Artificial Lipid Membranes: Past, Present, and Future
Abstract
:1. Introduction
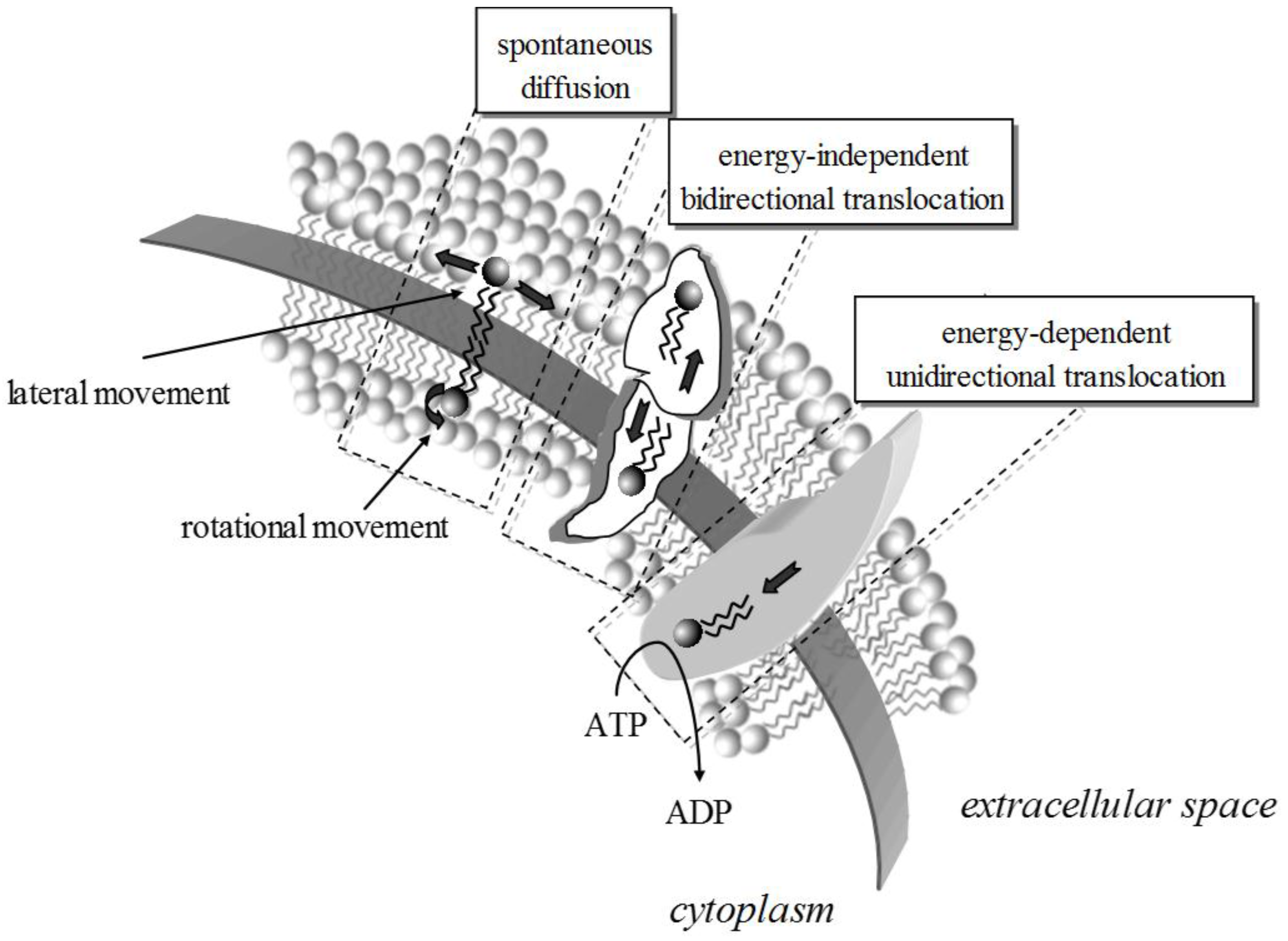
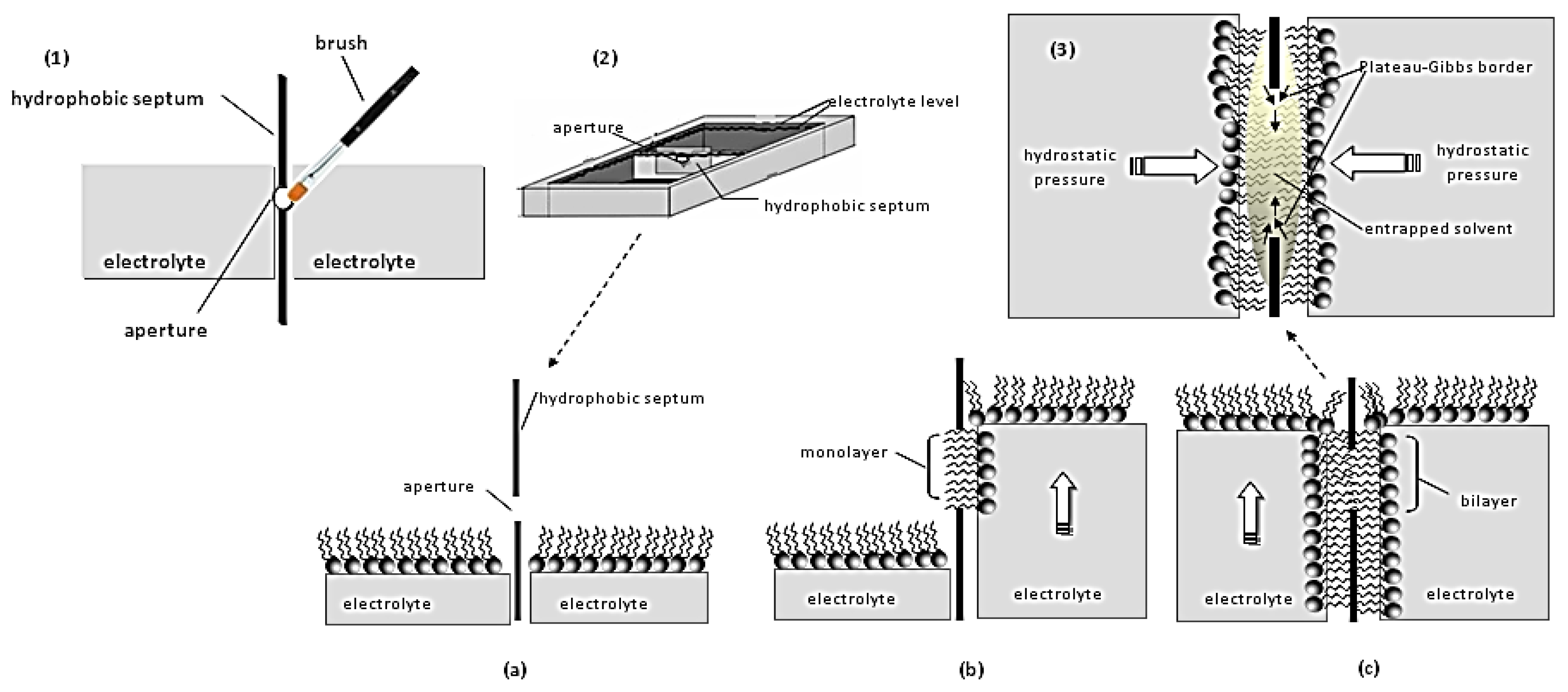
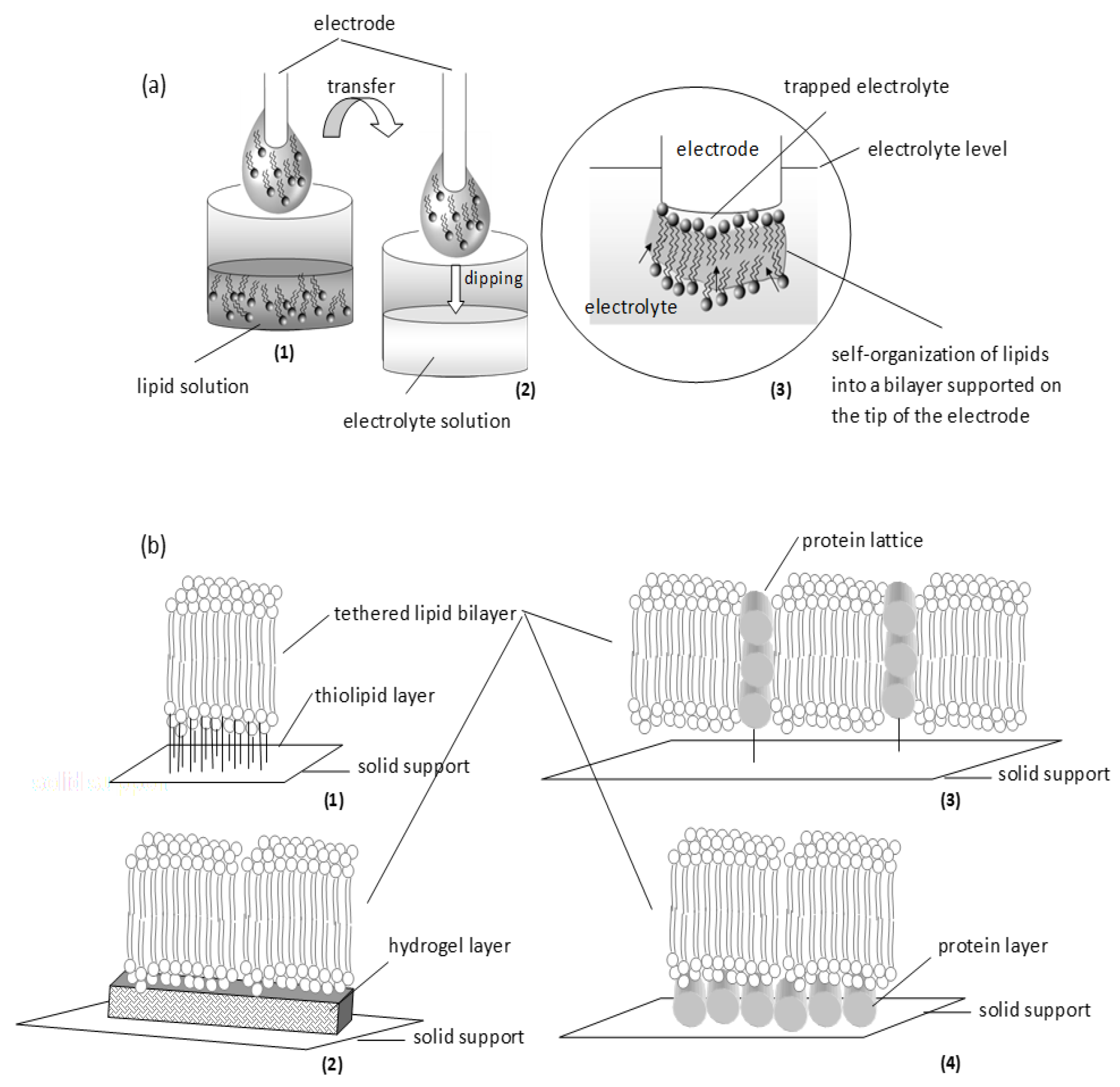
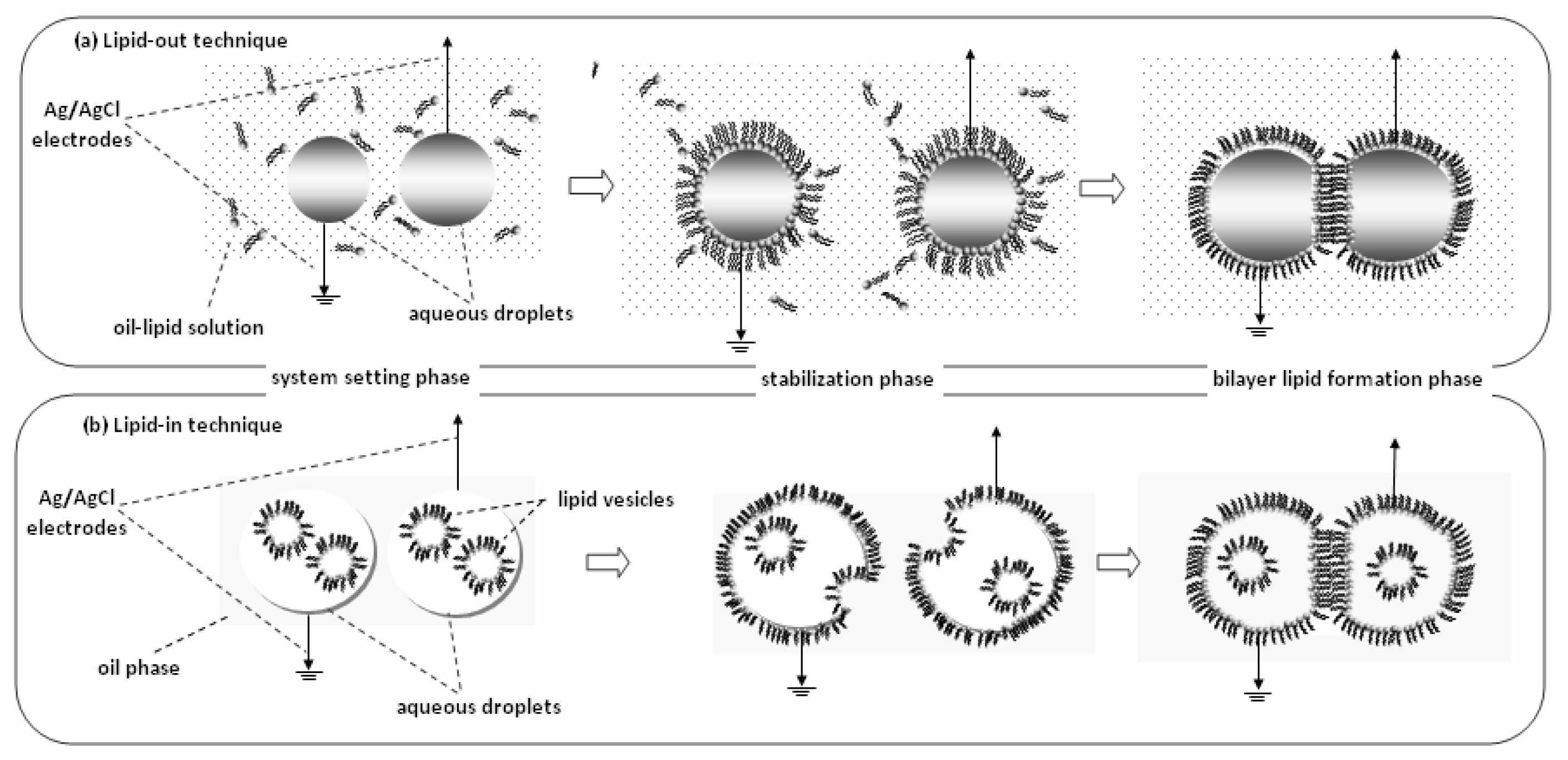
2. Lipid Membrane Platforms: Definitions, Configurations, and Stability
3. Applications, Applicability, and Trends
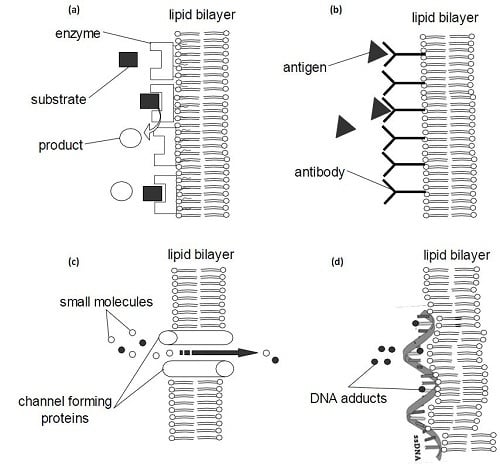
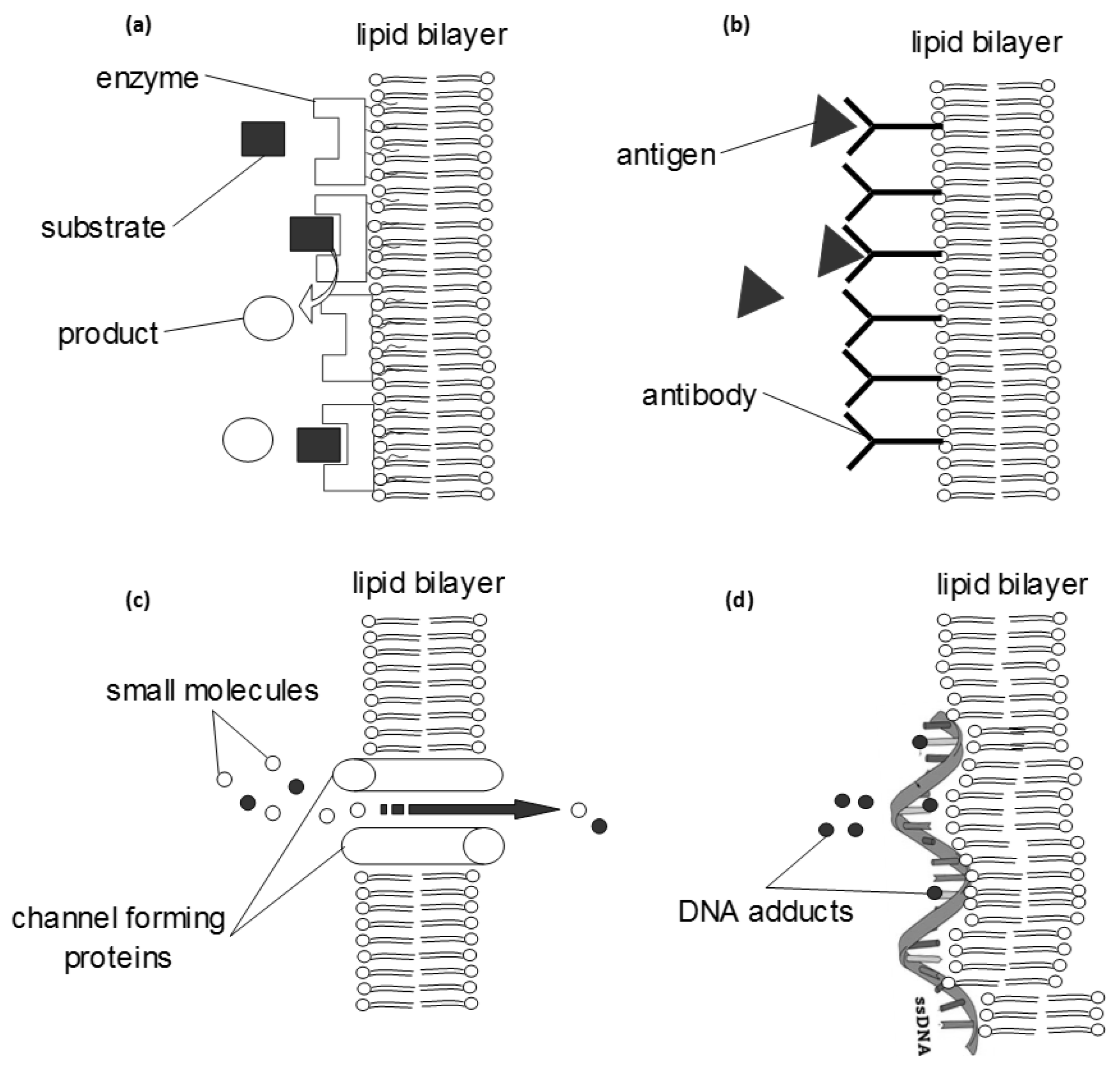
3.1. Biosensors
3.2. Drug Discovery, Delivery, and Testing
3.3. Tools in Research
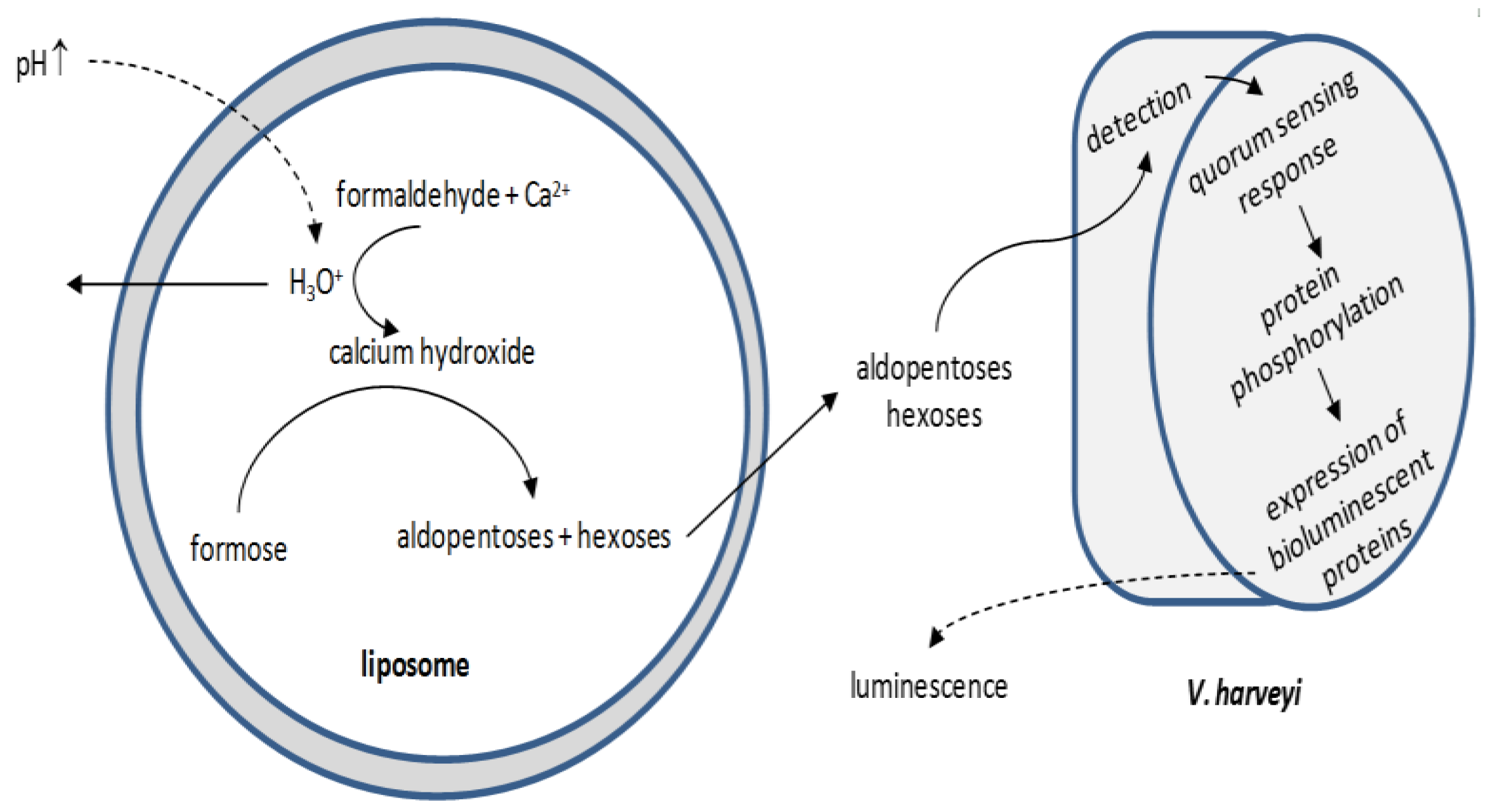
3.4. Current Trends and Future Perspectives
4. Concluding Remarks
Author Contributions
Conflicts of Interest
References
- Williamson, P.; Schlegel, R.A. Back and forth: The regulation and function of transbilayer phospholipid movement in eukaryotic cells. Mol. Membr. Biol. 1994, 11, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Manno, S.; Takakuwa, Y.; Monadas, N. Identification of a functional role for lipid asymmetry in biological membranes: Phosphatidylserine-skeletal protein interactions modulate membrane stability. Proc. Natl. Acad. Sci. USA 2002, 99, 1943–1948. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Goñi, F.M.; Alonso, A.; Bagatolli, L.A.; Brown, R.E.; Marsh, D.; Prieto, M.; Thewalt, J.L. Phase diagrams of lipid mixtures relevant to the study of membrane rafts. Biochim. Biophys. Acta 2008, 1781, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Veatch, S.L.; Soubias, O.; Keller, S.L.; Gawrisch, K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA 2007, 104, 17650–17655. [Google Scholar] [CrossRef] [PubMed]
- Korlach, J.; Schwille, P.; Webb, W.W.; Feigenson, G.W. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 8461–8466. [Google Scholar] [CrossRef] [PubMed]
- Bagatolli, L.A.; Gratton, E. Two photon fluorescence microscopy of coexisting lipid domains in giant unilamellar vesicles of binary phospholipid mixtures. Biophys. J. 2000, 78, 290–305. [Google Scholar] [CrossRef]
- Eeman, M.; Deleu, M. From biological membranes to biomimetic model membranes. Biotechnol. Agron. Soc. Environ. 2010, 14, 719–736. [Google Scholar]
- Madwar, C.; Gopalakrishnan, G.; Lennox, R.B. Interfacing living cells and spherically supported bilayer lipid membranes. Langmuir 2015, 31, 4704–4712. [Google Scholar] [CrossRef] [PubMed]
- Eghiaian, F. Lipid chirality revisited: A change in lipid configuration transforms membrane-bound protein domains. Biophys. J. 2015, 108, 2757–2758. [Google Scholar] [CrossRef] [PubMed]
- Deserno, M. Fluid lipid membranes: From differential geometry to curvature stresses. Chem. Phys. Lipids 2015, 185, 11–45. [Google Scholar] [CrossRef] [PubMed]
- Murate, M.; Kobayashi, T. Revisiting transbilayer distribution of lipids in the plasma membrane. Chem. Phys. Lipids 2016, 194, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G. Bilayer lipid membrane constructs: A strategic technology evaluation approach. In Advanced Bioelectronic Materials; Tiwari, A., Patra, H.K., Turner, A.P.F., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 311–354. [Google Scholar]
- Jin, H.; Jiao, F.; Daily, M.D.; Chen, Y.; Yan, F.; Ding, Y.-H.; Zhang, X.; Robertson, E.J.; Baer, M.D.; Chen, C.-L. Highly stable and self-repairing membrane-mimetic 2D nanomaterials assembled from lipid-like peptoids. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Terrettaz, S.; Ulrich, W.P.; Guerrini, R.; Verdini, A.; Vogel, H. Immunosensing by a synthetic ligand-gated ion channel. Angew. Chem. Int. Ed. 2001, 40, 1740–1743. [Google Scholar] [CrossRef]
- Steck, T.L.; Ye, J.; Lange, Y. Probing red cell membrane cholesterol movement with cyclodextrin. Biophys. J. 2002, 83, 2118–2125. [Google Scholar] [CrossRef]
- Pagano, R.E.; Cherry, R.J.; Chapman, D. Phase transitions and heterogeneity in lipid bilayers. Science 1973, 181, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, G.; Orädd, G. Lipid lateral diffusion and membrane heterogeneity. Biochim. Biophys. Acta 2009, 1788, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Griffin, K.L.; Cheng, C.Y.; Smith, E.A.; Dea, P.K. Effects of pentanol isomers on the phase behavior of phospholipid bilayer membranes. Biophys. Chem. 2010, 152, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.-H.M.; Boxer, S.G. Model membrane systems and their applications. Curr. Opin. Chem. Biol. 2007, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Siontorou, C.G.; Andreou, V.G.; Viras, K.G.; Krull, U.J. Bilayer-lipid membranes as electrochemical detectors for flow injection immunoanalysis. Electroanalysis 1995, 7, 1082–1089. [Google Scholar] [CrossRef]
- Siontorou, C.G.; Nikolelis, D.P.; Piunno, P.A.E.; Krull, U.J. Detection of DNA hybridization using self-assembled bilayer lipid membranes (BLMs). Electroanalysis 1997, 9, 1067–1071. [Google Scholar] [CrossRef]
- Marrink, S.J.; de Vries, A.H.; Tieleman, D.P. Lipids on the move: Simulations of membrane pores; domains; stalks and curves. Biochim. Biophys. Acta 2009, 1788, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Rudin, D.O.; Tien, R.T.; Westcott, W.C. Reconstitution of excitable cell membrane structure in vitro. Circ. Res. 1962, 26, 1167–1171. [Google Scholar] [CrossRef]
