Abstract
Background: Acute respiratory distress syndrome (ARDS) is an intense inflammatory process of the lungs. Most ARDS patients require mechanical ventilation (MV). Few studies have investigated the prediction of MV duration over time. We aimed at characterizing the best early scenario during the first two days in the intensive care unit (ICU) to predict MV duration after ARDS onset using supervised machine learning (ML) approaches. Methods: For model description, we extracted data from the first 3 ICU days after ARDS diagnosis from patients included in the publicly available MIMIC-III database. Disease progression was tracked along those 3 ICU days to assess lung severity according to Berlin criteria. Three robust supervised ML techniques were implemented using Python 3.7 (Light Gradient Boosting Machine (LightGBM); Random Forest (RF); and eXtreme Gradient Boosting (XGBoost)) for predicting MV duration. For external validation, we used the publicly available multicenter database eICU. Results: A total of 2466 and 5153 patients in MIMIC-III and eICU databases, respectively, received MV for >48 h. Median MV duration of extracted patients was 6.5 days (IQR 4.4–9.8 days) in MIMIC-III and 5.0 days (IQR 3.0–9.0 days) in eICU. LightGBM was the best model in predicting MV duration after ARDS onset in MIMIC-III with a root mean square error (RMSE) of 6.10–6.41 days, and it was externally validated in eICU with RMSE of 5.87–6.08 days. The best early prediction model was obtained with data captured in the 2nd day. Conclusions: Supervised ML can make early and accurate predictions of MV duration in ARDS after onset over time across ICUs. Supervised ML models might have important implications for optimizing ICU resource utilization and high acute cost reduction of MV.
1. Background
The acute respiratory distress syndrome (ARDS) is an important cause of morbidity, mortality, and costs in intensive care units (ICUs) worldwide [1]. It is a life-threatening form of acute respiratory failure characterized by inflammatory pulmonary edema leading to severe hypoxemia, requiring endotracheal intubation and mechanical ventilation (MV) in most cases [2]. The number of days on MV during the ICU stay is a major driver of high acute care costs [3,4,5]. We believe that an important intervention to mitigate these costs is timely recognition and treatment of conditions that can cause serious complications.
The Berlin definition of ARDS identifies three mutually exclusive categories of lung severity with PaO2/FiO2 ratios in the ranges >200–300 mmHg (mild ARDS), >100–200 mmHg (moderate ARDS), and ≤100 mmHg (severe ARDS) [6,7]. Some studies [8,9] have reported a progression of costs from mild, to moderate, to severe ARDS. Despite global acceptance of the Berlin criteria [10], some authors have questioned its ability to assess the “true” severity of lung injury [11]. A recent study argues that mild ARDS should be considered “severe in terms of level of care” [12]. This quality criterion (i.e., level of care) could be measured in terms of MV duration, but accurate predictions of MV duration are difficult for critical care physicians [13,14], particularly for patients requiring prolonged MV [14].
Predicting MV duration could influence important clinical decisions, such as timing of tracheostomy and initiation of oral nutrition [14]. In this context, one approach for an accurate prediction of MV duration is the use of artificial intelligence (AI) approaches, such as machine learning (ML). ML is a subset of AI in which machines extract knowledge from the data provided. ML is an exploratory process where there is no one-method-fits-all solution [15,16]. ML merges statistical analysis techniques with computer science to produce algorithms capable of “statistical learning” [17]. ML algorithms are divided into two categories: supervised and unsupervised [17]. Supervised learning algorithms, the ones used in our study, detect relationships between potential explanatory features and a known target outcome [16]. They are commonly used in ICUs to predict clinical outcomes [16,17,18,19,20,21]. Troché and Moine addressed the critical question on whether MV duration is predictable [22]. Herein, we present the use of three powerful supervised ML methods to develop novel models to predict MV duration in ARDS after onset over time, using the single-center MIMIC-III dataset under three different scenarios. Then, the eICU multicenter dataset was used to externally validate the best MIMIC-III prediction model.
