MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift
Abstract
1. Introduction
2. Current Insights into the Diagnosis and Management of LGGs
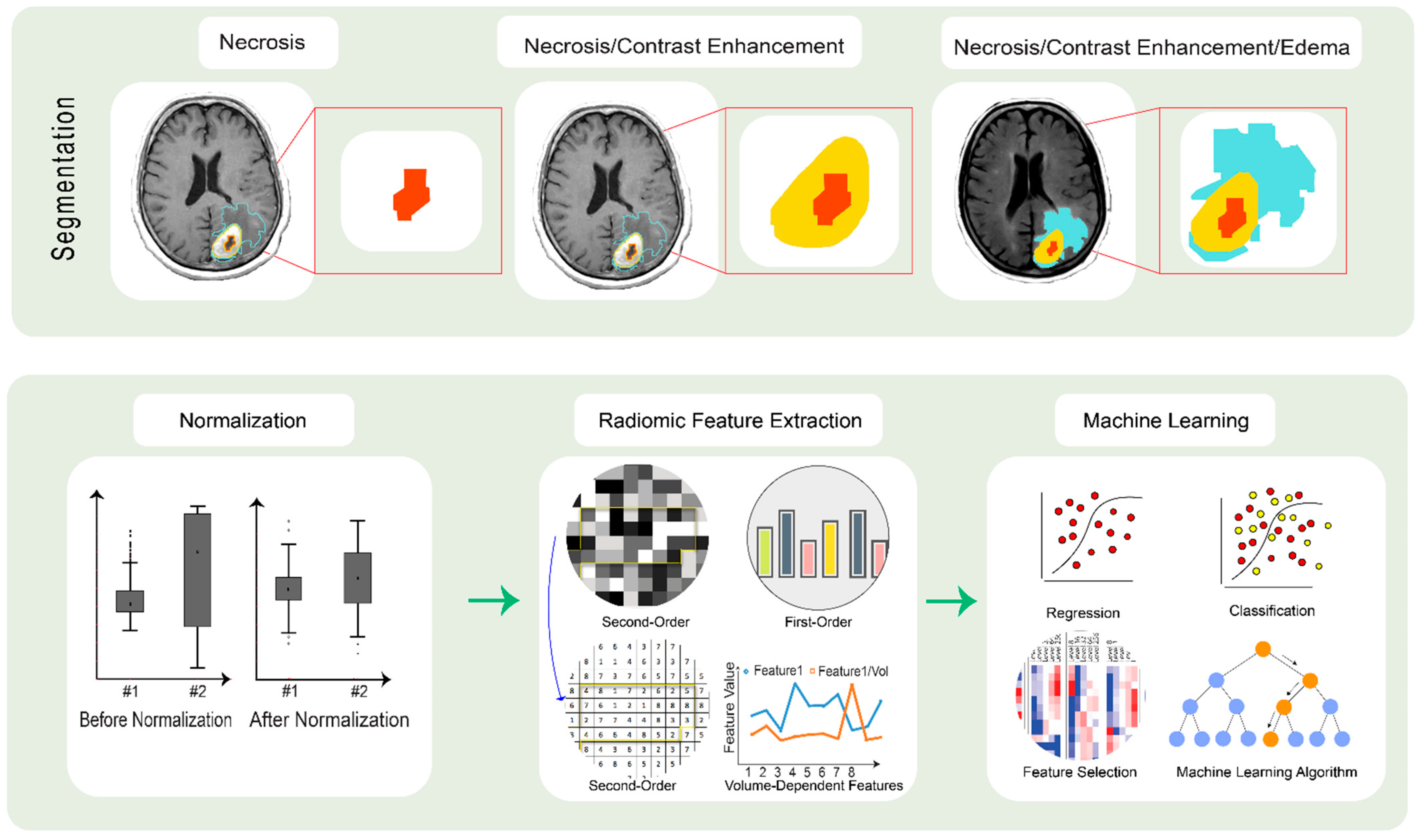
3. Implications of Radiomics in an LGG Diagnosis
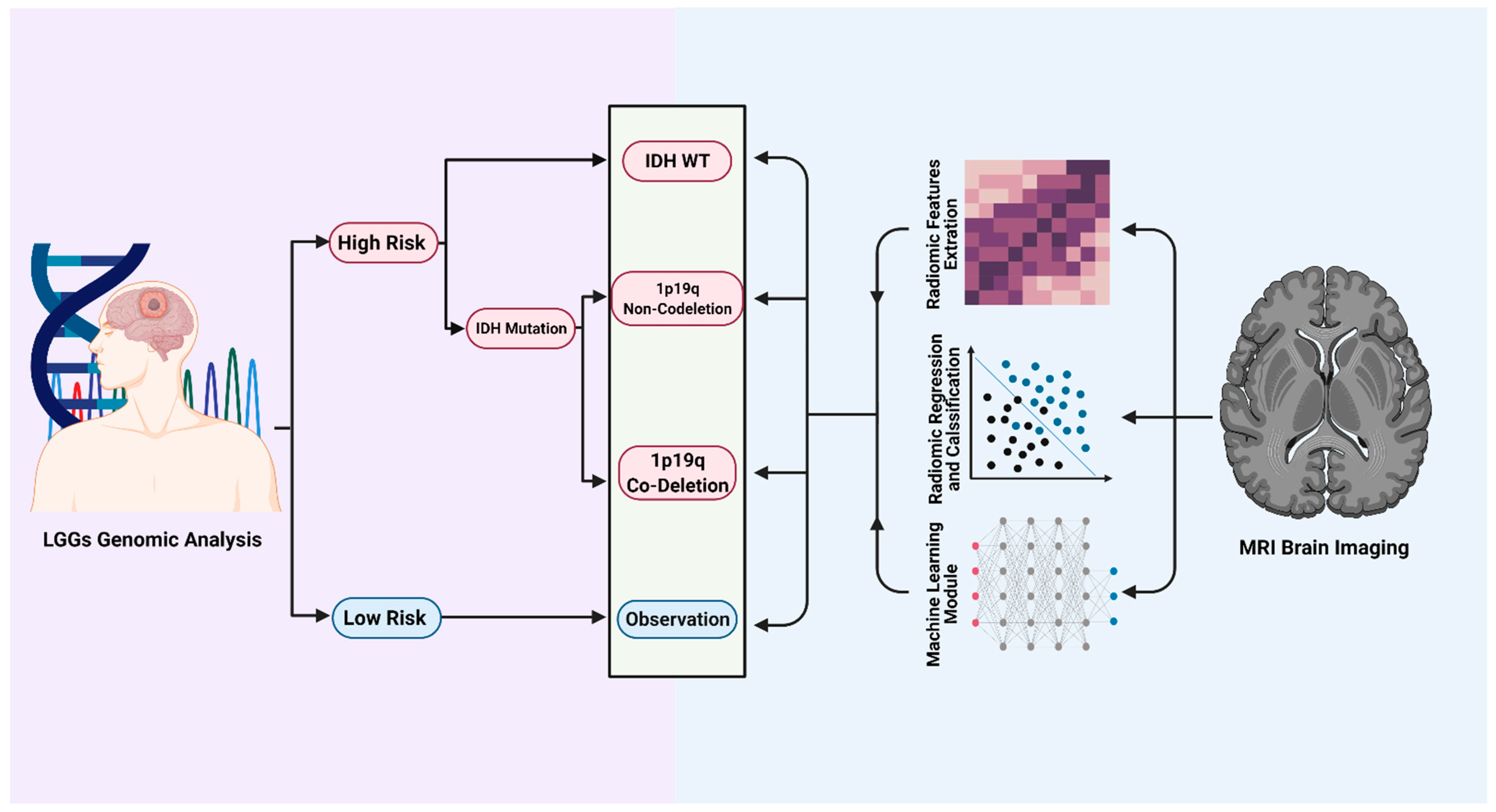
4. MRI-Based Radiomics Could Predict Molecular Markers and Overall Survival in Diffuse Lower Grade Gliomas
5. Predicting Clinical Outcomes in Patients with an LGG
6. Radiomics as a Prognostic Tool in an LGG
7. Pushing the Envelope and Future Directions for Radiomics in Neurooncology
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Forst, D.A.; Nahed, B.V.; Loeffler, J.S.; Batchelor, T.T. Low-grade gliomas. Oncologist 2014, 19, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncol. 2017, 19, v1–v88. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.; Giannini, C.; Eckel-Passow, J.; Lachance, D.; Parney, I.; Laack, N.; Jenkins, R. Management of diffuse low-grade gliomas in adults—Use of molecular diagnostics. Nat. Rev. Neurol. 2017, 13, 340–351. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Morshed, R.A.; Young, J.S.; Hervey-Jumper, S.L.; Berger, M.S. The management of low-grade gliomas in adults. J. Neurosurg. Sci. 2019, 63, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda-Sanchez, J.M.; Munoz Langa, J.; Arraez, M.A.; Fuster, J.; Hernandez Lain, A.; Reynes, G.; Rodriguez Gonzalez, V.; Vicente, E.; Vidal Denis, M.; Gallego, O. SEOM clinical guideline of diagnosis and management of low-grade glioma (2017). Clin. Transl. Oncol. 2018, 20, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Viaccoz, A.; Lekoubou, A.; Ducray, F. Chemotherapy in low-grade gliomas. Curr. Opin. Oncol. 2012, 24, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Pouratian, N.; Schiff, D. Management of low-grade glioma. Curr. Neurol. Neurosci. Rep. 2010, 10, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Tom, M.C.; Cahill, D.P.; Buckner, J.C.; Dietrich, J.; Parsons, M.W.; Yu, J.S. Management for Different Glioma Subtypes: Are All Low-Grade Gliomas Created Equal? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 133–145. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, W.; Liu, Z.; Liang, Y.; Liu, X.; Li, Y.; Tang, Z.; Jiang, T.; Tian, J. Predicting the Type of Tumor-Related Epilepsy in Patients With Low-Grade Gliomas: A Radiomics Study. Front Oncol. 2020, 10, 235. [Google Scholar] [CrossRef]
