Differential Expression of the Sphingolipid Pathway Is Associated with Sensitivity to the PP2A Activator FTY720 in Colorectal Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. RNA Extraction
2.3. Gene Expression Assay
2.4. FTY720 Preparation
2.5. Cell Viability Assays
2.6. RNA Sequencing
2.7. Statistical Analysis
3. Results
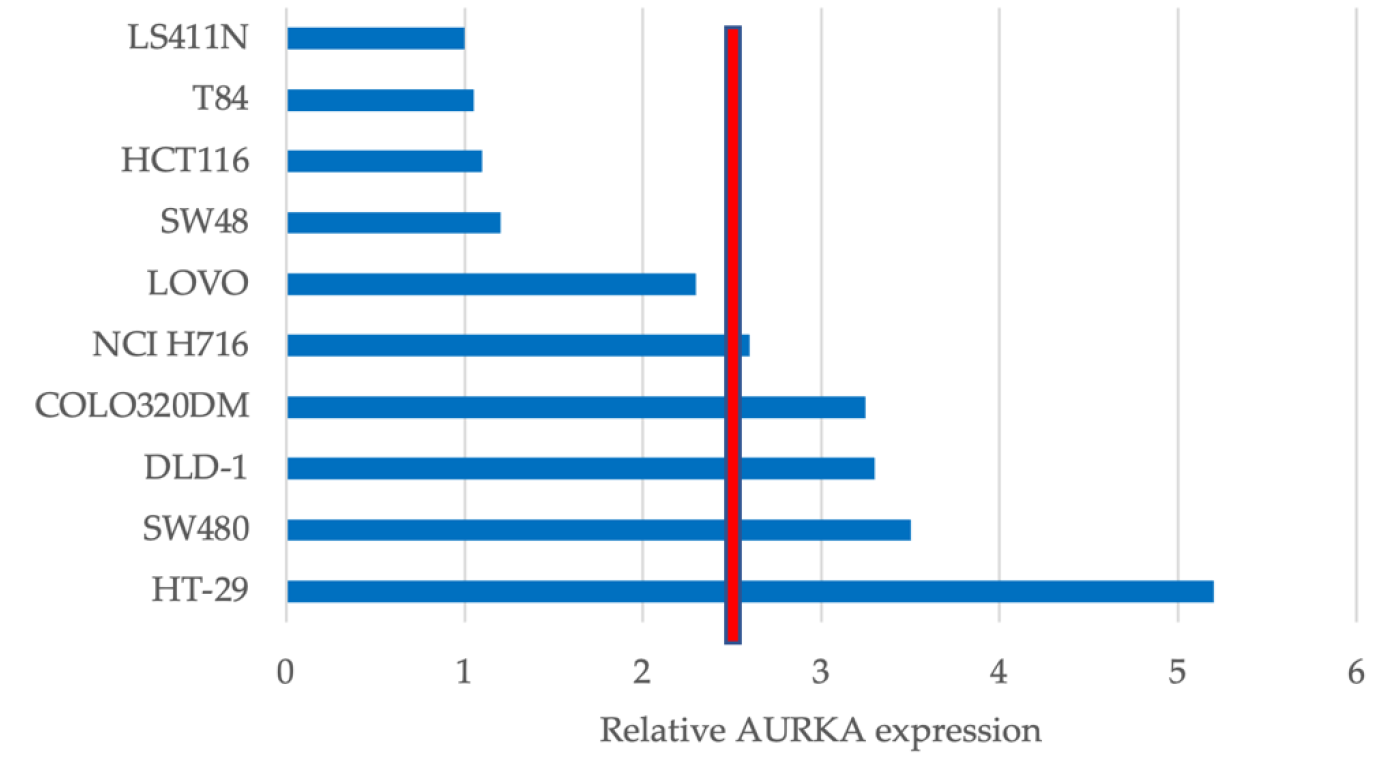
3.1. AURKA Is Differentially Expressed in CRC Cell Lines
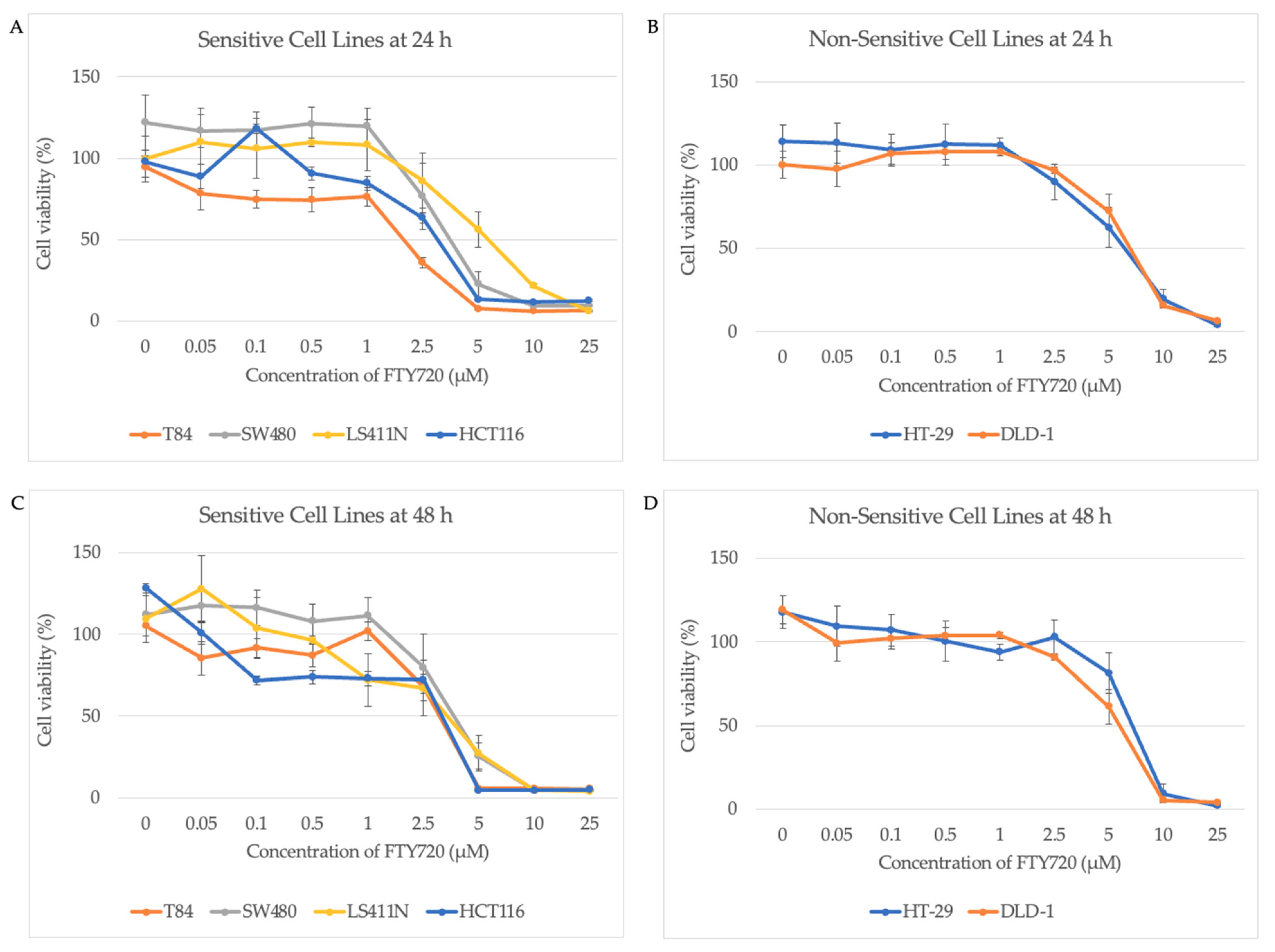
3.2. AURKA Expression Does Not Predict Sensitivity to FTY720
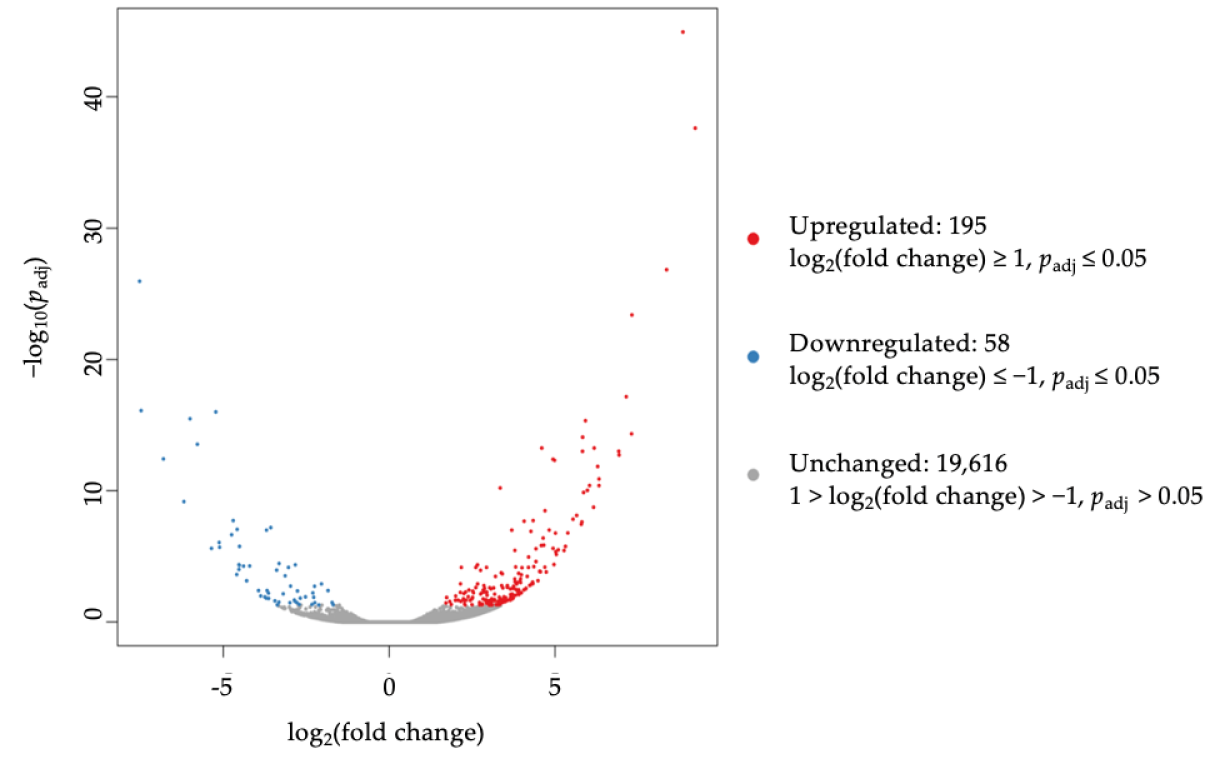
3.3. Differential Expression of Sphinogolipid Pathway Genes Is Enriched in FTY-Sensitive CRC Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Wong, M.C.; Huang, J.; Lok, V.; Wang, J.; Fung, F.; Ding, H.; Zheng, Z.-J. Differences in Incidence and Mortality Trends of Colorectal Cancer Worldwide Based on Sex, Age, and Anatomic Location. Clin. Gastroenterol. Hepatol. 2021, 19, 955–966.e61. [Google Scholar] [CrossRef]
- Sharma, R. An examination of colorectal cancer burden by socioeconomic status: Evidence from GLOBOCAN 2018. EPMA J. 2020, 11, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Grech, G.; Zhan, X.; Yoo, B.C.; Bubnov, R.; Hagan, S.; Danesi, R.; Vittadini, G.; Desiderio, D.M. EPMA position paper in cancer: Current overview and future perspectives. EPMA J. 2015, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Yeo, S.-G. Clinical utility of pretreatment prediction of chemoradiotherapy response in rectal cancer: A review. EPMA J. 2017, 8, 61–67. [Google Scholar] [CrossRef]
- Alyabsi, M.; Alhumaid, A.; Allah-Bakhsh, H.; Alkelya, M.; Aziz, M.A. Colorectal cancer in Saudi Arabia as the proof-of-principle model for implementing strategies of predictive, preventive, and personalized medicine in healthcare. EPMA J. 2020, 11, 119–131. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; De Vriendt, V.; Normanno, N.; Ciardiello, F.; Tejpar, S. KRAS, BRAF, PIK3CA, and PTEN mutations: Implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011, 12, 594–603. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Gustavsson, B.; Carlsson, G.; Machover, D.; Petrelli, N.; Roth, A.; Schmoll, H.-J.; Tveit, K.-M.; Gibson, F. A Review of the Evolution of Systemic Chemotherapy in the Management of Colorectal Cancer. Clin. Color. Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef]
