Abstract
In recent years, aquaporin biomimetic membranes (ABMs) for water separation have gained considerable interest. Although the first ABMs are commercially available, there are still many challenges associated with further ABM development. Here, we discuss the interplay of the main components of ABMs: aquaporin proteins (AQPs), block copolymers for AQP reconstitution, and polymer-based supporting structures. First, we briefly cover challenges and review recent developments in understanding the interplay between AQP and block copolymers. Second, we review some experimental characterization methods for investigating AQP incorporation including freeze-fracture transmission electron microscopy, fluorescence correlation spectroscopy, stopped-flow light scattering, and small-angle X-ray scattering. Third, we focus on recent efforts in embedding reconstituted AQPs in membrane designs that are based on conventional thin film interfacial polymerization techniques. Finally, we describe some new developments in interfacial polymerization using polyhedral oligomeric silsesquioxane cages for increasing the physical and chemical durability of thin film composite membranes.
1. Introduction
Aquaporin biomimetic membranes (ABMs) have attracted interest recently due to their potentially superior performance in terms of water flux and solute rejection as compared to conventional membranes. Their superior performance has been demonstrated experimentally [1,2,3]. The number of Web of Science entries for biomimetic membranes has increased by a factor of four from 2000 to 2011. The general ABM approach has been reviewed recently [4,5,6]. Basically, the ABMs developed until now are made from combining three components: Aquaporins (AQPs) which are membrane protein water channels; amphiphilic molecules in which the AQPs are embedded; and a polymer support structure. The amphiphilic molecules generally can be either lipids or polymers. Due to superior performance in terms of stability and flexibility [7], block copolymers (di- or triblock) have been predominantly investigated resulting in aquaporin-based biomimetic polymeric membranes (ABPMs), but a number of studies also address aquaporin-based biomimetic lipidic membranes (ABLMs).
Here, we attempt to provide a broad overview of how AQPs, block copolymers and polymer support structures interact and how these interactions can be characterized. In the first section, we will discuss the interplay between AQPs and block copolymers including general membrane protein incorporation into block copolymers, resulting in AQP-block copolymer complexes in vesicular (proteopolymersomes) or planar form. Many aspects of AQP incorporation were lessons learnt from the study of incorporation of other membrane proteins. We then review characterization methods for studying proteopolymersomes including freeze-fracture transmission electron microscopy (FF-TEM), stopped-flow light scattering (SFLS), fluorescence correlation spectroscopy (FCS) and small-angle X-ray scattering (SAXS). In order to fabricate membranes, the reconstituted AQPs need to be integrated in a suitable supporting matrix. Here, we will describe recent advances with emphasis on how to understand the interplay between proteopolymersomes and polymer-based supporting structures in the form of polyamide active layer (PA-AL) formation. PA-based thin film composite membranes represent a classical approach to membrane fabrication. However, it remains a challenge to control stability, surface roughness and other material properties of the PA-AL. We have therefore investigated if proteo- and polymersomes can be integrated in PA-AL containing polyhedral oligomeric silsesquioxanes (POSS) and how the AL is influenced by this integration in terms of physical and chemical stability and surface roughness. POSS is a well-defined nano-scale organic-inorganic structure that allows for constructing nano-structured hybrid materials and nanocomposites. With respect to membrane technology, POSS has been investigated in terms of creating membranes for molecular separation at elevated temperatures [8] and membranes with anti-fouling properties [9]. However, it is not clear if POSS is compatible with proteopolymersome incorporation. Here, we briefly describe methods to investigate POSS-proteopolymersome interactions using a microfluidic approach for membrane formation [10].
2. Interactions between Aquaporin Proteins and Block Copolymer Matrixes
Although most work on membrane protein incorporation has been performed with lipids as host matrix components (first proteoliposomes publication appeared in 1971 [11]), polymer-based incorporation has gained considerable interest since the first proteopolymersomes publication appeared in 2000 [12]. The early work focused on incorporation of membrane-spanning proteins including ATPases and bacteriorhodopsin into polymethyloxazoline-polydimethylsiloxane-polymethyloxazoline (PMOXA-PDMS-PMOXA) triblock copolymer bilayers in planar [13] or vesicular form [14,15,16].
