Role of lncRNAs in the Development of an Aggressive Phenotype in Gallbladder Cancer
Abstract
1. Introduction
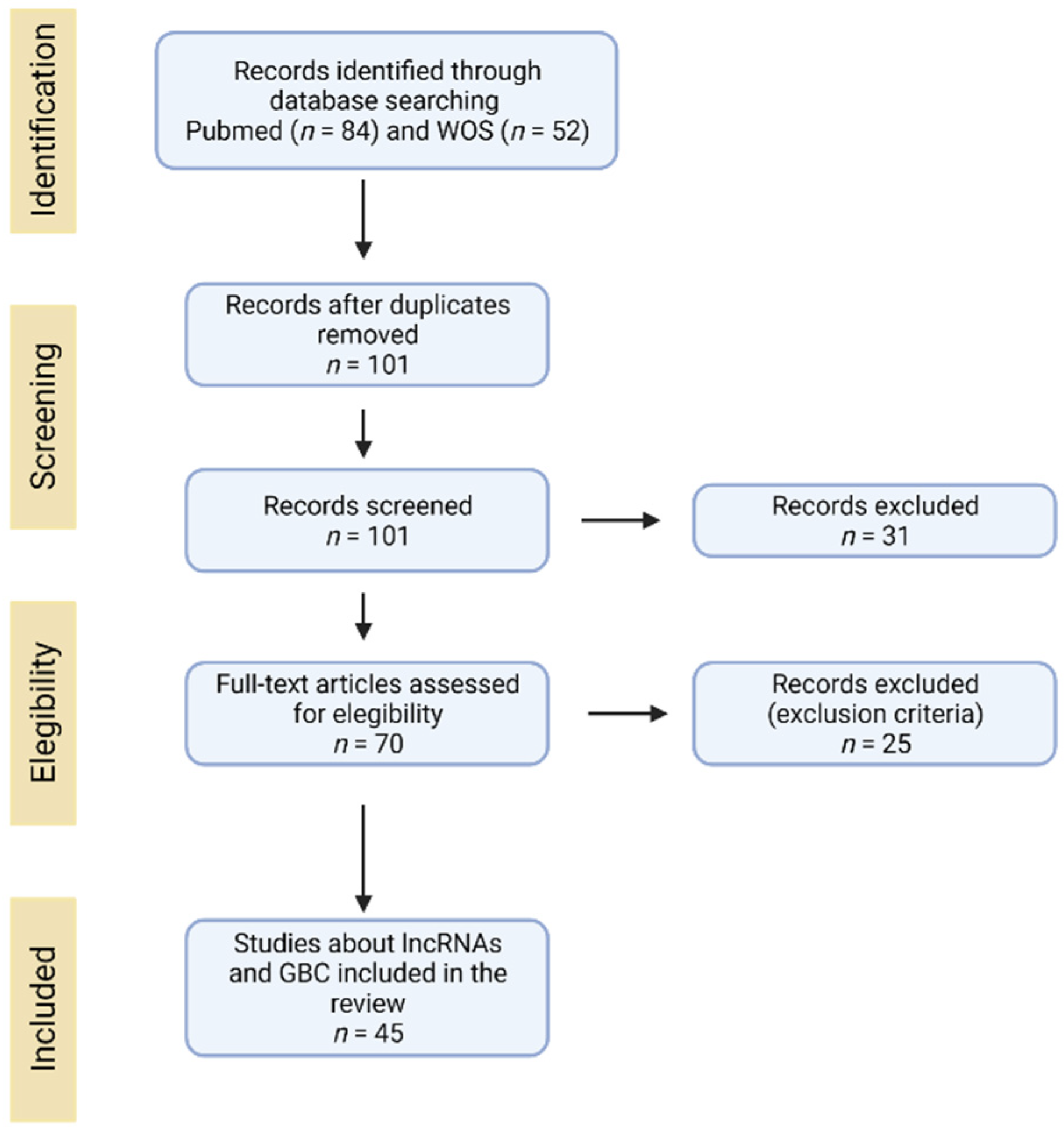
2. Search and Selection of Literature
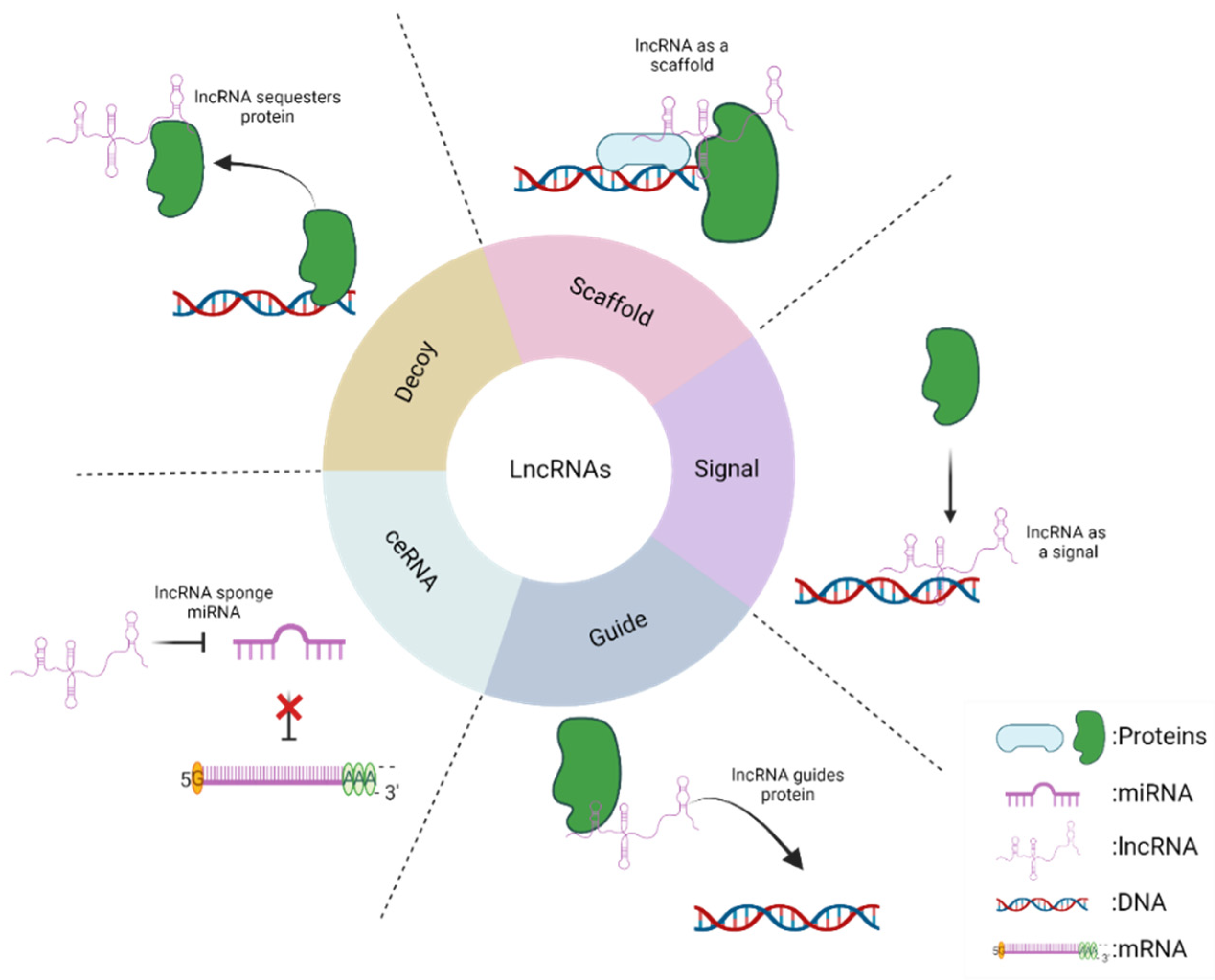
3. Long Non-Coding RNAs: Mechanisms of Action
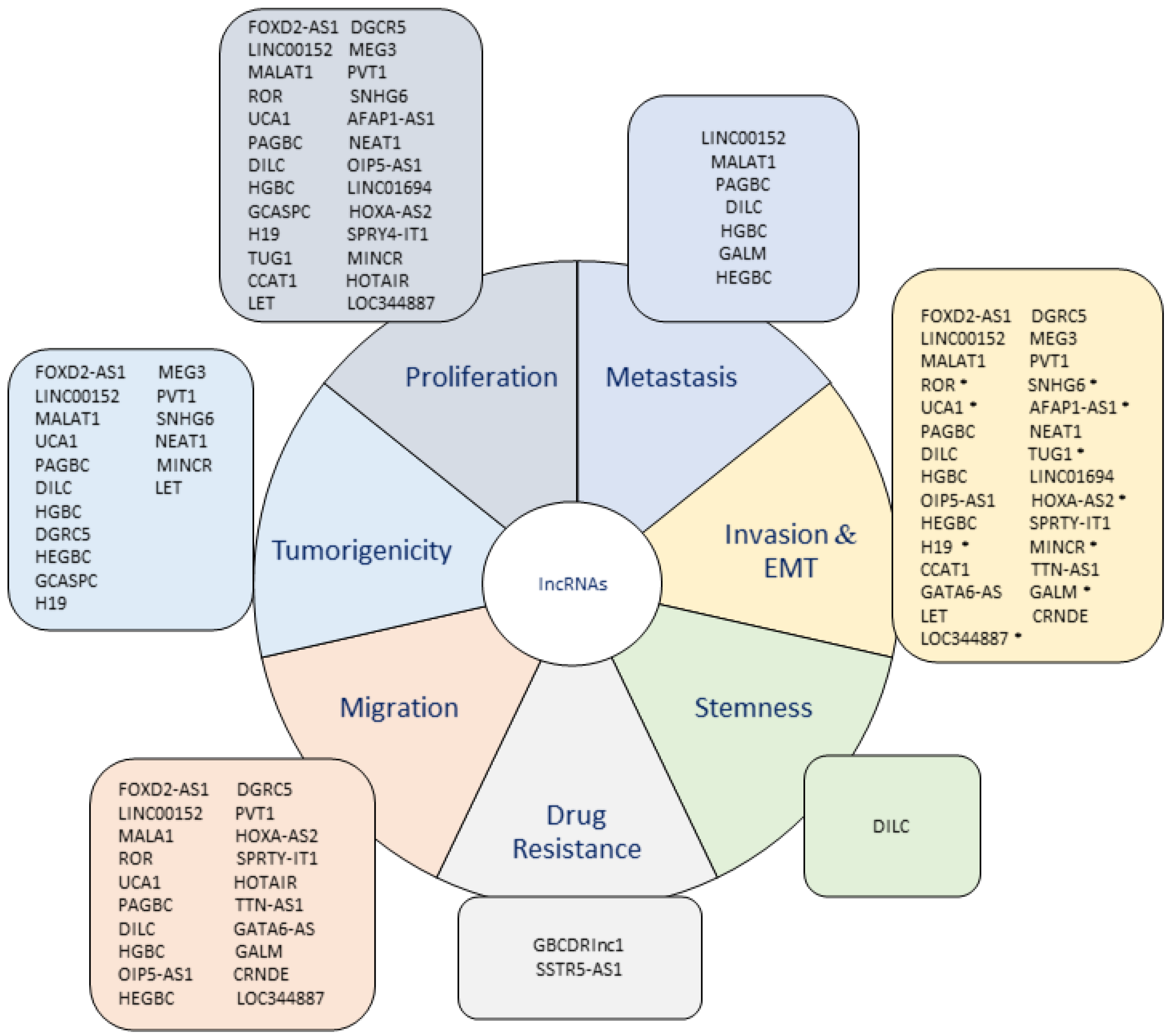
4. LncRNAs in Gallbladder Cancer
4.1. Upregulated lncRNAs in GBC
4.2. Downregulated lncRNAs in GBC
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Geng, L.; Wang, K.; Sun, J.; Xu, W.; Gong, S.; Zhu, Y. Long Noncoding RNA Expression Signatures of Colon Cancer Based on the ceRNA Network and Their Prognostic Value. Dis. Markers 2019, 2019, 7636757. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Wang, Z.; Zhang, Z.; Lu, M.; Wang, Y. Long Non-coding RNA LINC01787 Drives Breast Cancer Progression via Disrupting miR-125b Generation. Front. Oncol. 2019, 9, 1140. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Bekric, D.; Neureiter, D.; Ritter, M.; Jakab, M.; Gaisberger, M.; Pichler, M.; Kiesslich, T.; Mayr, C. Long Non-Coding RNAs in Biliary Tract Cancer-An Up-to-Date Review. J. Clin. Med. 2020, 9, 1200. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Cai, Q.; Jin, L.; Wang, S.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 2017, 8, 47957–47968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, S.H.; Zhang, M.D.; Wu, X.C.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumor Biol. 2016, 37, 12867–12875. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Yan, B.; Wang, Z. Long noncoding RNA: Its physiological and pathological roles. DNA Cell Biol. 2012, 31 (Suppl. 1), S34–S41. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci 2016, 41, 761–772. [Google Scholar] [CrossRef]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.P.; Jin, Y.P.; Wu, X.S.; Yang, Y.; Li, Y.S.; Li, H.F.; Xiang, S.S.; Song, X.L.; Jiang, L.; Zhang, Y.J.; et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol. Cancer 2019, 18, 167. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hundal, R.; Shaffer, E.A. Gallbladder cancer: Epidemiology and outcome. Clin. Epidemiol. 2014, 6, 99–109. [Google Scholar] [CrossRef]
- Bertran, E.; Heise, K.; Andia, M.E.; Ferreccio, C. Gallbladder cancer: Incidence and survival in a high-risk area of Chile. Int. J. Cancer 2010, 127, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Isin, M.; Dalay, N. LncRNAs and neoplasia. Clin. Chim. Acta 2015, 444, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wang, S.H.; Cai, Q.; Zhang, M.D.; Yang, Y.; Ding, J. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed. Pharm. 2016, 84, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shen, E.D.; Liao, M.M.; Hu, Y.B.; Wu, K.; Yang, P.; Zhou, L.; Chen, W.D. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumor Biol. 2016, 37, 9875–9886. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Q.; Hann, S.S. The functions and oncogenic roles of CCAT1 in human cancer. Biomed. Pharm. 2019, 115, 108943. [Google Scholar] [CrossRef]