- White, S.H. Analysis of the torus surrounding planar lipid bilayer membranes. Biophys. J. 1972, 12, 432–445. [Google Scholar] [CrossRef]
- Montal, M.; Mueller, P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef] [PubMed]
- Hirano-Iwata, A.; Niwano, M.; Sugawara, M. The design of molecular sensing interfaces with lipid-bilayer assemblies. TrAC Trends Anal. Chem. 2008, 27, 512–520. [Google Scholar] [CrossRef]
- Bartsch, P.; Walter, C.; Selenschik, P.; Honigmann, A.; Wagner, R. Horizontal bilayer for electrical and optical recordings. Materials 2012, 5, 2705–2730. [Google Scholar] [CrossRef]
- Warshaviak, D.T.; Muellner, M.J.; Chachisvilis, M. Effect of membrane tension on the electric field and dipole potential of lipid bilayer. Biochim. Biophys. Acta 2011, 1808, 2608–2617. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Batzias, F.A. Innovation in biotechnology: Moving from academic research to product development—The case of biosensors. Crit. Rev. Biotechnol. 2010, 30, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Bavi, N.; Nakayama, Y.; Bavi, O.; Cox, C.D.; Qin, Q.-H.; Martinac, B. Biophysical implications of lipid bilayer rheometry for mechanosensitive channels. Proc. Natl. Acad. Sci. USA 2014, 111, 13864–13869. [Google Scholar] [CrossRef] [PubMed]
- Boucher, P.-A.; Joós, B.; Zuckermann, M.J.; Fournier, L. Pore formation in a lipid bilayer under a tension ramp: Modeling the distribution of rupture tensions. Biophys. J. 2007, 92, 4344–4355. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; de Joannis, J.; Jiang, Y.; Gaulding, J.C.; Albrecht, B.; Yin, F.; Khanna, K.; Kindt, J.T. Bilayer edge and curvature effects on partitioning of lipids by tail length: Atomistic simulations. Biophys. J. 2008, 95, 2647–2657. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Batzias, F.A. A methodological combined framework for roadmapping biosensor research: A fault tree analysis approach within a strategic technology evaluation frame. Crit. Rev. Biotechnol. 2013, 34, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Šnejdárková, M.; Rehák, M.; Passechnik, V.I.; Sokolíková, L.; Sivák, B.; Ivanov, S.A. Electrostriction of lipid bilayers on a solid support and peculiarity of membranes from Archaeal lipids. Thin Solid Films 1996, 284–285, 817–821. [Google Scholar] [CrossRef]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef]
- Orrit, M. From Langmuir–Blodgett films to single molecules. Colloids Surf. B Biointerfaces 2009, 74, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Basu, J.K.; Sanyal, M.K. Ordering and growth of Langmuir–Blodgett films: X-ray scattering studies. Phys. Rep. 2002, 363, 1–84. [Google Scholar] [CrossRef]
- Charitat, T.; Bellet-Amalric, E.; Fragneto, G.; Graner, F. Adsorbed and free lipid bilayers at the solid-liquid interface. Eur. Phys. J. B 1999, 8, 583–593. [Google Scholar] [CrossRef]
- Fragneto, G.; Charitat, T.; Daillant, J. Floating lipid bilayers: Models for physics and biology. Eur. Biophys. J. 2012, 41, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Rebaud, S.; Maniti, O.; Girard-Egrot, A.P. Tethered bilayer lipid membranes (tBLMs): Interest and applications for biological membrane investigations. Biochimie 2014, 107, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Kongsuphol, O.; Fang, K.B.; Ding, Z. Lipid bilayer technologies in ion channel recordings and their potential in drug screening assay. Sens. Actuators B Chem. 2013, 185, 530–542. [Google Scholar] [CrossRef]
- Schuster, B.; Pum, D.; Sleytr, U.B. S-layer stabilized lipid membranes. Biointerphases 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Wilk, S.J.; Petrossian, L.; Goryll, M.; Thornton, T.J.; Goodnick, S.M.; Tang, J.M.; Eisenberg, R.S. Integrated electrodes on a silicon based ion channel measurement platform. Biosens. Bioelectr. 2008, 23, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tabata, K.V.; Noji, H.; Takeuchi, S. Electrophysiological recordings of single ion channels in planar lipid bilayers using a polymethyl methacrylate microfluidic chip. Biosens. Bioelectr. 2007, 22, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Mitev, D.J.; Ivanova, T.; Vassilieff, C.S. Kinetics of lipid layer formation at interfaces. Colloids Surf. B 2002, 24, 185–192. [Google Scholar] [CrossRef]
- Capitani, D.; Segre, A.L.; Dreher, F.; Walde, P.; Luisi, P.L. Multinuclear NMR investigation of phosphatidylcholine organogels. J. Phys. Chem. 1996, 100, 15211–15217. [Google Scholar] [CrossRef]
- Bayley, H.; Cronin, B.; Heron, A.; Holden, M.A.; Hwang, W.; Syeda, R.; Thompson, J.; Wallace, M. Droplet interface bilayers. Mol. Biosyst. 2008, 4, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.L.; Chen, M.; Cronin, B.; Holden, M.A.; Bayley, H. Asymmetric droplet interface bilayers. J. Am. Chem. Soc. 2008, 130, 5878–5879. [Google Scholar] [CrossRef] [PubMed]
- Szabo, M.; Wallace, M.I. Imaging potassium-flux through individual electropores in droplet interface bilayers. Biochim. Biophys. Acta 2016, 1858, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.A.; Needham, D.; Bayley, H. Functional bionetworks from nanoliter water droplets. J. Am. Chem. Soc. 2007, 129, 8650–8655. [Google Scholar] [CrossRef] [PubMed]