It is intriguing that membrane proteins can be incorporated functionally in polymeric bilayers (e.g., based on PMOXA-PDMS-PMOXA) that can be up to 10 times thicker than their lipidic counterparts [17]. In fact, proteopolymersomes have been observed with protein densities that exceed proteoliposomes by far [18].
A theoretical approach has been established for general membrane protein incorporation into amphiphilic structures. In this approach, the membrane protein incorporation efficiency depends on its hydrophobicity and its coupling to the host membrane, which is directly related to hydrophobic mismatch. To minimize the mismatch, the host membrane has to deform to match the hydrophobic length of the transmembrane segment of the membrane protein about 3–4 nm. The alternative mode of adaption, a host membrane-induced membrane protein deformation is unlikely as far as the compressibility of membrane proteins is generally one to two orders of magnitude higher than lipids [19]. For polymers, the compression-expansion modulus is assumed to rise linearly with increasing molecular weight (Mw), in which chain compression is favorable over chain stretching. This linear increase is consistent with the notion that the hydrophobic mismatch energy can be balanced with a decrease in stretching energy in the polymer chains around the incorporated membrane protein [20]. Srinivas and Discher found by using coarse-grain simulations that flexible hydrophobic chains can allow protein incorporation, even when the hydrophobic mismatch between membrane protein and hydrophobic interior of the chain region is greater than 22% [21,22]. Thus, membrane proteins can be incorporated more effectively if the hydrophobic chains are flexible [20]. Because flexible chains may however block the channel, no functionality of proteopolymersomes might be observed, even if the membrane protein has been incorporated functionally [21]. Moreover, high polydispersity can as well lead to a higher incorporation efficiency because the shorter chains can gather around the membrane protein and compensate for the hydrophobic mismatch. The good incorporation observed with PMOXA-PDMS-PMOXA could therefore also be attributed to their significantly high polydispersity index (P DI). In contrast, for natural lipid environment, the annual lipids around the incorporated protein can be selected in part by affinity to the protein surface and lateral diffusion [23]. The effect of hydrophobic mismatch is significant for ATPases, co-transporter proteins and ion channels [19], whereas for AQPs, the effects appear smaller—likely because the protein itself is structurally more rigid [24].
The first incorporation of AQPs in polymer bilayer was done in 2004 by Stoenescu and coworkers [25]. They incorporated AQP0 that is derived from the mammalian eye-lens, in polymersomes of three different block architectures (ABA, ABC, CBA, where A stands for PMOXA, B for PDMS and C for polyethylene oxide, PEO). The block configuration dictates the orientation of the incorporated AQP0. Where ABA had 50% of incorporated AQP0 with an orientation similar to that observed in liposomes, CBA had only 20% and ABC 80%, as evidenced by antibody labeling. In all cases, incorporation is achieved by adding AQP0 in detergent during the polymersome formation and removing the non-incorporated protein by size exclusion chromatography (SEC) [25].