- Ma, M.Z.; Chu, B.F.; Zhang, Y.; Weng, M.Z.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef]
- Ma, Y.C.; Yang, J.Y.; Yan, L.N. Relevant markers of cancer stem cells indicate a poor prognosis in hepatocellular carcinoma patients: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.; Arshad, M.; Kurozomi, S.; Althobiti, M.; Miligy, I.M.; Al-Izzi, S.; Toss, M.S.; Goh, F.Q.; Johnston, S.J.; Martin, S.G.; et al. Overexpression of the cancer stem cell marker CD133 confers a poor prognosis in invasive breast cancer. Breast Cancer Res. Treat. 2019, 174, 387–399. [Google Scholar] [CrossRef]
- Han, Y.; Xue, X.; Jiang, M.; Guo, X.; Li, P.; Liu, F.; Yuan, B.; Shen, Y.; Zhi, Q.; Zhao, H. LGR5, a relevant marker of cancer stem cells, indicates a poor prognosis in colorectal cancer patients: A meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 267–273. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Liao, M.; Deng, Z.; Gong, L.; Jiang, J.; Lynn, L.; Wu, K.; Miao, X. Clinicopathologic and prognostic significance of CD24 in gallbladder carcinoma. Pathol. Oncol. Res. 2011, 17, 45–50. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Yang, P.; Han, T.; Wang, R.Y.; Xing, X.L.; Si, A.F.; Ma, Q.Y.; Chen, Z.; Li, H.Y.; Zhang, B. Long non-coding RNA DILC promotes the progression of gallbladder carcinoma. Gene 2019, 694, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Chen, C.; Gu, X.; Li, L. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am. J. Cancer Res. 2021, 11, 1–13. [Google Scholar]
- Wang, R.; Dong, H.X.; Zeng, J.; Pan, J.; Jin, X.Y. LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330-5p/CD44 in NSCLC. J. Cell Physiol. 2018, 233, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.X.; Wang, R.; Jin, X.Y.; Zeng, J.; Pan, J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J. Cell Physiol. 2018, 233, 4126–4136. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Shi, M.; Xiang, T.; Bu, Y.Z. Long noncoding RNA DGCR5 represses hepatocellular carcinoma progression by inactivating Wnt signaling pathway. J. Cell Biochem. 2019, 120, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Wang, C.; He, J. Long non-coding RNA DGCR5 incudes tumorigenesis of triple-negative breast cancer by affecting Wnt/β-catenin signaling pathway. J. BUON 2020, 25, 702–708. [Google Scholar] [PubMed]