- Reimhult, E.; Baumann, M.; Kaufmann, S.; Kumar, K.; Spycher, P. Advances in nanopatterned and nanostructured supported lipid membranes and their applications. Biotechnol. Genet. Eng. Rev. 2010, 27, 185–216. [Google Scholar] [CrossRef] [PubMed]
- Kam, L.C. Capturing the nanoscale complexity of cellular membranes in supported lipid bilayers. J. Struct. Biol. 2009, 168, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bratakou, S.; Nikoleli, G.-P.; Siontorou, C.G.; Nikolelis, D.P.; Karapetis, S.; Tzamtzis, N. Development of an electrochemical biosensor for the rapid detection of saxitoxin based on air stable lipid films with incorporated anti-STX using graphene electrodes. Electroanalysis 2017, 29, 990–997. [Google Scholar] [CrossRef]
- Karapetis, S.; Nikoleli, G.-P.; Siontorou, C.G.; Nikolelis, D.P.; Tzamtzis, N.; Psaroudakis, N. Development of an electrochemical biosensor for the rapid detection of cholera toxin based on air stable lipid films with incorporated ganglioside GM1 using graphene electrodes. Electroanalysis 2016, 28, 1584–1590. [Google Scholar] [CrossRef]
- Shashi, K.; Satinder, K.; Bharat, P. A complete review on: Liposomes. Int. Res. J. Pharm. 2012, 3, 10–16. [Google Scholar]
- Patil, Y.P.; Jadhav, S. Novel methods for liposome preparation. Chem. Phys. Lipids 2014, 177, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, E.; Gomes, A.C.; Preto, A.; Cavaco-Paulo, A. Design of liposomal formulations for cell targeting. Colloids Surf. B 2015, 136, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Ollivon, M.; Lesieur, S.; Grabielle-Madelmont, C.; Paternostre, M. Vesicle reconstitution from lipid–Detergent mixed micelles. Biochim. Biophys. Acta 2000, 1508, 34–50. [Google Scholar] [CrossRef]
- Lesoin, L.; Crampon, C.; Boutin, O.; Badens, E. Preparation of liposomes using the supercritical anti-solvent (SAS) process and comparison with a conventional method. J. Supercrit. Fluids 2011, 57, 162–174. [Google Scholar] [CrossRef]
- Wagner, A.; Vorauer-Uhl, K. Liposome technology for industrial purposes. J. Drug Deliv. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Gubernator, J. Active methods of drug loading into liposomes: Recent strategies for stable drug entrapment and increasedin vivo activity. Expert Opin. Drug Deliv. 2011, 8, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Mayer, L.D.; Hope, M.J.; Cullis, P.R.; Janoff, A.S. Solute distributions and trapping efficiencies observed in freeze-thawed multilamellar vesicles. Biochim. Biophys. Acta 1985, 817, 193–196. [Google Scholar] [CrossRef]
- Bally, M.B.; Mayer, L.D.; Loughrey, H.; Redelmeier, T.; Madden, T.D.; Wong, K.; Harrigan, P.R.; Hope, M.J.; Cullis, P.R. Dopamine accumulation in large unilamellar vesicle systems induced by transmembrane ion gradients. Chem. Phys. Lipids 1988, 47, 97–107. [Google Scholar] [CrossRef]
- Chou, T.H.; Chen, S.C.; Chu, I.M. Effect of composition on the stability of liposomal irinotecan prepared by a pH gradient method. J. Biosci. Bioeng. 2003, 95, 405–408. [Google Scholar] [CrossRef]
- Abraham, S.A.; McKenzie, C.; Masin, D.; Ng, R.; Harasym, T.O.; Mayer, L.D.; Bally, M.B. In vitro andin vivo characterization of doxorubicin and vincristine coencapsulated within liposomes through use of transition metal ion complexation and pH gradient loading. Clin. Cancer Res. 2004, 10, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Vabbilisetty, P.; Sun, X.-L. Liposome surface functionalization based on different anchoring lipids via Staudinger ligation. Org. Biomol. Chem. 2014, 12, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Nobs, L.; Buchegger, F.; Gurny, R.; Allémann, E. Current methods for attaching targeting ligands to liposomes and nanoparticles. J. Pharm. Sci. 2004, 93, 1980–1992. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.P.; Berat, R.; Brisson, A.R. Formation of solid-supported lipid bilayers: An integrated view. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef] [PubMed]
- Troutier, A.-L.; Ladavière, C. An overview of lipid membrane supported by colloidal particles. Adv. Colloid Interface Sci. 2007, 133, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, G.; Rouiller, I.; Colman, D.R.; Lennox, R.B. Supported bilayers formed from different phospholipids on spherical silica substrates. Langmuir 2009, 25, 5455–5458. [Google Scholar] [CrossRef] [PubMed]
- Mornet, S.; Lambert, O.; Duguet, E.; Brisson, A. The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy. Nano Lett. 2005, 5, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Zhang, Z.; Taniguchi, T.; Sohgawa, M.; Yamashita, K.; Noda, M. A high-sensitive detection of several tens of nM of amyloid-β by cantilever-type biosensor immobilized DPPC liposome incorporated with cholesterol. Proc. Eng. 2016, 168, 565–568. [Google Scholar] [CrossRef]
- Zhang, Z.; Sohgawa, M.; Yamashita, K.; Noda, M. Real-time characterization of fibrillization process of amyloid-beta on phospholipid membrane using a new label-free detection technique based on a cantilever-based liposome biosensor. Sens. Actuators B Chem. 2016, 236, 893–899. [Google Scholar] [CrossRef]