The first demonstration of functional AQP incorporation was presented by Kumar in 2007 who incorporated bacterial AqpZ from E.coli in PMOXA-PDMS-PMOXA polymersomes [17] and proved their functionality within SFLS. SFLS is a common permeability characterization method, in which the polymersome shrinkage due to a response to osmolarity change is monitored over time by light scattering. Incorporation of AqpZ led to 800 times higher osmotic response of proteopolymersomes compared to empty polymersomes and showed that the activation energy, meaning the barrier for water to pass through the AqpZ, was comparable to that of AQP reconstituted in proteoliposomes and frog oocytes. The molar protein-to-amphiphile-ratio (mPAR) for optimal AqpZ performance in the triblock copolymer system was found to be 1:50 which would correspond to 1:100 in a (diblock- or lipid) bilayer system [17]. The high density reconstitution of AQP is further exemplified by the formation of 2D AQP crystals to achieve structural (crystallographic) information about AQP—analogous to what has been done with lipid based 2D AQP crystals [26]. For this purpose, a monolayer of nickel-functionalized polybutadiene-polyethylene oxide (PB-PEO) is accumulated at the water-interface, where in the aqueous solution, mixed micelles of detergent, histidine-tagged AqpZ and PDMS-PMOXA-PDMS were present [27]. The nickel affinity to the histidine binds the AqpZ to the PB-PEO layer [28], facilitating a high packing of AqpZ in this layer. After removing the detergent via biobeads and the PB-PEO via imidazole, densely packed AqpZ PMOXA-PDMS-PMOXA crystals were left, unfortunately not of sufficient quality to obtain any structural information [29,30].
2D crystals can in fact be used to investigate the influence of AQP on polymer self-assembly in general. AQP0 is known to easily form 2D crystals due to its natural occurrence in stacks in the eye lens [31]. The findings here were that AQP0 dictates the self-assembling behavior of both polymers in a reciprocal way to the hydrophilic volume ratio f. With increasing mPAR, the interfacial curvature decreases and polymersomes turn into membrane sheets and partly crystals (see Figure 1 and Figure 2). In the case of PB-PEO, formation of polymersomes only occurred by adding AQP0, whereas without AQP0 only cylindrical structures are observed. The highest measured packing densities of functional AQPs into vesicular structures are observed at PB-PEO polymersomes with an mPAR of 1:15, which is significantly higher than what has been achieved in proteoliposomes or frog oocytes. Although not all AQP0 protein was incorporated, the seven-fold increase in osmotic response is consistent with a high-packing density given the relatively low permeability of AQP0 [32]. In this case incorporation was done via mixing detergent-solubilized polymers with detergent-solubilized AQP0 and subsequently dialyzing out the detergent [18,33].
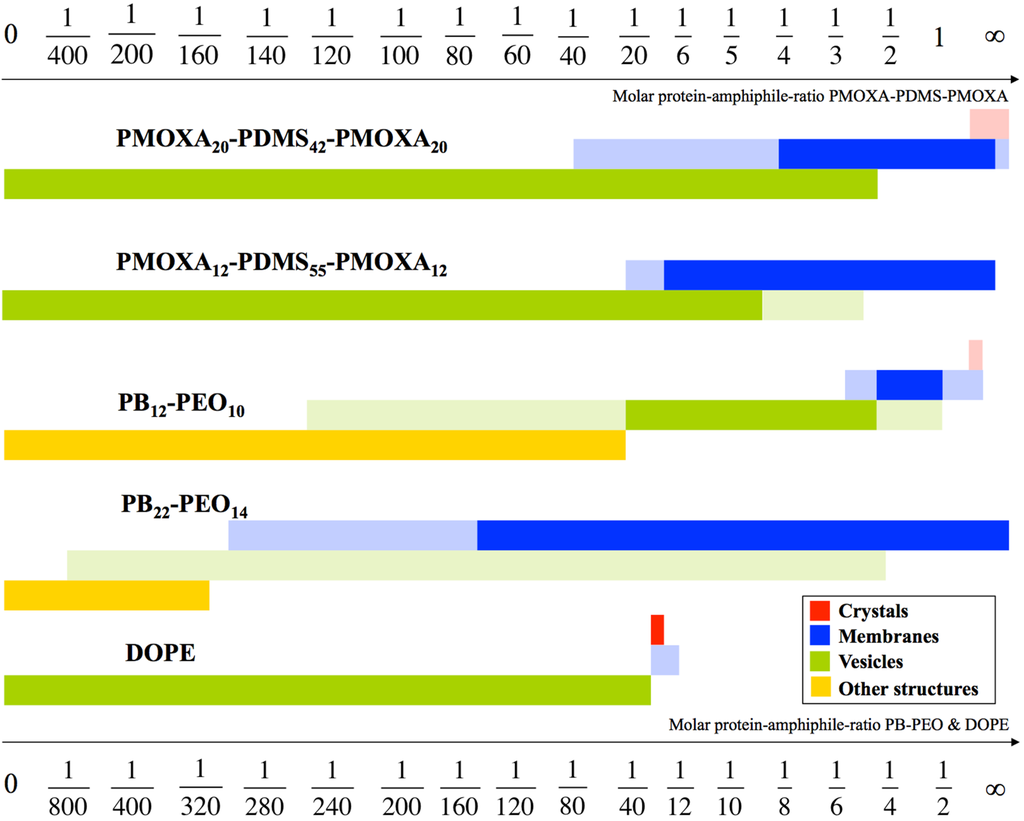
Figure 1.