- Shofty, B.; Artzi, M.; Ben Bashat, D.; Liberman, G.; Haim, O.; Kashanian, A.; Bokstein, F.; Blumenthal, D.T.; Ram, Z.; Shahar, T. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Kinoshita, M.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; Okita, Y.; Nonaka, M.; Moriuchi, S.; et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018, 8, 11773. [Google Scholar] [CrossRef] [PubMed]
- Colen, R.R.; Ologun, G.O.; Zinn, P.; AK, M.; Arora, R.; Burton, E.M.; Glitza, I.C.; Tawbi, H.A.-H.; Patel, S.P.; Diab, A.; et al. Radiomic signatures to predict response to targeted therapy and immune checkpoint blockade in melanoma patients (pts) on neoadjuvant therapy. J. Clin. Oncol. 2020, 38, 10067. [Google Scholar] [CrossRef]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Tian, Q.; Yan, L.F.; Zhang, X.; Zhang, X.; Hu, Y.C.; Han, Y.; Liu, Z.C.; Nan, H.Y.; Sun, Q.; Sun, Y.Z.; et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Zinn, P.O.; Singh, S.K.; Kotrotsou, A.; Hassan, I.; Thomas, G.; Luedi, M.M.; Elakkad, A.; Elshafeey, N.; Idris, T.; Mosley, J.; et al. A Coclinical Radiogenomic Validation Study: Conserved Magnetic Resonance Radiomic Appearance of Periostin-Expressing Glioblastoma in Patients and Xenograft Models. Clin. Cancer Res. 2018, 24, 6288–6299. [Google Scholar] [CrossRef]
- Pouratian, N.; Asthagiri, A.; Jagannathan, J.; Shaffrey, M.E.; Schiff, D. Surgery Insight: The role of surgery in the management of low-grade gliomas. Nat. Clin. Pract. Neurol. 2007, 3, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, G.; Barresi, V.; Castellano, A.; Tabouret, E.; Pasqualetti, F.; Salvalaggio, A.; Cerretti, G.; Caccese, M.; Padovan, M.; Zagonel, V.; et al. Clinical Management of Diffuse Low-Grade Gliomas. Cancers 2020, 12, 3008. [Google Scholar] [CrossRef]
- Tabrizi, S.; Shih, H.A. The path forward for radiation therapy in the management of low-grade gliomas. Neuro-Oncol. 2020, 22, 748–749. [Google Scholar] [CrossRef]
- Smith, J.S.; Chang, E.F.; Lamborn, K.R.; Chang, S.M.; Prados, M.D.; Cha, S.; Tihan, T.; Vandenberg, S.; McDermott, M.W.; Berger, M.S. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J. Clin. Oncol. 2008, 26, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Jaeckle, K.A.; Ballman, K.V.; van den Bent, M.; Giannini, C.; Galanis, E.; Brown, P.D.; Jenkins, R.B.; Cairncross, J.G.; Wick, W.; Weller, M.; et al. CODEL: Phase III study of RT, RT + Temozolomide (TMZ), or TMZ for newly-diagnosed 1p/19q Codeleted Oligodendroglioma. Analysis from the initial study design. Neuro-Oncol. 2020. [Google Scholar] [CrossRef]