- Liu, S.; Chu, B.; Cai, C.; Wu, X.; Yao, W.; Wu, Z.; Yang, Z.; Li, F.; Liu, Y.; Dong, P.; et al. DGCR5 Promotes Gallbladder Cancer by Sponging MiR-3619-5p via MEK/ERK1/2 and JNK/p38 MAPK Pathways. J. Cancer 2020, 11, 5466–5477. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Dai, C.; Yu, C.; Yin, X.B.; Liao, W.-y.; Huang, Y.; Zhou, F. Silencing of long non-coding RNA FOXD2-AS1 inhibits the progression of gallbladder cancer by mediating methylation of MLH1. Gene Ther. 2020, 28, 306–318. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82. [Google Scholar] [CrossRef]
- Li, H.; Hu, Y.; Jin, Y.; Zhu, Y.; Hao, Y.; Liu, F.; Yang, Y.; Li, G.; Song, X.; Ye, Y.; et al. Long noncoding RNA lncGALM increases risk of liver metastasis in gallbladder cancer through facilitating N-cadherin and IL-1β-dependent liver arrest and tumor extravasation. Clin. Transl. Med. 2020, 10, e201. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gao, Q.; Wu, X.; Feng, F.; Xu, K. Long noncoding RNA HEGBC promotes tumorigenesis and metastasis of gallbladder cancer via forming a positive feedback loop with IL-11/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 186. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, Z.; Lu, S.; Fu, W.; Liu, Z.; Jiang, X.; Tai, S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta 2018, 485, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cao, P.; Zhu, X.; Pan, M.; Zhong, K.; He, R.; Li, Y.; Jiao, X.; Gao, Y. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget 2017, 8, 33137–33143. [Google Scholar] [CrossRef]
- Ma, M.Z.; Li, C.X.; Zhang, Y.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 2014, 13, 156. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Esmaeili, M.; Taheri, M. H19 lncRNA: Roles in tumorigenesis. Biomed. Pharm. 2020, 123, 109774. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumor Biol. 2016, 37, 9721–9730. [Google Scholar] [CrossRef]
- Wang, S.H.; Ma, F.; Tang, Z.H.; Wu, X.C.; Cai, Q.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 160. [Google Scholar] [CrossRef]
- Wang, S.H.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Quan, Z.W. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. Am. J. Cancer Res. 2016, 6, 15–26. [Google Scholar]
- Liu, L.; Yan, Y.; Zhang, G.; Chen, C.; Shen, W.; Xing, P. Knockdown of LINC01694 inhibits growth of gallbladder cancer cells via miR-340-5p/Sox4. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Wu, X.C.; Wang, S.H.; Ou, H.H.; Zhu, B.; Zhu, Y.; Zhang, Q.; Yang, Y.; Li, H. The NmrA-like family domain containing 1 pseudogene Loc344887 is amplified in gallbladder cancer and promotes epithelial-mesenchymal transition. Chem. Biol. Drug Des. 2017, 90, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.; Kim, D.; Kim, W. Long non-coding RNA linc00152 acting as a promising oncogene in cancer progression. Genom. Inf. 2019, 17, e36. [Google Scholar] [CrossRef]