- Molinari, C.; Marisi, G.; Passardi, A.; Matteucci, L.; De Maio, G.; Ulivi, P. Heterogeneity in Colorectal Cancer: A Challenge for Personalized Medicine? Int. J. Mol. Sci. 2018, 19, 3733. [Google Scholar] [CrossRef]
- Hammond, W.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef] [PubMed]
- Linnekamp, J.F.; Hooff, S.R.V.; Prasetyanti, P.R.; Kandimalla, R.; Buikhuisen, J.Y.; Fessler, E.; Ramesh, P.; Lee, K.; Bochove, G.G.W.; de Jong, J.H.; et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 2018, 25, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Bruun, J.; Eide, P.W.; Eilertsen, I.A.; Ramirez, L.; Murumägi, A.; Arjama, M.A.; Danielsen, S.A.; Kryeziu, K.; Elez, E.; et al. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin. Cancer Res. 2018, 24, 794–806. [Google Scholar] [CrossRef]
- Briffa, R.; Grech, G. Molecular classification of colorectal cancer. J. Malta Coll. Pharm. Pract. 2016, 22, 35–40. [Google Scholar]
- Crigna, A.T.; Samec, M.; Koklesova, L.; Liskova, A.; Giordano, F.A.; Kubatka, P.; Golubnitschaja, O. Cell-free nucleic acid patterns in disease prediction and monitoring—hype or hope? EPMA J. 2020, 11, 603–627. [Google Scholar] [CrossRef]
- Mihajlovic, M.; Ninic, A.; Sopic, M.; Miljkovic, M.; Stefanovic, A.; Vekic, J.; Spasojevic-Kalimanovska, V.; Zeljkovic, D.; Trifunovic, B.; Stjepanovic, Z.; et al. Association among resistin, adenylate cyclase-associated protein 1 and high-density lipoprotein cholesterol in patients with colorectal cancer: A multi-marker approach, as a hallmark of innovative predictive, preventive, and personalized medicine. EPMA J. 2019, 10, 307–316. [Google Scholar] [CrossRef]
