of miR-125a with Respect to Its Variant Allele in the Pathogenesis of Recurrent Pregnancy Loss: A Study in a High Incidence Zone. Journal of Clinical Medicine, 11(13), 3834. https://doi.org/10.3390/jcm11133834