- Nikolelis, D.P.; Mitrokotsa, M. Stabilized lipid film based biosensor for atenolol. Biosens. Bioelectron. 2002, 17, 565–572. [Google Scholar] [CrossRef]
- Weingart, O.G.; Loessner, M.J. Nerve cell-mimicking liposomes as biosensor for botulinum neurotoxin complete physiological activity. Toxicol. Appl. Pharm. 2016, 313, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Raftopoulou, G.; Simantiraki, M.; Psaroudakis, N.; Nikoleli, G.-P.; Hianik, T. Preparation of a selective receptor for carbofuran for the development of a simple optical spot test for its rapid detection using stabilized in air lipid films with incorporated receptor. Anal. Chim. Acta 2008, 620, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Bratakou, S.; Nikoleli, G.-P.; Nikolelis, D.P.; Psaroudakis, N. Development of a potentiometric chemical sensor for the rapid detection of carbofuran based on air stable lipid films with incorporated calix [4] arene phosphoryl receptor using graphene electrodes. Electroanalysis 2015, 27, 2608–2613. [Google Scholar] [CrossRef]
- Nikolelis, D.P.; Simantiraki, M.; Siontorou, C.G. Flow injection analysis of carbofuran in foods using air stable lipid film based acetylcholinesterase biosensor. Anal. Chim. Acta 2005, 537, 169–177. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Jiang, J.-H.; Wu, H.L.; Shen, G.L.; Yu, R.Q. An ultrasensitive chemiluminescence biosensor for cholera toxin based on ganglioside-functionalized supported lipid membrane and liposome. Biosens. Bioelectron. 2008, 24, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Nikoleli, G.-P.; Ibupoto, Z.H.; Nikolelis, D.P.; Likodimos, V.; Psaroudakis, N.; Tzamtzis, N.; Willander, M.; Hianik, T. Potentiometric cholesterol biosensing application of graphene electrode with stabilized polymeric lipid membrane. Cent. Eur. J. Chem. 2013, 11, 1554–1561. [Google Scholar] [CrossRef]
- Nikoleli, G.-P.; Nikolelis, D.P.; Tzamtzis, N.; Psaroudakis, N. A selective immunosensor for d-dimer based on antibody immobilized on a graphene electrode with incorporated lipid films. Electroanalysis 2014, 26, 1522–1527. [Google Scholar] [CrossRef]
- Guan, H.; Zhang, H.; Yu, J.; Chi, D. The novel acetylcholinesterase biosensors based on liposome bioreactors–Chitosan nanocomposite film for detection of organophosphates pesticides. Food Res. Int. 2012, 49, 15–21. [Google Scholar] [CrossRef]
- Fritzen-Garcia, M.B.; Zoldan, V.C.; Oliveira, I.R.W.; Soldi, V.; Pasa, A.A.; Creczynski-Pasa, T.B. Peroxidase immobilized on phospholipid bilayers supported on au (111) by DTT self-assembled monolayers: Application to dopamine determination. Biotechnol. Bioeng. 2013, 110, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Raftopoulou, G.; Chatzigeorgiou, P.; Nikoleli, G.-P.; Viras, K. Optical portable biosensors based on stabilized lipid membrane for the rapid detection of doping materials in human urine. Sens. Actuators B Chem. 2008, 130, 577–582. [Google Scholar] [CrossRef]
- Jiao, T.; Leca-Bouvier, B.D.; Boullanger, P.; Blum, L.J.; Girard-Egrot, A.P. A chemiluminescent Langmuir–Blodgett membrane as the sensing layer for the reagentless monitoring of an immobilized enzyme activity. Colloids Surf. A Physicochem. Eng. Asp. 2010, 354, 284–290. [Google Scholar] [CrossRef]
- Graça, J.S.; de Oliveira, R.F.; de Moraes, M.L.; Ferreira, M. Amperometric glucose biosensor based on layer-by-layer films of microperoxidase-11 and liposome-encapsulated glucose oxidase. Bioelectrochemistry 2014, 96, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Luna, D.M.N.; Oliveira, M.D.L.; Nogueira, M.L.; Andrade, C.A. Biosensor based on lectin and lipid membranes for detection of serum glycoproteins in infected patients with dengue. Chem. Phys. Lipids 2014, 180, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Nikolelis, D.P.; Krull, U.J. Flow injection monitoring and analysis of mixtures of hydrazine compounds using filter-supported bilayer lipid membranes with incorporated DNA. Anal. Chem. 2000, 72, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Xiong, L.; Zheng, D.; Li, Y.; Liu, Q.; Han, K.; Liu, W.; Tao, K.; Yang, S.; Xia, J. Bilayer lipid membrane biosensor with enhanced stability for amperometric determination of hydrogen peroxide. Talanta 2011, 85, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Bratakou, S.; Nikoleli, G.-P.; Siontorou, C.G.; Karapetis, S.; Nikolelis, D.P.; Tzamtzis, N. Electrochemical biosensor for naphthalene acetic acid in fruits and vegetables based on lipid films with incorporated auxin-binding protein receptor using graphene electrodes. Electroanalysis 2016, 28, 2171–2177. [Google Scholar] [CrossRef]
- Siontorou, C.G.; Georgopoulos, K.N. A biosensor platform for soil management: The case of nitrites. J. Clean. Prod. 2016, 111, 133–142. [Google Scholar] [CrossRef]
- Michaloliakos, A.I.; Nikoleli, G.-P.; Siontorou, C.G.; Nikolelis, D.P. Rapid flow injection electrochemical detection of Arochlor 1242 using stabilized lipid membranes with incorporated sheep anti-PCB antibody. Electroanalysis 2012, 24, 495–501. [Google Scholar] [CrossRef]