- Buckner, J.C.; Shaw, E.G.; Pugh, S.L.; Chakravarti, A.; Gilbert, M.R.; Barger, G.R.; Coons, S.; Ricci, P.; Bullard, D.; Brown, P.D.; et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N. Engl. J. Med. 2016, 374, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Mizobuchi, Y.; Nakajima, K.; Fujihara, T.; Matsuzaki, K.; Mure, H.; Nagahiro, S.; Takagi, Y. The risk of hemorrhage in stereotactic biopsy for brain tumors. J. Med. Investig. 2019, 66, 314–318. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, L.; Armocida, D.; Sampirisi, L.; Paglia, F.; Berra, L.V.; Santoro, A. Role of endoscopic surgical biopsy in diagnoses of intraventricular/periventricular tumors: Review of literature including a monocentric case series. Acta Neurol. Belg. 2020, 120, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.S.; Carter, B.S.; Chen, C.C. Role of Biopsies in the Management of Intracranial Gliomas. Prog. Neurol. Surg. 2018, 30, 232–243. [Google Scholar] [CrossRef]
- Lara-Almunia, M.; Hernández-Vicente, J. Related factors with diagnostic yield and intracranial hemorrhagic complications in frame-based stereotactic biopsy. Review. Neurocirugia 2021. [Google Scholar] [CrossRef]
- Bhandari, A.P.; Liong, R.; Koppen, J.; Murthy, S.V.; Lasocki, A. Noninvasive Determination of IDH and 1p19q Status of Lower-grade Gliomas Using MRI Radiomics: A Systematic Review. AJNR Am. J. Neuroradiol. 2021, 42, 94–101. [Google Scholar] [CrossRef]
- Leenstra, S.; Troost, D.; Hulsebos, T.J.; Bosch, D.A. Genetic versus histological grading in stereotactic biopsies. Stereotact. Funct. Neurosurg. 1994, 63, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Shofty, B.; Artzi, M.; Shtrozberg, S.; Fanizzi, C.; DiMeco, F.; Haim, O.; Peleg Hason, S.; Ram, Z.; Bashat, D.B.; Grossman, R. Virtual biopsy using MRI radiomics for prediction of BRAF status in melanoma brain metastasis. Sci. Rep. 2020, 10, 6623. [Google Scholar] [CrossRef] [PubMed]
- Tselikas, L.; Sun, R.; Ammari, S.; Dercle, L.; Yevich, S.; Hollebecque, A.; Ngo-Camus, M.; Nicotra, C.; Deutsch, E.; Deschamps, F.; et al. Role of image-guided biopsy and radiomics in the age of precision medicine. Chin. Clin. Oncol. 2019, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.H.; Lee, S.H.; Kim, J.; Park, H. Classification of the glioma grading using radiomics analysis. PeerJ 2018, 6, e5982. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Jiang, J.; Zhang, S.; Shi, J.; Xu, K.; Shen, N.; Zhang, J.; Li, L.; Zhao, L.; Qin, Y.; et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur. Radiol. 2019, 29, 1986–1996. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Morizane, A.; Doi, D.; Magotani, H.; Onoe, H.; Hayashi, T.; Mizuma, H.; Takara, S.; Takahashi, R.; Inoue, H.; et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 2017, 548, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchian, S.; Kilinc, O.; Onyewadume, L.; Tippareddy, C.; McGivney, D.; Ma, D.; Griswold, M.; Sunshine, J.; Gulani, V.; Barnholtz-Sloan, J.S.; et al. Radiomic analysis of magnetic resonance fingerprinting in adult brain tumors. Eur. J. Nucl. Med. Mol. Imaging 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, S.; Qin, Y.; Zhang, Y.; Wang, N.; Liu, H. High-order radiomics features based on T2 FLAIR MRI predict multiple glioma immunohistochemical features: A more precise and personalized gliomas management. PLoS ONE 2020, 15, e0227703. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Vallieres, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncol. 2017, 19, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Wang, K.; Li, S.W.; Wang, J.F.; Ma, J.; Jiang, T.; Dai, J.P. Patterns of Tumor Contrast Enhancement Predict the Prognosis of Anaplastic Gliomas with IDH1 Mutation. AJNR Am. J. Neuroradiol. 2015, 36, 2023–2029. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Y.; Fan, X.; Wang, J.; Li, G.; Ma, J.; Jiang, T.; Dai, J. Radiological features combined with IDH1 status for predicting the survival outcome of glioblastoma patients. Neuro-Oncol. 2016, 18, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Li, S.; Fan, X.; Sun, Z.; Yang, Z.; Wang, K.; Zhang, Z.; Jiang, T.; Liu, Y.; et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging 2019, 11, 673–696. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.H.; Poisson, L.M.; Brat, D.J.; Zhou, Y.; Cooper, L.; Snuderl, M.; Thomas, C.; Franceschi, A.M.; Griffith, B.; Flanders, A.E.; et al. T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin. Cancer Res. 2017, 23, 6078–6085. [Google Scholar] [CrossRef] [PubMed]
- Broen, M.P.G.; Smits, M.; Wijnenga, M.M.J.; Dubbink, H.J.; Anten, M.H.M.E.; Schijns, O.E.M.G.; Beckervordersandforth, J.; Postma, A.A.; van den Bent, M.J. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: A validation study. Neuro-Oncol. 2018, 20, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Lasocki, A.; Gaillard, F.; Gorelik, A.; Gonzales, M. MRI Features Can Predict 1p/19q Status in Intracranial Gliomas. AJNR Am. J. Neuroradiol. 2018, 39, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Tay, K.L.; Tsui, A.; Phal, P.M.; Drummond, K.J.; Tress, B.M. MR imaging characteristics of protoplasmic astrocytomas. Neuroradiology 2011, 53, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Hanzély, Z.; Polgár, C.; Fodor, J.; Brucher, J.M.; Vitanovics, D.; Mangel, L.C.; Afra, D. Role of early radiotherapy in the treatment of supratentorial WHO Grade II astrocytomas: Long-term results of 97 patients. J. Neurooncol. 2003, 63, 305–312. [Google Scholar] [CrossRef] [PubMed]