- Zhong, Y.; Wu, X.; Li, Q.; Ge, X.; Wang, F.; Wu, P.; Deng, X.; Miao, L. Long noncoding RNAs as potential biomarkers and therapeutic targets in gallbladder cancer: A systematic review and meta-analysis. Cancer Cell Int. 2019, 19, 169. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Z.Q.; Wang, S.H.; Li, C.; Zhu, Z.G.; Quan, Z.W.; Zhang, W.J. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am. J. Transl. Res. 2016, 8, 4068–4081. [Google Scholar]
- Cai, Q.; Wang, Z.; Wang, S.; Weng, M.; Zhou, D.; Li, C.; Wang, J.; Chen, E.; Quan, Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Li, Z.X.; Zhu, Q.N.; Zhang, H.B.; Hu, Y.; Wang, G.; Zhu, Y.S. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 2018, 10, 6757–6768. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.S.; Wang, X.A.; Wu, W.G.; Hu, Y.P.; Li, M.L.; Ding, Q.; Weng, H.; Shu, Y.J.; Liu, T.Y.; Jiang, L.; et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol. Ther. 2014, 15, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Li, Q.; Wang, X.; Jiao, X.; Zheng, J.; Li, Z.; Pan, X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget 2017, 8, 46993–47006. [Google Scholar] [CrossRef]
- Chen, Q.; Su, Y.; He, X.; Zhao, W.; Wu, C.; Zhang, W.; Si, X.; Dong, B.; Zhao, L.; Gao, Y.; et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol. Lett. 2016, 12, 1361–1366. [Google Scholar] [CrossRef]
- Luan, C.; Li, Y.; Liu, Z.; Zhao, C. Long Noncoding RNA MALAT1 Promotes the Development of Colon Cancer by Regulating. OncoTargets Ther. 2020, 13, 3653–3665. [Google Scholar] [CrossRef]
- Jen, J.; Tang, Y.A.; Lu, Y.H.; Lin, C.C.; Lai, W.W.; Wang, Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Schäffers, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.S.; Chi, Y.Y.; Xue, J.Y.; Liu, M.Y.; Huang, S.; Mo, M.; Zhou, S.L.; Wu, J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 2016, 7, 37957–37965. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Yao, Z.; Xu, M.; Chen, J.; Lu, Y.; Yuan, L.; Zhou, S.; Zou, X.; Xu, R. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 244. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhang, W.J.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 2016, 7, 37857–37867. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, L.; Xu, X.; Shen, C. Knockdown of Long Noncoding RNA. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Wang, S.H.; Zhang, W.J.; Wu, X.C.; Weng, M.Z.; Zhang, M.D.; Cai, Q.; Zhou, D.; Wang, J.D.; Quan, Z.W. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J. Cell Mol. Med. 2016, 20, 2299–2308. [Google Scholar] [CrossRef]