2. Methods
2.1. Study Design and Patient Population
We used two publicly available clinical databases for development and external validation of the best ML predictive model: MIMIC- III [23] and eICU, respectively [24]. Data of the first 3 ICU days (day 1 for representative data within the first 24 h after ARDS onset, day 2 for data within 24–48 h after onset, and day 3 for data within 48–72 h after onset) (n = 2466, 1445, and 1278 patients, respectively) were extracted from the single-center dataset MIMIC-III (MetaVision, 2008–2012) [23]. Similarly, data of the first 3 ICU days after ARDS onset (n = 5153, 2981, and 2326 patients, respectively) were extracted from the multicenter dataset eICU (2014–2015) [24]. Patients <18 years were excluded. Data extraction from both datasets was performed using Python 3.7. The selection of clinical variables was based on prior studies [9,19,25,26,27]. All extracted patients from both datasets fulfilled the Berlin definition for ARDS [6]. For the purpose of this study, prolonged MV was defined as being ventilated for >48 h [22,28]. Disease progression in each dataset was tracked along those 3 ICU days.
2.2. MIMIC-III
Medical Information Mart for Intensive Care III (MIMIC-III) is a large single-center database containing de-identified health-related data of about 60,000 ICUs patients admitted to the Beth Israel Deaconess Medical Center (Boston, MA, USA) between 2001 and 2012 [23]. There were six predictors: baseline demographic information (age); ventilator parameters including PEEP; blood gas parameters including FiO2, PaO2, PaO2/FiO2, and PaCO2. The main target variable was MV duration.
2.3. eICU
eICU is a multicenter ICU database and it has a high granularity of data of more than 200,000 ICU admissions [24]. We used this database for external validation of the best prediction model obtained from MIMIC-III in order to obtain the MV duration prediction in the eICU database.
2.4. Predictive Models
During the first 24 h of ARDS onset, misdiagnosis can occur if clinicians consider qualifying PaO2 values resulting from acute events unrelated to the disease process (such as endotracheal tube obstruction, barotrauma, or hemodynamic instability), instead of considering only PaO2 values while patients are clinically stable. It is also well established that changes in PEEP and FiO2 within the first few hours of routine intensive care management alter the PaO2/FiO2 ratio in ARDS patients [11]. Since in a substantial proportion of patients diagnosed as having ARDS did not meet ARDS criteria within the first 24 h of care, we decided to examine supervised ML models in the following three scenarios during the first two ICU days: (i) scenario I: predicting MV duration using information captured in the 1st ICU day; (ii) scenario II: predicting MV duration using information captured in the 2nd ICU day; (iii) scenario III: predicting MV duration using information captured in the 1st and 2nd ICU days, then comparing these three scenarios with scenario IV for predicting MV duration using the information captured in the 3rd ICU day exclusively.
We implemented three robust supervised ML algorithms via Python 3.7, including Light Gradient Boosting Machine (LightGBM) [29], Random Forest (RF) [30], and eXtreme Gradient Boosting (XGBoost) [31] to generate predictive models for MV duration after ARDS onset over time in the development database. For external validation purposes, we used the multicenter eICU dataset, as these three methods sacrifice the explicitness of the model in favor of predictive quality, and the generated models should be seen as “black box” with a high predictive robustness. For the development database, we optimized each model’s parameters through a grid search over the respective model’s hyperparameter space and the quality of all prediction models was computed based on a 10-fold cross-validation approach, which means that the dataset was divided into 10 folds, and in each run, 9 were used for training, and the remaining 1 was used for testing. Root-mean-square error (RMSE) was used to assess the predictive quality of the models. RMSE flags more significant differences between the predicted and the actual patient readings when they occur [32]. MV duration was expressed in days.
3. Results
For development and validation databases, mean values and 95% confidence intervals (CI) of baseline parameters during the first three ICU days after ARDS onset are reported in Table 1. The median and interquartile range (IQR) of MV duration are reported in Table 2.

Table 1.
Predictors and their descriptive statistics in MIMIC-III and eICU at 24 h, 48 h, and 72 h.

Table 2.
MV Duration in ARDS across MIMIC-III and eICU.
Table 3 shows the performance of the three supervised ML methods for the predictive scenarios in the development database. Table 4 shows the results of external validation of the best prediction model obtained from MIMIC-III to obtain the MV duration prediction in the eICU database.

Table 3.
Performances of LightGBM, RF, and XGBoost models to predict MV duration over time in MIMIC-III.

Table 4.
External validation of the best prediction model (LightGBM) obtained from MIMIC-III to obtain the MV duration prediction in the eICU database.
For the development database, the best early ML model for predicting MV duration was obtained by scenario II with RMSE = 6.10 days, using LightGBM algorithm. Figure 1a represents the Bland–Altman plot for LightGBM prediction and truth values in scenario II.