- van den Bent, M.J.; Afra, D.; de Witte, O.; Ben Hassel, M.; Schraub, S.; Hoang-Xuan, K.; Malmström, P.O.; Collette, L.; Piérart, M.; Mirimanoff, R.; et al. Long-term efficacy of early versus delayed radiotherapy for low-grade astrocytoma and oligodendroglioma in adults: The EORTC 22845 randomised trial. Lancet 2005, 366, 985–990. [Google Scholar] [CrossRef]
- Dhawan, S.; Patil, C.G.; Chen, C.; Venteicher, A.S. Early versus delayed postoperative radiotherapy for treatment of low-grade gliomas. Cochrane Database Syst. Rev. 2020, 1, CD009229. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yang, G.; Hao, X.; Gu, D.; Tan, Y.; Wang, X.; Dong, D.; Zhang, S.; Wang, L.; Zhang, H.; et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur. Radiol. 2019, 29, 877–888. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Zhang, J.; Xue, H.; Wang, L.; Jing, R.; Chen, S.; Che, F.; Heng, X.; Li, G.; et al. An MRI-based radiomics signature as a pretreatment noninvasive predictor of overall survival and chemotherapeutic benefits in lower-grade gliomas. Eur. Radiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Jain, R.; Lee, S.-K. Machine learning and radiomic phenotyping of lower grade gliomas: Improving survival prediction. Eur. Radiol. 2020, 30, 3834–3842. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Muragaki, Y.; Maruyama, T.; Komori, T.; Tamura, M.; Nitta, M.; Tsuzuki, S.; Kawamata, T. Calcification on CT is a simple and valuable preoperative indicator of 1p/19q loss of heterozygosity in supratentorial brain tumors that are suspected grade II and III gliomas. Brain Tumor. Pathol. 2016, 33, 175–182. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Liu, X.; Du, Y.; Tang, Z.; Wang, K.; Wei, J.; Dong, D.; Zang, Y.; Dai, J.; et al. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. Neuroimage Clin. 2018, 19, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Li, Y.; Sun, Z.; Fan, X.; Xu, K.; Wang, K.; Li, S.; Zhang, Z.; Jiang, T.; Liu, X.; et al. Radiogenomics of lower-grade gliomas: A radiomic signature as a biological surrogate for survival prediction. Aging 2018, 10, 2884–2899. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, Y.; Liu, X.; Liang, Y.; Fan, Z.; Jiang, T.; Wang, Y.; Wang, L. Molecular profiles for insular low-grade gliomas with putamen involvement. J. Neurooncol. 