- Walter, P.; Kristi, K.; Silvia, S.; Jasmin, H.; Marko, B.; Thomas, B.H.; Andreas, W. Multiplexed DNA Methylation Analysis in Colorectal Cancer Using Liquid Biopsy and Its Diagnostic and Predictive Value. Curr. Issues Mol. Biol. 2021, 43, 1419–1435. [Google Scholar]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Grech, G.; Baldacchino, S.; Saliba, C.; Grixti, M.P.; Gauci, R.; Petroni, V.; Fenech, A.; Scerri, C. Deregulation of the protein phosphatase 2A, PP2A in cancer: Complexity and therapeutic options. Tumor Biol. 2016, 37, 11691–11700. [Google Scholar] [CrossRef]
- Cristóbal, I.; Manso, R.; Rincón, R.; Caramés, C.; Senin, C.; Borrero, A.; Martínez-Useros, J.; Rodriguez, M.; Zazo, S.; Aguilera, O.; et al. PP2A Inhibition Is a Common Event in Colorectal Cancer and Its Restoration Using FTY720 Shows Promising Therapeutic Potential. Mol. Cancer Ther. 2014, 13, 938–947. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Z.; Jiang, C.; Ding, Y. PP2A-Mediated Anticancer Therapy. Gastroenterol. Res. Pract. 2013, 2013, 675429. [Google Scholar] [CrossRef]
- Fujita, T.; Inoue, K.; Yamamoto, S.; Ikumoto, T.; Sasaki, S.; Toyama, R.; Chiba, K.; Hoshino, Y.; Okumoto, T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J. Antibiot. 1994, 47, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-H.; Hla, T.; Ferrer, F. FTY720 inhibits tumor growth and enhances the tumor-suppressive effect of topotecan in neuroblastoma by interfering with the sphingolipid signaling pathway. Pediatr. Blood Cancer 2013, 60, 1418–1423. [Google Scholar] [CrossRef] [PubMed]
- Pitman, M.; Woodcock, J.; Lopez, A.F.; Pitson, S. Molecular Targets of FTY720 (Fingolimod). Curr. Mol. Med. 2012, 12, 1207–1219. [Google Scholar] [CrossRef]
- White, C.; Alshaker, H.; Cooper, C.; Winkler, M.; Pchejetski, D. The emerging role of FTY720 (Fingolimod) in cancer treatment. Oncotarget 2016, 7, 23106–23127. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, Y.; Otsuki, K.; Fujita, T.; Uesato, S. Effects of Phosphorylation of Immunomodulatory Agent FTY720 (Fingolimod) on Antiproliferative Activity against Breast and Colon Cancer Cells. Biol. Pharm. Bull. 2008, 31, 1177–1181. [Google Scholar] [CrossRef]