Schematic drawing of aggregate morphologies as a function of mPAR. PB12-PEO10 undergoes four transitions. Surprisingly, the vesicle shape remained at significantly higher densities at block copolymers, compared to a standard lipid like 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). The mPAR of the one-molecule-bilayer-forming ABA triblock copolymers was divided by two enabling direct comparison with the PB-PEO diblock copolymers and DOPE, both forming bilayers. The morphologies in full color are the main morphologies, pale colors denote coexisting morphologies. Adapted from [34].
With respect to fabrication of biomimetic membranes for technological purposes the first protein incorporation approaches from 2009–2011 were mainly lipid based [35,36], but also planar polymeric membranes have been demonstrated with functional incorporation of gramicidin A [37]. These efforts were pioneered by the Danish company Aquaporin A/S. Their later achievements in fabricating biomimetic membrane will be discussed in the next sections, as well as the work coming out of the groups at the Singapore Membrane Technology Center (SMTC) at Nanyang Technological University (NTU) and the National University of Singapore (NUS).
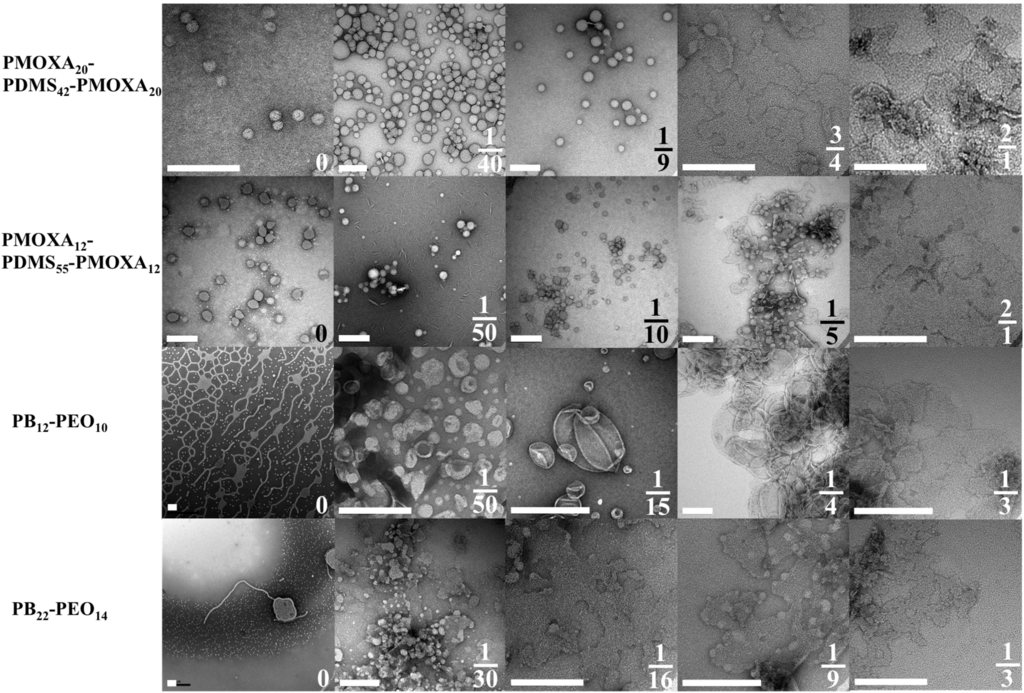
Figure 2.