2018, 138, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Young, R.J.; Babb, J.S.; Peccerelli, N.; Chheang, S.; Gruber, M.L.; Miller, D.C.; Golfinos, J.G.; Zagzag, D.; Johnson, G. Gliomas: Predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008, 247, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Galldiks, N.; Zadeh, G.; Lohmann, P. Artificial Intelligence, Radiomics, and Deep Learning in Neuro-Oncology. Neurooncol. Adv. 2020, 2, iv1–iv2. [Google Scholar] [CrossRef] [PubMed]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Radiomics for Gleason Score Detection through Deep Learning. Sensors 2020, 20, 5411. [Google Scholar] [CrossRef]
- Parekh, V.S.; Jacobs, M.A. Deep learning and radiomics in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, K.; Zhao, J.; Liu, L.; Feng, J. FOXO4 expression is associated with the occurrence and outcome of seizures: An RNA-sequencing analysis of low-grade gliomas. Seizure 2017, 52, 41–45. [Google Scholar] [CrossRef]
- Park, C.J.; Choi, Y.S.; Park, Y.W.; Ahn, S.S.; Kang, S.G.; Chang, J.H.; Kim, S.H.; Lee, S.K. Diffusion tensor imaging radiomics in lower-grade glioma: Improving subtyping of isocitrate dehydrogenase mutation status. Neuroradiology 2020, 62, 319–326. [Google Scholar] [CrossRef]
- Wang, T.; Yang, Y.; Xu, X.; Niu, X.; Yang, R.; Gao, T.; Kong, L.; Mao, Q.; Qiu, Y. An Integrative Survival Analysis for Multicentric Low-Grade Glioma. World Neurosurg. 2020, 134, e189–e195. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, R.; Dictus, C.; Hartmann, C.; Zurn, O.; Edler, L.; Hartmann, M.; Combs, S.; Herold-Mende, C.; Wirtz, C.R.; Unterberg, A. Long-term outcome and survival of surgically treated supratentorial low-grade glioma in adult patients. Acta Neurochir. 2009, 151, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habib, A.; Jovanovich, N.; Hoppe, M.; Ak, M.; Mamindla, P.; R. Colen, R.; Zinn, P.O. MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. J. Clin. Med. 2021, 10, 1411. https://doi.org/10.3390/jcm10071411
Habib A, Jovanovich N, Hoppe M, Ak M, Mamindla P, R. Colen R, Zinn PO. MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. Journal of Clinical Medicine. 2021; 10(7):1411. https://doi.org/10.3390/jcm10071411
Chicago/Turabian StyleHabib, Ahmed, Nicolina Jovanovich, Meagan Hoppe, Murat Ak, Priyadarshini Mamindla, Rivka R. Colen, and Pascal O. Zinn. 2021. "MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift" Journal of Clinical Medicine 10, no. 7: 1411. https://doi.org/10.3390/jcm10071411
APA StyleHabib, A., Jovanovich, N., Hoppe, M., Ak, M., Mamindla, P., R. Colen, R., & Zinn, P. O. (2021). MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. Journal of Clinical Medicine, 10(7), 1411. https://doi.org/10.3390/jcm10071411