- Li, J.; Wang, S.-W.; Zhang, D.-S.; Sun, Y.; Zhu, C.-Y.; Fei, Q.; Hu, J.; Zhang, C.; Sun, Y.-M. FTY720-induced enhancement of autophagy protects cells from FTY720 cytotoxicity in colorectal cancer. Oncol. Rep. 2016, 35, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, H.-D.; Ji, X.-J.; Cong, Z.-X.; Zhu, J.-H.; Zhou, Y. FTY720 for cancer therapy (Review). Oncol. Rep. 2013, 30, 2571–2578. [Google Scholar] [CrossRef]
- Cristóbal, I.; Rincón, R.; Manso, R.; Caramés, C.; Zazo, S.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Deregulation of the PP2A Inhibitor SET Shows Promising Therapeutic Implications and Determines Poor Clinical Outcome in Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2015, 21, 347–356. [Google Scholar] [CrossRef]
- Baldacchino, S.; Saliba, C.; Scerri, J.; Scerri, C.; Grech, G. Optimization of a Multiplex RNA-based Expression Assay Using Breast Cancer Archival Material. J. Vis. Exp. 2018, 138, e57148. [Google Scholar] [CrossRef] [PubMed]
- Baldacchino, S.; Saliba, C.; Petroni, V.; Fenech, A.G.; Borg, N.; Grech, G. Deregulation of the phosphatase, PP2A is a common event in breast cancer, predicting sensitivity to FTY720. EPMA J. 2014, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Morad, S.A.F.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, P.P.; Deng, X.; Ito, T.; Carr, B.K.; May, W.S. Ceramide Induces Bcl2 Dephosphorylation via a Mechanism Involving Mitochondrial PP2A. J. Biol. Chem. 1999, 274, 20296–20300. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Park, W.; Min, H.; Kwon, T.; Baek, S.; Hwang, I.; Kim, S.; Park, J. Altered mRNA expression levels of the major components of sphingolipid metabolism, ceramide synthases and their clinical implication in colorectal cancer. Oncol. Rep. 2018, 40, 3489–3500. [Google Scholar] [CrossRef]
- Liang, J.; Nagahashi, M.; Kim, E.Y.; Harikumar, K.; Yamada, A.; Huang, W.-C.; Hait, N.C.; Allegood, J.C.; Price, M.M.; Avni, D.; et al. Sphingosine-1-Phosphate Links Persistent STAT3 Activation, Chronic Intestinal Inflammation, and Development of Colitis-Associated Cancer. Cancer Cell 2013, 23, 107–120. [Google Scholar] [CrossRef]