- Siontorou, C.G.; Nikolelis, D.P.; Krull, U.J.; Chiang, K.-L. A triazine herbicide minisensor based on surface-stabilized bilayer lipid membranes. Anal. Chem. 1997, 69, 3109–3114. [Google Scholar] [CrossRef] [PubMed]
- Nikoleli, G.-P.; Israr, M.Q.; Tzamtzis, N.; Nikolelis, D.P.; Willander, M.; Psaroudakis, N. Structural characterization of graphene nanosheets for miniaturization of potentiometric urea lipid film based biosensors. Electroanalysis 2012, 24, 1285–1295. [Google Scholar] [CrossRef]
- Tzamtzis, N.; Psychoyios, V.N.; Nikoleli, G.-P.; Nikolelis, D.P.; Psaroudakis, N.; Willander, M.; Israr, M.Q. Flow potentiometric injection analysis of uric acid using lipid stabilized films with incorporated uricase on ZnO nanowires. Electroanalysis 2012, 24, 1719–1725. [Google Scholar] [CrossRef]
- Nikolelis, D.P.; Theoharis, G. Rapid detection of vanillin in alcoholic beverages using stabilized polymerized lipid film based biosensors. Electroanalysis 2002, 14, 1661–1667. [Google Scholar] [CrossRef]
- Majd, S.; Yusko, E.C.; Billeh, Y.N.; Macrae, M.X.; Yang, J.; Mayer, M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010, 21, 439–476. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J.; Bezrukov, S.M. Protonation dynamics of the alpha-toxin ion channel from spectral analysis of pH-dependent current fluctuations. Biophys. J. 1995, 69, 94–105. [Google Scholar] [CrossRef]
- Gurnev, P.A.; Nestorovich, E.M. Channel-forming bacterial toxins in biosensing and macromolecule delivery. Toxins 2014, 6, 2483–2540. [Google Scholar] [CrossRef] [PubMed]
- Nikolelis, D.P.; Siontorou, C.G.; Krull, U.J.; Katrivanos, P.L. Ammonium ion minisensors from self-assembled bilayer lipid membranes using gramicidin as an ionophore. Modulation of ammonium selectivity by platelet-activating factor. Anal. Chem. 1996, 68, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef] [PubMed]
- Michot, J.M.; Seral, C.; Van Bambeke, F.; Tulkens, P.M. Influence of efflux transporters on the accumulation and efflux of four quinolones (ciprofloxacin; levofloxacin; garenoxacin; and moxifloxacin) in J774 macrophages. Antimicrob. Agents Chemother. 2005, 49, 2429–2437. [Google Scholar] [CrossRef] [PubMed]
- Bensikaddour, H.; Snoussi, K.; Lins, L.; Van Bambeke, F.; Tulkens, P.M.; Brasseur, R.; Goormaghtigh, E.; Mingeot-Leclercq, M.-P. Interactions of ciprofloxacin with DPPC and DPPG: Fluorescence anisotropy; ATR-FTIR and 31P NMR spectroscopies and conformational analysis. Biochim. Biophys. Acta 2008, 1778, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Fa, N.; Ronkart, S.; Schanck, A.; Deleu, M.; Gaigneaux, A.; Goormaghtigh, E.; Mingeot-Leclercq, M.P. Effect of the antibiotic azithromycin on thermotropic behavior of DOPC or DPPC bilayers. Chem. Phys. Lipids 2006, 144, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Fa, N.; Lins, L.; Courtoy, P.J.; Dufrêne, Y.; van der Smissen, P.; Brasseur, R.; Tyteca, D.; Mingeot-Leclercq, M.P. Decrease of elastic moduli of DOPC bilayers induced by a macrolide antibiotic, azithromycin. Biochim. Biophys. Acta 2007, 1768, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Klopman, G.; Zhu, H. Recent methodologies for the estimation of n-octanol/water partition coefficients and their use in the prediction of membrane transport properties of drugs. Mini Rev. Med. Chem. 2005, 5, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Gameiro, P.; Reis, S.; Lima, J.L.; de Castro, B. Derivative spectrophotometry as a tool for the determination of drug partition coefficients in water/dimyristoyl-L-alpha-phosphatidylglycerol (DMPG) liposomes. Biophys. Chem. 2001, 94, 97–106. [Google Scholar] [CrossRef]
- Baciu, M.; Sebai, S.C.; Ces, O.; Mulet, X.; Clarke, J.A.; Shearman, G.C.; Law, R.V.; Templer, R.H.; Plisson, C.; Parker, C.A.; et al. Degradative transport of cationic amphiphilic drugs across phospholipid bilayers. Philos. Trans. R. Soc. A 2006, 364, 2597–2614. [Google Scholar] [CrossRef] [PubMed]
- Pavinatto, F.J.; Caseli, L.; Pavinatto, A.; dos Santos, D.S., Jr.; Nobre, T.M.; Zaniquelli, M.E.D.; Silva, H.S.; Miranda, P.B.; de Oliveira, O.N., Jr. Probing chitosan and phospholipid interactions using Langmuir and Langmuir-Blodgett films as cell membrane models. Langmuir 2007, 23, 7666–7671. [Google Scholar] [CrossRef] [PubMed]
- Yusupov, M.; Van der Paal, J.; Neyts, E.C.; Bogaerts, A. Synergistic effect of electric field and lipid oxidation on the permeability of cell membranes. Biochim. Biophys. Acta 2017, 1861, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; Gran, M.L.; Peppas, N.A. Targeted nanodelivery of drugs and diagnostics. Nano Today 2010, 5, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Huynh, R.; Chaubet, F.; Jozefonvicz, J. Anticoagulant properties of dextranmethylcarboxylate benzylamide sulfate (DMCBSu); a new generation of bioactive functionalized dextran. Carbohydr. Res. 2001, 332, 75–83. [Google Scholar] [CrossRef]
- Barrera, C.; Herrera, A.P.; Rinaldi, C. Colloidal dispersions of monodisperse magnetite nanoparticles modified with poly(ethylene glycol). J. Colloid Interface Sci. 2009, 329, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, G.; Danelon, C.; Izewska, P.; Prummer, M.; Bolinger, P.Y.; Geissbühler, I.; Demurtas, D.; Dubochet, J.; Vogelm, H. Multifunctional lipid/quantum-dot hybrid nanocontainers for controlled targeting of live cells. Angew. Chem. Int. Ed. 2006, 45, 5478–5483. [Google Scholar] [CrossRef] [PubMed]