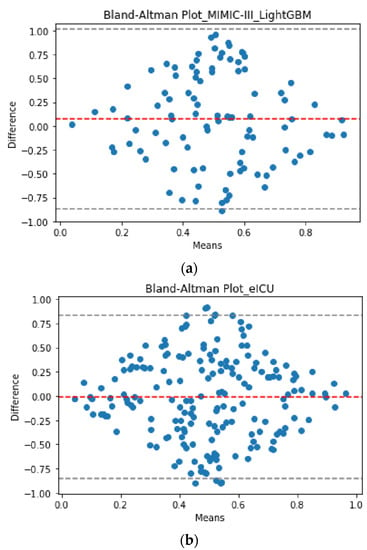
Figure 1.
Bland–Altman plot for the truth vs. the predicted values of MV duration using LightGBM (the best validated model) in Scenario II (the best early scenario). (a) Development database; (b) validation database. The X- and Y-axes stand for the mean and the difference of the two measurements, respectively. Please note that the values shown in the Bland–Altman plot are normalized in the interval (0, 1) (i.e., values are scaled to have corresponding values between 0 and 1).
For the validation database, the best early ML predictive model for MV duration was also observed for scenario II with RMSE = 5.87 days. This finding reinforces the idea that the best early approach for predicting MV duration is to consider the condition of the patient in the second ICU day after ARDS onset, rather than the first ICU day, or both. Figure 1b represents the Bland–Altman plot for prediction and truth values in scenario II using the external validation of LightGBM. The Bland–Altman plots illustrate agreement between the LightGBM models using the development and validation databases.
4. Discussion
Comparing the difference of RMSE means in the best early scenario (scenario II) with the prediction based on the data of patients in their third ICU day (scenario IV), yields minor RMSE differences (development database: 0.18 day (6.10–5.92) for LightGBM, and validation database: 0.16 day (5.87–5.71)). According to these low differences for both the development and validation datasets, our major finding was that the prediction results of LightGBM models based on the data of the second ICU day (scenario II) are very close to those corresponding results of LightGBM models based on the data of the third ICU day (scenario IV). Consequently, the LightGBM model can accurately predict MV duration without considering/waiting for the data of the third ICU day. This means that MV duration can be predicted earlier, and this could lead to better allocation of MV resources, reducing high acute costs of MV in ARDS, and improving patient care.
MV duration beyond 48 h in patients with ARDS provides information about risk factors in those patients [28] and has a direct correlation with ICU costs [4,5]. An early predictive model for MV duration can optimize ICU-level resource utilization [5,33]. Previous attempts to predict MV duration using conventional ICU scores or traditional statistical regression based techniques have proven to be difficult and failed to deal with the diversity of big data in the modern ICU databases [22]. ML is reliable, and it is a non-invasive modality to generate models for predicting MV duration. Most previous works considered a discriminatory prediction model to determine if a patient will remain intubated after a fixed number of days (e.g., 7 days) [22]. By contrast, our approach is numerical, and it predicts the number of MV days earlier by using commonly accessible clinical variables during the first two ICU days. Furthermore, to strengthen the evidence of our results, we used a multicenter database (eICU) for external validation, in which the best model obtained from a single-center database (MIMIC-III) was used to obtain the MV duration prediction in the eICU database. Our findings could be used to facilitate optimal triage, more timely management, and ICU resource utilization [34]. They may also affect some important clinical decisions, including timing of tracheostomy and, potentially, transfers to long-term ventilator weaning units or referral to other centers [13].
Herein, the main objective of using ML was to show that the application of ML is a promising approach to predict MV duration early. The ML contribution in this large study is to demonstrate the applicability of this approach, while not trying to choose the most proper ML model. Furthermore, we believe that the results of an efficient ML technique can yield accurate results for predicting MV duration. In terms of clinical relevance, our ML findings showed that using clinical data from the first ICU day is less predictive than data from the second ICU day. Previous studies showed that the accuracy of intensivists to predict MV duration is limited [13]. However, comparison to other published ML prediction of MV duration is difficult, as we aimed at predicting MV duration for MV >48 h and prior studies predicted for different outcomes under different time frames, in different populations, and using different ML metrics. A recent ML study showed that RMSE for predicting MV duration in ARDS patients for MV >48 h, was 6.23 days [9]. However, this study in [9] had several weaknesses: (1) it ignored the temporal dependency of the longitudinal predictor and treated each observed data point independently, and (2) it was only based on the single-center MIMIC-III database without external validation. Hence, those findings have serious limitations for the generalizability in the context of assessing the prediction of ARDS outcome.