- Mandala, S.; Hajdu, R.; Bergstrom, J.; Quackenbush, E.; Xie, J.; Milligan, J.; Thornton, R.; Shei, G.-J.; Card, D.; Keohane, C.; et al. Alteration of Lymphocyte Trafficking by Sphingosine-1-Phosphate Receptor Agonists. Science 2002, 296, 346–349. [Google Scholar] [CrossRef]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the sphingolipid rheostat: Evolving concepts in cancer therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef]
- Pyne, S.; Bittman, R.; Pyne, N. Sphingosine Kinase Inhibitors and Cancer: Seeking the Golden Sword of Hercules. Cancer Res. 2011, 71, 6576–6582. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Su, Y.-J.; Qin, M.-B.; Mao, Y.-B.; Huang, J.-A.; Tang, G.-D. Sphingosine kinase 1 promotes tumor progression and confers malignancy phenotypes of colon cancer by regulating the focal adhesion kinase pathway and adhesion molecules. Int. J. Oncol. 2012, 42, 617–626. [Google Scholar] [CrossRef]
- Liu, S.; Xu, C.; Wu, W.; Fu, Z.; He, S.; Qin, M.; Huang, J. Sphingosine kinase 1 promotes the metastasis of colorectal cancer by inducing the epithelial-mesenchymal transition mediated by the FAK/AKT/MMPs axis. Int. J. Oncol. 2018, 54, 41–52. [Google Scholar] [CrossRef]
- Bae, G.; Do, S.-I.; Kim, K.; Park, J.H.; Cho, S.; Kim, H.-S. Increased Sphingosine Kinase 1 Expression Predicts Distant Metastasis and Poor Outcome in Patients With Colorectal Cancer. Anticancer Res. 2019, 39, 663–670. [Google Scholar] [CrossRef]
- Kawamori, T.; Kaneshiro, T.; Okumura, M.; Maalouf, S.; Uflacker, A.; Bielawski, J.; Hannun, Y.A.; Obeid, L. Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J. 2008, 23, 405–414. [Google Scholar] [CrossRef]
- Shida, D.; Inoue, S.; Yoshida, Y.; Kodaka, A.; Tsuji, T.; Tsuiji, M. Sphingosine kinase 1 is upregulated with lysophosphatidic acid receptor 2 in human colorectal cancer. World J. Gastroenterol. 2016, 22, 2503–2511. [Google Scholar] [CrossRef]