- Thanh, N.T.K.; Green, L.A.W. Functionalisation of nanoparticles for biomedical applications. Nano Today 2010, 5, 213–230. [Google Scholar] [CrossRef]
- Matsuno, R.; Ishihara, K. Integrated functional nanocolloids covered with artificial cell membranes for biomedical applications. Nano Today 2011, 6, 61–74. [Google Scholar] [CrossRef]
- Geissbuehler, I.; Hovius, R.; Martinez, K.L.; Adrian, M.; Thampi, K.R.; Vogel, H. Lipid-coated nanocrystals as multifunctionalized luminescent scaffolds for supramolecular biological assemblies. Angew. Chem. Int. Ed. 2005, 44, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Dubertret, B.; Skourides, P.; Norris, D.J.; Noireaux, V.; Brivanlou, A.H.; Libchaber, A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 2002, 298, 1759–1762. [Google Scholar] [CrossRef] [PubMed]
- Zakharian, E. Recording of ion channel activity in planar lipid bilayer experiments. Methods Mol. Biol. 2013, 998, 109–118. [Google Scholar] [PubMed]
- Kullman, L.; Winterhalter, M.; Bezrukov, S.M. Transport of maltodextrins through maltoporin: A single-channel study. Biophys. J. 2002, 82, 803–812. [Google Scholar] [CrossRef]
- White, R.J.; Ervin, E.N.; Yang, T.; Chen, X.; Daniel, S.; Cremer, P.S.; White, H.S. Single ion-channel recordings using glass nanopore membranes. J. Am. Chem. Soc. 2007, 129, 11766–11775. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Tsuji, Y.; Sato, K.; Osaki, T.; Kamiya, K.; Hirano, M.; Ide, T.; Miki, N.; Takeuchi, S. Automated parallel recordings of topologically identified single ion channels. Sci. Rep. 2013, 3, 1995. [Google Scholar] [CrossRef] [PubMed]
- Sadler, E.E.; Kapanidis, A.N.; Tucker, S.J. Solution-based single-molecule FRET studies of K+ channel gating in a lipid bilayer. Biophys. J. 2016, 110, 2663–2670. [Google Scholar] [CrossRef] [PubMed]
- Volpe, D.A. Application of method suitability for drug permeability classification. AAPS J. 2010, 12, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Wohnsland, F.; Faller, B. High-throughput permeability pH profile and high-throughput alkane/water log P with artificial membranes. J. Med. Chem. 2001, 44, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Flaten, G.E.; Dhanikula, A.B.; Luthman, K.; Brandl, M. Drug permeability across a phospholipid vesicle based barrier: A novel approach for studying passive diffusion. Eur. J. Pharm. Sci. 2006, 27, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Corti, G.; Maestrelli, F.; Cirri, M.; Zerrouk, N.; Mura, P. Development and evaluation of an in vitro method for prediction of human drug absorption II. Demonstration of the method suitability. Eur. J. Pharm. Sci. 2006, 27, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Hamill, O.P.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers. Arch. 1981, 391, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, M.; Smart, T.G. Single-channel recording of ligand-gated ion channels. Nat. Protoc. 2007, 2, 2826–2841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, J.; Huang, W. Study of the ion-channel behavior on glassy carbon electrode supported bilayer lipid membranes stimulated by perchlorate anion. Mater. Sci. Eng. C 2015, 55, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Stankowski, S.; Rizzo, V. Thermodynamics analysis of incorporation and aggregation in a membrane: Application to the pore-forming peptide alamethicin. Biochim. Biophys. Acta 1986, 861, 141–151. [Google Scholar] [CrossRef]
- Tosteson, M.T.; Alvarez, O.; Hubbell, W.; Bieganski, R.M.; Attenbach, C.; Caporales, L.H.; Levy, J.J.; Nutt, R.F.; Rosenblatt, M.; Tosteson, D.C. Primary structure of peptides and ion channels. Role of amino acid side chains in voltage gating of melittin channels. Biophys. J. 1990, 58, 1367–1375. [Google Scholar] [CrossRef]
- Guidelli, R.; Becucci, L. Mechanism of voltage-gated channel formation in lipid membranes. Biochim. Biophys. Acta 2016, 1858, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Dosoky, N.S.; Williams, J.D. Engineering lipid bilayer membranes for protein studies. Int. J. Mol. Sci. 2013, 14, 21561–21597. [Google Scholar] [CrossRef] [PubMed]
- Otera, H.; Ishihara, N.; Mihara, K. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 2013, 1833, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Kultz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005, 67, 225–257. [Google Scholar] [CrossRef] [PubMed]
- Török, Z.; Crul, T.; Maresca, B.; Schütz, G.J.; Viana, F.; Dindia, L.; Piotto, S.; Brameshuber, M.; Balogh, G.; Péter, M.; et al. Plasma membranes as heat stress sensors: From lipid-controlled molecular switches to therapeutic applications. Biochim. Biophys. Acta 2014, 1838, 1594–1618. [Google Scholar] [CrossRef] [PubMed]