From the cost perspective, the mean incremental cost of MV in ICU patients in the US was $1522 per day [4]. For instance, if we compare our findings with the result of the best ML method used in [9], which had a RMSE of 6.23 days, we see that LightGBM approach (the best approach) improved the current state of the art. This improvement can be quantified in terms 0.13 day (6.23–6.10) and about US $198 per patient according to [4]. Developing early predictive models using ML could assist to implement policies for the reduction of high acute care costs in ARDS [3,4,5]. Previous clinical studies showed acute costs incurred by mechanically ventilated ICU patients, but there is a significant difference in costs between ventilated ARDS patients and those without ARDS [35]. More specifically, ARDS diagnosis increases total ICU and hospital costs for mechanically ventilated ICU patients, suggesting higher total costs due to more days on a ventilator, although there is no clear severity-dependent relationship between ARDS severity and incurred costs [35]. The benchmarking of ML algorithms is possible through publicly available databases such as MIMIC-III [19,27] or eICU [19,36].
We acknowledge that our study has several strengths. First, we have analyzed a large population of over 7000 ARDS patients from two ICU databases within the first three ICU days after ARDS onset. Second, we have implemented and externally validated the best ML model (LightGBM) that can predict MV duration early and accurately using commonly accessible clinical variables. Third, early prediction of MV duration can inform population-level ICU resource allocation. Despite its strengths, we also acknowledge some limitations. First, our study is based on a retrospective analysis of data and should be confirmed through further prospective studies. Second, one could argue that the outcome of MV duration is somewhat subjective and could be a function of local practice or intrinsic bias inherent in such critical care decisions. However, our ability to predict a clinically relevant and difficult-to-predict outcome (MV duration) early supports the value of the proposed supervised ML models.
5. Conclusions
Predicting MV duration after ARDS onset over time is complex and cannot be adequately performed by critical care physicians. Our findings showed that the ML-based early prediction of MV duration is more accurate when predictive models are based on the clinical features of ARDS patients in the second ICU day after ARDS onset.
Author Contributions
M.S. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. M.S., D.R. and J.V., participated in the forming of the research question. M.S. was responsible for developing the software and obtaining the results. M.S. and D.R. performed the analysis of the results. M.S., D.R. and J.V. were responsible for drafting the first and subsequent versions of the manuscript. J.V. provided critical appraisal during data analysis and interpretation. All authors have read and agreed to the published version of the manuscript.
Funding
No funding was received for the access, development and analysis of the data. M. Sayed and D. Riaño are funded by the Spanish Ministry of Science and Innovation, Madrid, Spain (PID2019-105789RBI00), and the State Research Agency (AEI, Spain) under the grant agreement (RED2018-102312-T). J. Villar is funded by Instituto de Salud Carlos III, Madrid, Spain (#CB06/06/1088; #PI16/00049; #PI19/00141), and the European Regional Developments’ Funds (FEDER).
Institutional Review Board Statement
The datasets used for the analysis in this study are publicly available.
Informed Consent Statement
The datasets for the analysis are de-identified.
Data Availability Statement
By reasonable request to M.S. and D.R.
Acknowledgments
The authors want to thank Arthur S. Slutsky for his contribution in reviewing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest in relation to this manuscript.