- Tan, S.S.L.; Khin, L.W.; Wong, L.; Yan, B.; Ong, C.W.; Datta, A.; Salto-Tellez, M.; Lam, Y.; Yap, C.T. Sphingosine Kinase 1 Promotes Malignant Progression in Colon Cancer and Independently Predicts Survival of Patients With Colon Cancer by Competing Risk Approach in South Asian Population. Clin. Transl. Gastroenterol. 2014, 5, e51. [Google Scholar] [CrossRef]
- Rosa, R.; Marciano, R.; Malapelle, U.; Formisano, L.; Nappi, L.; D’Amato, C.; D’Amato, V.; Damiano, V.; Marfè, G.; Del Vecchio, S.; et al. Sphingosine Kinase 1 Overexpression Contributes to Cetuximab Resistance in Human Colorectal Cancer Models. Clin. Cancer Res. 2013, 19, 138–147. [Google Scholar] [CrossRef]
- Paugh, B.S.; Paugh, S.W.; Bryan, L.; Kapitonov, D.; Wilczynska, K.M.; Gopalan, S.M.; Rokita, H.; Milstien, S.; Spiegel, S.; Kordula, T. EGF regulates plasminogen activator inhibitor-1 (PAI-1) by a pathway involving c-Src, PKCdelta, and sphingosine kinase 1 in glioblastoma cells. FASEB J. 2008, 22, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Shida, D.; Fang, X.; Kordula, T.; Takabe, K.; Lépine, S.; Alvarez, S.E.; Milstien, S.; Spiegel, S. Cross-talk between LPA1 and Epidermal Growth Factor Receptors Mediates Up-regulation of Sphingosine Kinase 1 to Promote Gastric Cancer Cell Motility and Invasion. Cancer Res. 2008, 68, 6569–6577. [Google Scholar] [CrossRef] [PubMed]
- Sukocheva, O.; Wadham, C.; Holmes, A.; Albanese, N.; Verrier, E.; Feng, F.; Bernal, A.; Derian, C.K.; Ullrich, A.; Vadas, M.A.; et al. Estrogen transactivates EGFR via the sphingosine 1-phosphate receptor Edg-3: The role of sphingosine kinase-1. J. Cell Biol. 2006, 173, 301–310. [Google Scholar] [CrossRef]
- Gu, X.; Jiang, Y.; Xue, W.; Song, C.; Wang, Y.; Liu, Y.; Cui, B. SPNS 2 promotes the malignancy of colorectal cancer cells via regulating Akt and ERK pathway. Clin. Exp. Pharmacol. Physiol. 2019, 46, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Zuo, Z.; Hao, C.; Ma, Y. Sphingosine kinase 2 promotes colorectal cancer cell proliferation and invasion by enhancing MYC expression. Tumor Biol. 2016, 37, 8455–8460. [Google Scholar] [CrossRef]
- Xiao, M.; Liu, Y.; Zou, F. Sensitization of human colon cancer cells to sodium butyrate-induced apoptosis by modulation of sphingosine kinase 2 and protein kinase D. Exp. Cell Res. 2012, 318, 43–52. [Google Scholar] [CrossRef]