- Školová, B.; Kováčik, A.; Tesař, O.; Opálka, L.; Vávrová, K. Phytosphingosine; sphingosine and dihydrosphingosine ceramides in model skin lipid membranes: Permeability and biophysics. Biochim. Biophys. Acta 2017, 1859, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Baksh, M.M.; Dean, C.; Pautot, S.; DeMaria, S.; Isacoff, E.; Groves, J.T. Neuronal activation by GPI-linked neuroligin-1 displayed in synthetic lipid bilayer membranes. Langmuir 2005, 21, 10693–10698. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, G.; Thostrup, P.; Rouiller, I.; Lucido, A.L.; Belkaid, W.; Colman, D.R.; Lennox, R.B. Lipid bilayer membrane-triggered presynaptic vesicle assembly. ACS Chem. Neurosci. 2010, 1, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Madwar, C.; Gopalakrishnan, G.; Lennox, R.B. Lipid Microdomains in Synapse Formation. ACS Chem. Neurosci. 2016, 7, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hu, S.; Chen, X. Artificial cells: From basic science to applications. Mater. Today 2016, 19, 516–532. [Google Scholar] [CrossRef] [PubMed]
- Oberholzer, T.; Albrizio, M.; Luisi, P.L. Polymerase chain reaction in liposomes. Chem. Biol. 1995, 2, 677–682. [Google Scholar] [CrossRef]
- Oberholzer, T.; Nierhaus, K.H.; Luisi, P.L. Protein expression in liposomes. Biochem. Biophys. Res. Commun. 1999, 261, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Sato, K.; Wakabayashi, M.; Nakaishi, T.; Ko-Mitamura, E.P.; Shima, Y.; Urabe, I.; Yomo, T. Synthesis of functional protein in liposome. J. Biosci. Bioeng. 2001, 92, 590–593. [Google Scholar] [CrossRef]
- Kuruma, Y.; Stano, P.; Ueda, T.; Luisi, P.L. A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim. Biophys. Acta 2009, 1788, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Noga, M.J.; de Graaf, P.; Westerlaken, I.; Yildirim, E.; Danelon, C. Cell-free phospholipid biosynthesis by gene-encoded enzymes reconstituted in liposomes. PLoS ONE 2016, 11, e0163058. [Google Scholar] [CrossRef] [PubMed]
- Nourian, Z.; Roelofsen, W.; Danelon, C. Triggered gene expression in fed-vesicle microreactors with a multifunctional membrane. Angew. Chem. Int. Ed. 2012, 51, 3114–3118. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.M.; Winzer, K.; Davis, B.G. Sugar synthesis in a protocellular model leads to a cell signalling response in bacteria. Nat. Chem. 2009, 1, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Lentini, R.; Santero, S.P.; Chizzolini, F.; Cecchi, D.; Fontana, J.; Marchioretto, M.; del Bianco, C.; Terrell, J.L.; Spencer, A.C.; Martini, L.; et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Berclaz, N.; Müller, M.; Walde, P.; Luisi, P.L. Growth and transformation of vesicles studied by ferritin labeling and cryo-transmission electron microscopy. J. Phys. Chem. B 2001, 105, 1056–1064. [Google Scholar] [CrossRef]
- Zhu, T.F.; Szostak, J.W. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 2009, 131, 5705–5713. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K.; Tamura, M.; Shohda, K. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 2011, 3, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Lentini, R.; Martín, N.Y.; Mansy, S.S. Communicating artificial cells. Curr. Opin. Chem. Biol. 2016, 34, 53–61. [Google Scholar] [CrossRef] [PubMed]
| Method | Description | Types of Liposomes Produced | Ref. |
|---|---|---|---|
| Physical dispersion: lipid film hydration by shaking (Bangham method) | Lipids are dissolved in a mixture of solvents in a round bottom flask; solvent evaporation leaves a thin film at the bottom that subsequently is rehydrated with an aqueous buffer. The compounds to be encapsulated can be added either at the solvent mixture or the aqueous buffer. | multilamellar and giant unilamellar vesicles Size reduction (as post-treatment): small unilamellar vesicles (micro-emulsification, bath or probe sonication followed by ultra centrifugation); oligolamellar and/or large unilamellar vesicles (membrane extrusion); small unilamellar vesicles of complex architecture (freeze-thaw sonication) | [56] |
| Physical dispersion: lipid film hydration by non-shaking | Lipids dissolved in organic solvent are freeze dried prior to addition of aqueous buffer. Alternatively, the film is deposited on electrodes and subsequently hydrated in the presence of anelectric field. | multilamellar and giant unilamellar vesicles Size reduction: as above | [56,57] |
| Solvent dispersion: ethanol or ether injection | Lipids in solvent are mixed with the aqueous phase that contains the components to be encapsulated. | small unilamellar vesicles | [58] |
| Solvent dispersion: reverse phase evaporation | A water-in-oil emulsion is formed; the evaporation of the organic phase produces an aqueous suspension of vesicles. | small and large unilamellar vesicles | [57] |
| Detergent solubilization: micelle–vesicle transition | Detergents are used for the solubilization of lipids in micellar systems; the vesicles are released through dilution, gel chromatography, hollow fiber dialysis, membrane filtration, or adsorption to hydrophobic matrix (resins or dextrins). | multilamellar, oligolamellar, large unilamellar vesicles (dialysis); small unilamellar vesicles (gel chromatography, filtration, adsorption) | [59] |