- Wang, S.H.; Yang, Y.; Wu, X.C.; Zhang, M.D.; Weng, M.Z.; Zhou, D.; Wang, J.D.; Quan, Z.W. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett. 2016, 380, 122–133. [Google Scholar] [CrossRef]
- Yang, F.; Tang, Z.; Duan, A.; Yi, B.; Shen, N.; Bo, Z.; Yin, L.; Zhu, B.; Qiu, Y.; Li, J. Long Noncoding RNA. OncoTargets Ther. 2020, 13, 2357–2367. [Google Scholar] [CrossRef]
- Meng, X.; Ma, J.; Wang, B.; Wu, X.; Liu, Z. Long non-coding RNA OIP5-AS1 promotes pancreatic cancer cell growth through sponging miR-342-3p via AKT/ERK signaling pathway. J. Physiol. Biochem. 2020, 76, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.S.; Chu, K.J.; Ling, C.C.; Wu, T.M.; Zhu, X.C.; Liu, J.B.; Yu, F.; Li, Z.Z.; Wang, J.H.; Gao, Q.X.; et al. Long Noncoding RNA OIP5-AS1 Promotes the Progression of Liver Hepatocellular Carcinoma via Regulating the hsa-miR-26a-3p/EPHA2 Axis. Mol. Ther. Nucleic Acids 2020, 21, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dou, L.; Qin, Y.; Yang, H.; Yan, P. OIP5-AS1 contributes to tumorigenesis in hepatocellular carcinoma by miR-300/YY1-activated WNT pathway. Cancer Cell Int. 2020, 20, 440. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.; Luo, H. Long Non-Coding RNA OIP5-AS1 Contributes to Gallbladder Cancer Cell Invasion and Migration by miR-143-3p Suppression. Cancer Manag. Res. 2020, 12, 12983–12992. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xu, Q. LncRNA PVT1 regulates gallbladder cancer progression through miR-30d-5p. J. Biol. Regul. Homeost. Agents 2020, 34, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cai, Q.; Wang, S.; Wang, J.; Quan, Z. Long noncoding RNA PVT1 promoted gallbladder cancer proliferation by epigenetically suppressing miR-18b-5p via DNA methylation. Cell Death Dis. 2020, 11, 871. [Google Scholar] [CrossRef]
- Wu, X.S.; Wang, F.; Li, H.F.; Hu, Y.P.; Jiang, L.; Zhang, F.; Li, M.L.; Wang, X.A.; Jin, Y.P.; Zhang, Y.J.; et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017, 18, 1837–1853. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, X.; Ge, N.; Guo, W.; Feng, F.; Wan, F. Long non-coding RNA SPRY4-IT1 promotes gallbladder carcinoma progression. Oncotarget 2017, 8, 3104–3110. [Google Scholar] [CrossRef]
- Liu, X.F.; Wang, K.; Du, H.C. LncRNA SNHG6 regulating Hedgehog signaling pathway and affecting the biological function of gallbladder carcinoma cells through targeting miR-26b-5p. Eur. Rev. Med. Pharm. Sci. 2020, 24, 7598–7611. [Google Scholar] [CrossRef]
- Xue, Z.; Yang, B.; Xu, Q.; Zhu, X.; Qin, G. Long non-coding RNA SSTR5-AS1 facilitates gemcitabine resistance via stabilizing NONO in gallbladder carcinoma. Biochem. Biophys. Res. Commun. 2020, 522, 952–959. [Google Scholar] [CrossRef]