- Xiao, M.; Liu, Y.G.; Zou, M.C.; Zou, F. Sodium butyrate induces apoptosis of human colon cancer cells by modulating ERK and sphingosine kinase 2. Biomed. Environ. Sci. 2014, 27, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Degagné, E.; Pandurangan, A.; Bandhuvula, P.; Kumar, A.; Eltanawy, A.; Zhang, M.; Yoshinaga, Y.; Nefedov, M.; De Jong, P.J.; Fong, L.G.; et al. Sphingosine-1-phosphate lyase downregulation promotes colon carcinogenesis through STAT3-activated microRNAs. J. Clin. Investig. 2014, 124, 5368–5384. [Google Scholar] [CrossRef] [PubMed]
- Oskouian, B.; Saba, J. Sphingosine-1-Phosphate Metabolism and Intestinal Tumorigenesis: Lipid Signaling Strikes Again. Cell Cycle 2007, 6, 522–527. [Google Scholar] [CrossRef]
- Qin, L.; Liu, S.; Qin, M.; Wu, W.; Qin, N.; Fu, Z.; Xu, C.; Huang, J.; Lai, M. Down-regulation of sphingosine kinase 1 (SphK1) enhances the chemosensitivity to cisplatin in human colon cancer RKO cells. Chin. J. Cell. Mol. Immunol. 2017, 33, 623–629. [Google Scholar]
- Ju, T.; Gao, D.; Fang, Z.-Y. Targeting colorectal cancer cells by a novel sphingosine kinase 1 inhibitor PF-543. Biochem. Biophys. Res. Commun. 2016, 470, 728–734. [Google Scholar] [CrossRef]
- Xun, C.; Chen, M.-B.; Qi, L.; Tie-Ning, Z.; Peng, X.; Ning, L.; Zhi-Xiao, C.; Li-Wei, W. Targeting sphingosine kinase 2 (SphK2) by ABC294640 inhibits colorectal cancer cell growth in vitro and in vivo. J. Exp. Clin. Cancer Res. 2015, 34, 94. [Google Scholar] [CrossRef] [PubMed]
- Golubnitschaja, O.; EMPA; Costigliola, V. General Report & Recommendations in Predictive, Preventive and Personalised Medicine 2012: White Paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012, 3, 14. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Yeghiazaryan, K.; Costigliola, V.; Trog, D.; Braun, M.; Debald, M.; Kuhn, W.; Schild, H.H. Risk assessment, disease prevention and personalised treatments in breast cancer: Is clinically qualified integrative approach in the horizon? EPMA J. 2013, 4, 6. [Google Scholar] [CrossRef][Green Version]