| Proliposomes: hydration | Proliposomes are formed by drying a lipid solution; solvent removal proceeding with rotary vacuum evaporation, fluidized bed adsorption or spray drying. When diluted in aqueous phase (along with the components to be encapsulated), a vesicle dispersion is produced; encapsulation efficiencies are high and the products can be sterilized. | multilamellar vesicles | [58] |
| Supercritical fluid technology: anti-solvent method and reverse phase evaporation | In the anti-solvent method, the lipids dissolve in supercritical CO2 and then precipitate in the form of ultra-fine particles. In reverse phase evaporation, supercritical CO2 is used instead of conventional solvents. | multilamellar and giant unilamellar vesicles (anti-solvent method); small and large unilamellar vesicles (reverse phase evaporation) | [60] |
| Microfluidic methods: hydrodynamic focusing, droplets, pulsed jet flow, thin film hydration | Microfluidics offer micro-to nanoliter volumes of vesicles dispersions and precise control over production. | small unilamellar vesicles (micro hydrodynamic focusing); giant unilamellar vesicles (microfluidic droplets and pulsed jet flow microfluidics); large unilamellar vesicles (thin film hydration in microtubes) | [57] |
| Analyte | Biological System/Membrane | Transducer Type | Detection Limit | Ref. |
|---|---|---|---|---|
| Amyloid-β protein | Cholesterol incorporated liposomes | Micro-cantilever with NiCr thin film strain gauge | 75 nM | [73] |
| Amyloid-β protein, real time continuous monitoring of fibrilization | Liposomes | Micro-cantilever with NiCr thin film strain gauge | 1 μΜ | [74] |
| Atenolol | Polymerized membranes | Ag/AgCl electrodes | 20 μM | [75] |
| Botulinum neurotoxin | Trisialoganglioside functionalized liposomes | Fluorescence | – | [76] |
| Carbofuran pesticidein foods | Resorcinarene receptor/glass filter supported membranes | Fluorescence | 1 nM | [77] |
| Carbofuran pesticide in foods | Calixarene receptor/polymerized membranes | Graphene-nanosheets- based electrodes | 1 μΜ | [78] |
| Carbofuran pesticide in foods | Acetylcholinesterase/polymerized membranes | Ag/AgCl electrodes | 1 nM | [79] |
| Cholera toxin | Ganglioside GM1/liposomes | Chemiluminescence | 0.8 pM | [80] |
| Cholera toxin in water | Ganglioside GM1/polymerized membranes | Graphene-nanosheets- based electrodes | 1 nM | [55] |
| Cholesterol | Cholesterol oxidase/polymerized membranes | Graphene-nanosheets-based electrodes | 0.1 μM | [81] |
| d-dimer | Antibody/polymerized membranes | Graphene-nanosheets- based electrodes | 1 μM | [82] |
| Dichlorvos pesticide | Acetylcholinesterase/liposome-chitosan nanocomposite | Ag/AgCl electrodes | 0.25 μM | [83] |
| Dopamine | Peroxidase/dithiotreitol supported membranes | Au electrode | 2 μM | [84] |
| Dopamine in human urine | Pirogallolarene receptor/polymerized membranes | Fluorescence | 1 nM | [85] |
| Enzyme activity, reagentless monitoring of | Langmuir-Blodgett membranes | Electro-chemiluminescence | – | [86] |
| Ephedrine in human urine | Permethoxy receptor/polymerized membranes | Fluorescence | 1 nM | [85] |
| Glucose | Glucose oxidase/microperoxidase functionalized liposomes | Indium-tin oxide (ITO) electrode | 8.6 μM | [87] |
| Glycoproteins in serum | Concanavalin A/liposomes | Electrochemical impedance spectroscopy | not reported | [88] |
| Hydrazine pesticides in water | DNA/glass filter supported membranes | Ag/AgCl electrodes | 78 pM | [89] |
| Hydrogen peroxide | Peroxidase/polymerized membrane | Electrochemical impedance spectroscopy | 0.1 μΜ | [90] |
| Naphthalene acetic acid in foods | Auxin-binding protein receptor/polymerized membranes | Graphene-nanosheets-based electrodes | 10 nM | [91] |
| Nitrites in soil | Methaemoglobin/metal- supported membranes | Ag/AgCl electrodes | 0.9 μg/L | [92] |
| Polychlorinated biphenyls (arochlor) | Antibody/polymerized membranes | Ag/AgCl electrodes | 10 pM | [93] |
| Saxitoxin in foods and water | Anti-saxitoxin receptor/polymerized membranes | Graphene-nanosheets-based electrodes | 1 nM | [54] |
| Triazine herbicides in water | Metal supported membranes | Ag/AgCl electrodes | 15 nM | [94] |
| Urea | Urease/polymerized membranes | Graphene-nanosheets-based electrodes | 1 μM | [95] |
| Uric acid | Uricase/polymerized membranes | ZnO nanowires-based electrodes | 1 μM | [96] |
| Vanillin in alcoholic beverages and wine | Polymerized membranes | Ag/AgCl electrodes | 0.3 μM | [97] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siontorou, C.G.; Nikoleli, G.-P.; Nikolelis, D.P.; Karapetis, S.K. Artificial Lipid Membranes: Past, Present, and Future. Membranes 2017, 7, 38. https://doi.org/10.3390/membranes7030038
Siontorou CG, Nikoleli G-P, Nikolelis DP, Karapetis SK. Artificial Lipid Membranes: Past, Present, and Future. Membranes. 2017; 7(3):38. https://doi.org/10.3390/membranes7030038
Chicago/Turabian StyleSiontorou, Christina G., Georgia-Paraskevi Nikoleli, Dimitrios P. Nikolelis, and Stefanos K. Karapetis. 2017. "Artificial Lipid Membranes: Past, Present, and Future" Membranes 7, no. 3: 38. https://doi.org/10.3390/membranes7030038
APA StyleSiontorou, C. G., Nikoleli, G.-P., Nikolelis, D. P., & Karapetis, S. K. (2017). Artificial Lipid Membranes: Past, Present, and Future. Membranes, 7(3), 38. https://doi.org/10.3390/membranes7030038