- Ma, F.; Wang, S.H.; Cai, Q.; Jin, L.Y.; Zhou, D.; Ding, J.; Quan, Z.W. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomed. Pharm. 2017, 88, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Y.; Shao, R.; Hu, Y.; Gao, H. LncRNA TTN-AS1 acts as a tumor promoter in gallbladder carcinoma by regulating miR-107/HMGA1 axis. World J. Surg. Oncol. 2021, 19, 163. [Google Scholar] [CrossRef]
- Zhang, L.; Geng, Z.; Meng, X.; Meng, F.; Wang, L. Screening for key lncRNAs in the progression of gallbladder cancer using bioinformatics analyses. Mol. Med. Rep. 2018, 17, 6449–6455. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Liu, H.; Wang, Y.; Wang, J.; Ni, X.; Ai, Z.; Pan, H.; Shao, Y. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget 2016, 7, 72833–72844. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Tang, J.; Hou, Y. LncRNA GATA6-AS inhibits cancer cell migration and invasion in gallbladder cancer by downregulating miR-421. OncoTargets Ther. 2019, 12, 8047–8053. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Z.; Zhang, Y.; Weng, M.Z.; Wang, S.H.; Hu, Y.; Hou, Z.Y.; Qin, Y.Y.; Gong, W.; Zhang, Y.J.; Kong, X.; et al. Long Noncoding RNA GCASPC, a Target of miR-17-3p, Negatively Regulates Pyruvate Carboxylase-Dependent Cell Proliferation in Gallbladder Cancer. Cancer Res. 2016, 76, 5361–5371. [Google Scholar] [CrossRef]
- Ma, M.Z.; Kong, X.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Zhang, W.J.; Quan, Z.W. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol. Carcinog. 2015, 54, 1397–1406. [Google Scholar] [CrossRef]
- Bao, D.; Yuan, R.X.; Zhang, Y. Effects of lncRNA MEG3 on proliferation and apoptosis of gallbladder cancer cells through regulating NF-κB signaling pathway. Eur. Rev. Med. Pharm. Sci. 2020, 24, 6632–6638. [Google Scholar] [CrossRef]
- Jin, L.; Cai, Q.; Wang, S.; Mondal, T.; Wang, J.; Quan, Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018, 9, 1017. [Google Scholar] [CrossRef]
- Tian, J.; Hu, X.; Gao, W.; Zhang, J.; Chen, M.; Zhang, X.; Ma, J.; Yuan, H. Identification of the long non-coding RNA LET as a novel tumor suppressor in gastric cancer. Mol. Med. Rep. 2017, 15, 2229–2234. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gu, L.; Zhang, J.; Shi, M.; Zhan, Q.; Shen, B.; Peng, C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed. Pharm. 2017, 89, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.K.; Hu, P.P.; Xu, F. Prognostic significance of long non-coding RNA MALAT1 for predicting the recurrence and metastasis of gallbladder cancer and its effect on cell proliferation, migration, invasion, and apoptosis. J. Cell Biochem. 2018, 119, 3099–3110. [Google Scholar] [CrossRef] [PubMed]
| Expression | LncRNA | Aggressive Phenotype | Mechanism | Ref. |
|---|---|---|---|---|
| Up-regulated | AFAP1-AS1 | P, I, E | ND | [34] |