References
- Rubenfeld, G.D.; Caldwell, E.; Peabody, E.; Weaver, J.; Martin, D.P.; Neff, M.; Stern, E.J.; Hudson, L.D. Incidence and outcomes of acute lung injury. N. Engl. J. Med. 2005, 353, 1685–1693. [Google Scholar] [CrossRef]
- Slutsky, A.S.; Villar, J.; Pesenti, A. Happy 50th birthday ARDS! Intensive Care Med. 2016, 42, 637–639. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bice, T.; Carson, S.S. Acute Respiratory Distress Syndrome: Cost (Early and Long-Term). Semin. Respir. Crit. Care Med. 2019, 40, 137–144. [Google Scholar] [CrossRef]
- Dasta, J.F.; McLaughlin, T.P.; Mody, S.H.; Piech, C.T. Daily cost of an intensive care unit day: The contribution of mechanical ventilation. Crit. Care Med. 2005, 33, 1266–1271. [Google Scholar] [CrossRef] [PubMed]
- Marti, J.; Hall, P.; Hamilton, P.; Lamb, S.; McCabe, C.; Lall, R.; Darbyshire, J.; Young, D.; Hulme, C. One-year resource utilisation, costs and quality of life in patients with acute respiratory distress syndrome (ARDS): Secondary analysis of a randomised controlled trial. J. Intensive Care 2016, 4. [Google Scholar] [CrossRef]
- ARDS Definition Task Force; Ranieri, V.M.; Rubenfeld, G.D.; Thompson, B.T.; Ferguson, N.; Caldwell, E.; Fan, E.; Camporota, L.; Slutsky, A.S. Acute respiratory distress syndrome: The Berlin definition. JAMA 2012, 307, 2526–2533. [Google Scholar] [CrossRef]
- Ferguson, N.D.; Fan, E.; Camporota, L.; Antonelli, M.; Anzueto, A.; Beale, R.; Brochard, L.; Brower, R.; Esteban, A.; Gattinoni, L.; et al. The Berlin definition of ARDS: An expanded rationale, justification, and supplementary material. Intensive Care Med. 2012, 38, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.; Kornblith, L.Z.; Hendrickson, C.M.; Howard, B.M.; Conroy, A.S.; Moazed, F.; Calfee, C.S.; Cohen, M.J.; Callcut, R.A. Health care utilization and the cost of posttraumatic acute respiratory distress syndrome care. J. Trauma Acute Care Surg. 2018, 85, 148–154. [Google Scholar] [CrossRef]
- Sayed, M.; Riaño, D. Modelling ICU Patients to Improve Care Requirements and Outcome Prediction of Acute Respiratory Distress Syndrome: A Supervised Learning Approach. In Artificial Intelligence in Medicine: Knowledge Representation and Transparent and Explainable Systems. Lecture Notes in Artificial Intelligence (LINAI), Proceedings of the AIME 2019 International Workshops KR4HC-ProHealth/TEAAM, Poznan, Poland, 26–29 June 2019; Marcos, M., Juarez, J.M., Lenz, R., Nalepa, G.J., Nowaczyk, S., Peleg, M., Stefanowski, J., Stiglic, G., Eds.; Springer: Cham, Switzerland, 2019; Volume 11979, pp. 39–49. [Google Scholar]
- Del Sorbo, L.; Ranieri, V.M.; Ferguson, N.D. The Berlin definition met our needs: Yes. Intensive Care Med. 2016, 42, 643–647. [Google Scholar] [CrossRef]
- Villar, J.; Pérez-Méndez, L.; Kacmarek, R.M. The Berlin definition met our needs: No. Intensive Care Med. 2016, 42, 648–650. [Google Scholar] [CrossRef]
- Pirracchio, R.; Gropper, M.A. Heterogeneity in intensive care: Low severity does not mean low risk! Anesthesiology 2019, 130, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Casas, J.B.; Connery, S.M.; Montoya, R.; Dwivedi, A.K.; Lee, S. Accuracy of early prediction of duration of mechanical ventilation by intensivists. Ann. Am. Thorac. Soc. 2014, 11, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Casas, J.B.; Dwivedi, A.K.; Connery, S.M.; Quansah, R.; Ellerbrook, L.; Galvis, J. Predictive models of prolonged mechanical ventilation yield moderate accuracy. J. Crit. Care 2015, 30, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Austin, P.C.; Tu, J.V.; Ho, J.E.; Levy, D.; Lee, D.S. Using methods from the data-mining and machine-learning literature for disease classification and prediction: A case study examining classification of heart failure subtypes. J. Clin. Epidemiol. 2013, 66, 398–407. [Google Scholar] [CrossRef]
- Cherifa, M.; Pirracchio, R. What every intensivist should know about Big Data and targeted machine learning in the intensive care unit. Rev. Bras. Ter. Intensive 2019, 31, 444–446. [Google Scholar] [CrossRef]
- Gutierrez, G. Artificial Intelligence in the Intensive Care Unit. Crit. Care 2020, 24, 101. [Google Scholar] [CrossRef]
- Greco, M.; Caruso, P.F.; Cecconi, M. Artificial Intelligence in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2021, 42, 2–9. [Google Scholar] [CrossRef]
- Sayed, M.; Riaño, D.; Villar, J. Novel criteria to classify ARDS severity using a machine learning approach. Crit. Care 2021, 25. [Google Scholar] [CrossRef]
- Pirracchio, R.; Petersen, M.L.; Carone, M.; Resche-Rigon, M.; Chevret, S.; van der Laan, M.J. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Respir. Med. 2015, 3, 42–52. [Google Scholar] [CrossRef]
- Troché, G.; Moine, P. Is the duration of mechanical ventilation predictable? Chest 1997, 112, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Parreco, J.; Hidalgo, A.; Parks, J.J.; Kozol, R.; Rattan, R. Using artificial intelligence to predict prolonged mechanical ventilation and tracheostomy placement. J. Surg. Res. 2018, 228, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Physionet.org, ‘MIMIC-III Critical Care Database’. Available online: https://mimic.physionet.org/about/mimic/ (accessed on 2 July 2020).