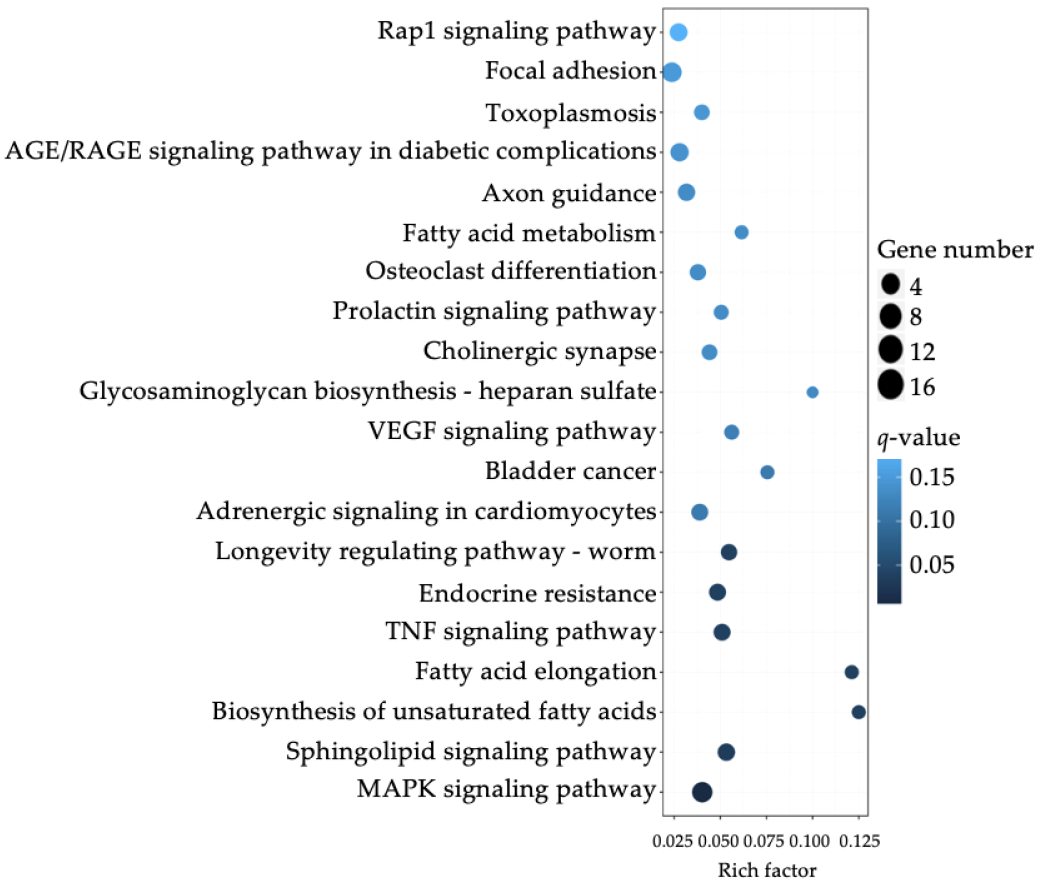
| Index | Pathway | p-Value | Adjusted p-Value (padj) | Odds Ratio | Combined Score |
|---|---|---|---|---|---|
| 1 | Biosynthesis of unsaturated fatty acids | 0.0000200 | 0.00286 | 18.08 | 195.57 |
| 2 | Bladder cancer | 0.00175 | 0.0502 | 8.56 | 54.31 |
| 3 | Fatty acid elongation | 0.00468 | 0.0912 | 9.86 | 52.91 |
| 4 | VEGF signaling pathway | 0.000893 | 0.0348 | 7.35 | 51.62 |
| 5 | Sphingolipid signaling pathway | 0.000138 | 0.0107 | 5.78 | 51.36 |
| 6 | AGE/RAGE signaling pathway in diabetic complications | 0.000281 | 0.0164 | 6.01 | 49.17 |
| 7 | MAPK signaling pathway | 0.0000245 | 0.00287 | 4.07 | 43.24 |
| 8 | Nitrogen metabolism | 0.0191 | 0.193 | 10.48 | 41.47 |
| 9 | Prolactin signaling pathway | 0.00193 | 0.0502 | 6.10 | 38.16 |
| 10 | Growth hormone synthesis, secretion and action | 0.000803 | 0.0348 | 4.99 | 35.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciberras, P.; Grech, L.; Grech, G. Differential Expression of the Sphingolipid Pathway Is Associated with Sensitivity to the PP2A Activator FTY720 in Colorectal Cancer Cell Lines. J. Clin. Med. 2021, 10, 4999. https://doi.org/10.3390/jcm10214999
Sciberras P, Grech L, Grech G. Differential Expression of the Sphingolipid Pathway Is Associated with Sensitivity to the PP2A Activator FTY720 in Colorectal Cancer Cell Lines. Journal of Clinical Medicine. 2021; 10(21):4999. https://doi.org/10.3390/jcm10214999
Chicago/Turabian StyleSciberras, Peter, Laura Grech, and Godfrey Grech. 2021. "Differential Expression of the Sphingolipid Pathway Is Associated with Sensitivity to the PP2A Activator FTY720 in Colorectal Cancer Cell Lines" Journal of Clinical Medicine 10, no. 21: 4999. https://doi.org/10.3390/jcm10214999
APA StyleSciberras, P., Grech, L., & Grech, G. (2021). Differential Expression of the Sphingolipid Pathway Is Associated with Sensitivity to the PP2A Activator FTY720 in Colorectal Cancer Cell Lines. Journal of Clinical Medicine, 10(21), 4999. https://doi.org/10.3390/jcm10214999