| ANRIL | P, T | ND | [33] | |
| CCAT1 | I, P | Increases BMI-1 expression through the sponging of miR-218-5p | [35] | |
| DILC | S, P, I, M, T, ME | Promotes Wnt/β-catenin pathway | [41] | |
| DGCR5 | P, T, I, M, E | Decreases ZO-1 expression and increases MEK/ERK1/2 and JNK/p38 MAPK pathways | [47] | |
| FENDRR | ND | ND | [94] | |
| FOXD2-AS1 | P, M, T, I | Promotes MLH1 methylation by recruiting DNMT1 | [48] | |
| GBCDRlnc1 | D | Increases autophagy activity by enhancing the conversion from LC3-I into LC3-II and by inhibiting the ubiquitination of phosphoglycerate kinase 1(PGK1) | [49] | |
| GALM | I, M, ME, E | Acts as a sponge by competitively binding to miR-200 family and binding to IL-1β mRNA | [50] | |
| HGBC | P, T, M, E, ME | Acts through the interaction among HGBC and HuR | [24] | |
| HEGBC | P, M, T, ME | Acts through a HEGBC/IL-11/STAT3 positive regulatory loop | [51] | |
| HOXA-AS2 | P, M, I, E | ND | [53] | |
| HOTAIR | M, P | Increases c-Myc expression through the sponging of miR-130a | [54] | |
| H19 | P, T, M, I, E | Acts through both H19/miR-194-5p/AKT2 and H19/miR-342-3p/FOXM1 axes | [56,57,58] | |
| LINC01694 | P, T, I | Acts via LIN01694/miR-340-5p/SOX4 axis | [59] | |
| LOC344887 | P, M, I, E | ND | [60] | |
| LINC00152 | P, T, E, M, I, ME | Acts through LINC00152/miR-138/HIF-1a axis activating PI3K pathway | [63,64] | |
| MALAT1 | P, T, M, I, ME | MALAT1 regulates MCL-1 expression as a competing endogenous RNA for miR-363-3p | [66,75,76,78] | |
| MINCR | P, M, T, I, E | Modulates the ability of miR-26a-5p to bind to EZH2 | [79] | |
| NEAT1 | P, M, I, T | Increases surviving expression and acts as a sponge of miR-335 | [80] | |
| OIP5-AS1 | P, M, I | Reduces miR-143-3p expression | [84] | |
| PVT1 | P, M, I, E | Upregulates HK2 through its competitive endogenous activity on miR-143 as well as miR-18b-5p and miR-30d-5p | [85,86,87] | |
| PAGBC | P, M, I, ME | Binds competitively miR-133b and miR-511 activating PI3K/mTOR pathway | [88] | |
| ROR | P, M, I, E | ND | [13] | |
| SPRY4-IT1 | P, I, M | ND | [89] | |
| SNHG6 | P, I, T, E | Decreases miR-26b-5p expression and regulates Hedgehog signaling pathway | [90] | |
| SSTR5-AS1 | D | NONO/SSTR5_AS1 interaction prevents the degradation of NONO | [91] | |
| TUG1 | P, I, E | Decreases miR-300 expression | [92] | |
| UCA1 | M, T, I, E | Recruits EZH2 and induces p21 repression | [12,77] | |
| TTN-AS1 | M, I | Acts as a sponge to miR-107 and upregulates HMGA1 expression | [93] | |
| Down-regulated | GATA6-AS | P, I | Decreases mir-421 expression through TMP-2 | [96] |
| GCASPC | P, T | Inhibition of pyruvate carboxylase by GCASPC and miR-17-3p | [97] | |
| LET | P, I, T | ND | [98] | |