TEM images of aggregate morphologies as a function mPAR. Where the PMOXA-PDMS-PMOXA copolymers self-assemble to vesicles, PB-PEO forms network- and sperm-like structures and only after incorporation of AQP0 vesicular structures are observed. Scale bar is 200nm. Adapted from [33].
Table 1 summarizes all experimental membrane protein (and peptide) incorporations in block copolymer membranes, including polymer chemistry and stochiometry, PDI, the number-average molecular weight (Mn), f, the incorporated membrane protein, the transport cargo (e.g., water for AQP), if there was functional incorporation, mPAR, the shape of the polymer self-assembled structure, how polymer and membrane protein were mixed and how the function incorporation was measured. Mn (which can be quantified using NMR) is related to Mw as PDI = Mw/Mn. The table excludes those incorporation studies which do not involve block copolymer-protein interactions, such as cell-free expression systems [38,39,40], encapsulation in hydrophobic interior [16], nanopores [41,42], non-amphiphilic polymers [43] and hydrogel approaches [44,45]. With this limitation, the table shows that most results were published by Wolfgang Meier and coworkers using on PMOXA-PDMS-PMOXA triblock copolymers.

Table 1.
Overview of studies of membrane protein incorporation into amphiphilic block copolymers. Most studies are done with the porin OmpF, followed by AqpZ. For explanations please refer to the list of abbreviations.
| Polymer | Mn | PDI | f | Membrane Protein | Transport Cargo | FI? | mPAR | S | Incorporation method Main Functional incorporation measurement | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PMOXA13–PDMS23–PMOXA13 | 3.9 | e− | X | 1:3300 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] | |||
| PMOXA13–PDMS23–PMOXA13 | 4.7 | NA | 0.44 | Alamethicin | X | 1:590 | P | MAq | Current change | [50] | |
| PMOXA13–PDMS23–PMOXA13 | 4.7 | NA | 0.44 | Hemolysin | X | 1:110,000,000 | P | MAq | Current change | [50] | |
| PMOXA13–PDMS23–PMOXA13 | 4.7 | NA | 0.44 | OmpG | X | 1:33,000,000 | P | MAq | Current change | [50] | |
| PMOXA20-PDMS41-PMOXA20 | 6.4 | 1.61 | 0.49 | NtAQP1 | CO2 | X | 1:360 | P | MOr | Cargo → Reaction inside vesicle → pH change | [51] |
| PMOXA20-PDMS41-PMOXA20 | 6.4 | 1.61 | 0.49 | NtPIP2:1 | CO2 | X | 1:360 | P | MOr | Cargo → Reaction inside vesicle → pH change | [51] |
| PMOXA20-PDMS41-PMOXA20 | 6.5 | <1.2 | 0.51 | AQP0 | H2O | ND | 10:1–1:1 | P | MAq & dialysis | [18] | |
| PMOXA20-PDMS41-PMOXA20 | 6.5 | <1.2 | 0.51 | AQP0 | H2O | ND | 10:1–1:50 | V | MAq & dialysis | [18] | |
| PMOXA20-PDMS41-PMOXA20 | 6.5 | <1.2 | 0.51 | AQP0 | H2O | - | 1:2.5–0 | V | MAq & dialysis | Vesicle size change | [18] |
| PMOXA12-PDMS54-PMOXA12 | 6.0 | 1.01 | 0.2 | AqpZ | H2O | X | 1:100–1:1600 | V | MAq & biobeads | Vesicle size change | [3,52] |
| PMOXA19-PDMS74-PMOXA19 | 8.7 | 1.46 | 0.23 | ||||||||