- Physionet.org, ‘eICU Collaborative Research Database’. Available online: https://eicu-crd.mit.edu/about/eicu/ (accessed on 19 October 2020).
- Villar, J.; Ambrós, A.; Soler, J.A.; Martínez, D.; Ferrando, C.; Solano, R.; Mosteiro, F.; Blanco, J.; Martín-Rodríguez, C.; Fernández, M.D.M.; et al. Age, PaO2/FIO2, and Plateau Pressure Score: A Proposal for a Simple Outcome Score in Patients With the Acute Respiratory Distress Syndrome. Crit. Care Med. 2016, 44, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Wang, S.; Liu, R.; Wang, H.; Zheng, J.; Yu, K. Risk factors for outcomes of acute respiratory distress syndrome patients: A retrospective study. J. Thorac. Dis. 2019, 11, 673–685. [Google Scholar] [CrossRef]
- Le, S.; Pellegrini, E.; Green-Saxena, A.; Summers, C.; Hoffman, J.; Calvert, J.; Das, R. Supervised machine learning for the early prediction of acute respiratory distress syndrome (ARDS). J. Crit. Care 2020, 60, 96–102. [Google Scholar] [CrossRef]
- Gong, M.N.; Schenk, L.; Gajic, O.; Mirhaji, P.; Sloan, J.; Dong, Y.; Festic, E.; Herasevich, V. Early intervention of patients at risk for acute respiratory failure and prolonged mechanical ventilation with a checklist aimed at the prevention of organ failure: Protocol for a pragmatic stepped-wedged cluster trial of PROOFCheck. BMJ Open 2016, 6. [Google Scholar] [CrossRef]
- Dehua, W.; Yang, Z.; Yi, Z. LightGBM: An effective miRNA classification method in breast cancer patients. In Proceedings of the 2017 International Conference on Computational Biology and Bioinformatics (ICCBB 2017), Newark, NJ, USA, 18–20 October 2017; pp. 7–11. [Google Scholar] [CrossRef]
- Boulesteix, A.-L.; Janitza, S.; Kruppa, J.; König, I.R. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 493–507. [Google Scholar] [CrossRef]
- Chen, T.; Carlos, G. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ′16), San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Hagan, R.; Gillan, C.J.; Spence, I.; McAuley, D.; Shyamsundar, M. Comparing regression and neural network techniques for personalized predictive analytics to promote lung protective ventilation in Intensive Care Units. Comput. Biol. Med. 2020, 126. [Google Scholar] [CrossRef]
- Marco, L. Intensive care resource allocation: When difficult choices have to be made. BJMP 2013, 6, 4–6. [Google Scholar]
- Seneff, M.G.; Zimmerman, J.E.; Knaus, W.A.; Wagner, D.P.; Draper, E.A. Predicting the Duration of Mechanical Ventilation. The importance of disease and patient characteristics. Chest 1996, 110, 469–479. [Google Scholar] [CrossRef]
- Marshall, Y.; Hayley, B.; Chao-Ping, W.; Michelle, N. Increased Economic Costs Associated with Acute Respiratory Distress Syndrome in Mechanically Ventilated Patients in the Intensive Care Unit. Am. J. Respir. Crit. Care Med. 2017, 195, A7579. [Google Scholar]
- Sheikhalishahi, S.; Balaraman, V.; Osmani, V. Benchmarking machine learning models on multi-centre eICU critical care dataset. PLoS ONE 2020, 15, e0235424. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).