| MEG3 | P, T, E, I | Promotes EZH2 degradation regulating LATS2 and NF-κb pathway | [33,99,100] |
| LncRNA | Overall Survival | Tumor Size | TNM Stage | Histological Grade | Distant Metastasis | Lymphatic Invasion | Ref. |
|---|---|---|---|---|---|---|---|
| AFAP1-AS1 | Decrease | Increase | NS | NS | ND | Negative | [34] |
| ANRIL | Decrease | NS | NS | NS | ND | Negative | [33] |
| CCAT1 | ND | Increase | Advanced | ND | ND | Positive | [35] |
| GBCDRlnc1 | Decrease | NS | Advanced | Poorer | ND | Negative | [49] |
| GCASPC | Decrease | Increase | Advanced | ND | NS | Positive | [97] |
| GALM | Decrease | NS | Advanced | Poorer | ND | Positive | [50] |
| HGBC | Decrease | ND | Advanced | NS | ND | Positive | [24] |
| HEGBC | Decrease | Increase | Advanced | NS | ND | Positive | [51] |
| HOTAIR | ND | ND | Advanced | ND | ND | Positive | [54] |
| H19 | Decrease | Increase | NS | NS | ND | Positive | [56,57,58] |
| LINC01694 | Decrease | NS | Advanced | Poorer | ND | ND | [59] |
| LET | Decrease | Increase | Advanced | Poorer | ND | Positive | [98] |
| LINC00152 | Decrease | Increase | Advanced | NS | ND | Positive | [63,64] |
| MALAT1 | Decrease | Increase | NS | NS | ND | Positive | [66,75,76,78,103] |
| MEG3 | Decrease | NS | Advanced | Poorer | ND | Positive | [33,99,100] |
| MINCR | Decrease | Increase | ND | ND | ND | Positive | [79] |
| PAGBC | Decrease | ND | Advanced | ND | ND | ND | [88] |
| PVT1 | Decrease | NS | Advanced | NS | Increase | ND | [85,86,87] |
| ROR | Decrease | Increase | NS | NS | ND | Positive | [13] |
| SNHG6 | ND | Increase | Advanced | Poorer | Increase | Positive | [90] |
| SSTR5-AS1 | Decrease | ND | ND | ND | ND | ND | [91] |
| UCA1 | Decrease | Increase | Advanced | NS | ND | Positive | [12,77] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Moreno, P.; Riquelme, I.; Brebi, P.; Roa, J.C. Role of lncRNAs in the Development of an Aggressive Phenotype in Gallbladder Cancer. J. Clin. Med. 2021, 10, 4206. https://doi.org/10.3390/jcm10184206
Pérez-Moreno P, Riquelme I, Brebi P, Roa JC. Role of lncRNAs in the Development of an Aggressive Phenotype in Gallbladder Cancer. Journal of Clinical Medicine. 2021; 10(18):4206. https://doi.org/10.3390/jcm10184206
Chicago/Turabian StylePérez-Moreno, Pablo, Ismael Riquelme, Priscilla Brebi, and Juan Carlos Roa. 2021. "Role of lncRNAs in the Development of an Aggressive Phenotype in Gallbladder Cancer" Journal of Clinical Medicine 10, no. 18: 4206. https://doi.org/10.3390/jcm10184206
APA StylePérez-Moreno, P., Riquelme, I., Brebi, P., & Roa, J. C. (2021). Role of lncRNAs in the Development of an Aggressive Phenotype in Gallbladder Cancer. Journal of Clinical Medicine, 10(18), 4206. https://doi.org/10.3390/jcm10184206