| PMOXA12-PDMS54-PMOXA12 | 6.0 | 1.01 | 0.30 | AqpZ | H2O | X | 1:50–1:400 | V | MAq & biobeads | Vesicle size change | [53,54] |
| PMOXA12-PDMS54-PMOXA12 | 6.0 | 1.01 | 0.30 | Hemolysin | - | 1:83,000,000 | P | MAq | Current change | [50] | |
| PMOXA20-PDMS54-PMOXA20 | 7.4 | NA | 0.42 | TsX | Nucleosides | X | 1:450 | V | MOr, SI & SEC | Cargo → Encapsulated enzyme activity → Color change | [55] |
| PMOXA8-PDMS55-PMOXA8 | 5.4 | NA | 0.22 | AqpZ | H2O | X | 1:3500 | V | PFR, biobeads & SEC | Vesicle size change | [56] |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | OmpF | ELF97 | X | 1:1200 | V | MAq & SEC Cargo | Precipitation inside vesicle → Color change | [57] |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | OmpF | Acridine orange | X | 1:9,100,000 | V | PPFR & SEC | Cargo release → Color change | [58] |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | OmpF | Paraquat. Pyocyanin | X | 1:640 | V | MAq & dialysis | No cargo → No detoxication of encapsulated enzyme→ Cell death | [59,60] |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | AQP0 | H2O | ND | 10:1–1:25 | P | MAq & dialysis | [18] | |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | AQP0 | H2O | - | 1:3–0 | V | MAq & dialysis | Vesicle size change | [18] |
| PMOXA12-PDMS55-PMOXA12 | 6.1 | 1.64 | 0.30 | AqpZ | H2O | ND | 1:4 | Cr. V | MAq & biobeads | [29] | |
| PMOXA7-PDMS60-PMOXA7 | 5.6 | NA | 0.19 | Gramicidin A | Monovalent cations | X | 1:81,000 | P | MOr | Current change | [37] |
| PMOXA8-PDMS60-PMOXA8 | 5.8 | NA | 0.21 | AqpZ | H2O | X | 1:3800 | V | PFR, biobeads & SEC | Vesicle size change | [56] |
| PMOXA13-PDMS62-PMOXA13 | 6.8 | 1.47 | 0.29 | NADH reductase | e− | X | 1:1900 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA15-PDMS62-PMOXA15 | 7.1 | 1.50 | 0.32 | NADH reductase | e− | X | 1:1800 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA12-PDMS65-PMOXA12 | 6.9 | 1.67 | 0.27 | MloK1 | Potassium | X | 1:390 | P | MAq & biobeads | Current change | [61] |
| PMOXA15-PDMS68-PMOXA15 | 7.6 | NA | 0.30 | LamB | Maltohexaose | X | NA | P | MAq | Current change at varying cargo concentrations | [62] |
| PMOXA15-PDMS68-PMOXA15 | 7.6 | NA | 0.30 | OmpF | Actylthiocholine | X | 1:10000 | V | PFR | Cargo → Encapsulated enzyme activity → Color change | [62] |
| PMOXA15-PDMS68-PMOXA15 | 7.6 | 1.20 | 0.30 | AqpZ | H2O | X | 1:10–1:1000 | V | PFR & biobeads | Vesicle size change | [63] |
| PMOXA15-PDMS68-PMOXA15 | 7.6 | 1.20 | 0.30 | Hemolysin | 1:66,000,000 | V | MAq | Current change | [50] | ||
| PMOXA21-PDMS69-PMOXA21 | 8.7 | 2.00 | 0.37 | NADH reductase | e− | X | 1:1500 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA16-PDMS72-PMOXA16 | 8.0 | 1.17 | 0.30 | OmpF | Enone | X | 1:220 | V | PPFR & dialysis | Cargo → Encapsulated enzyme activity → Color change | [64] |
| PMOXA-PDMS-PMOXA | 8.8 | NA | NA | OmpF | ELF97 | X | 1:50 | V | MAq & SEC | Cargo → Precipitation inside vesicle → Color change | [65] |
| PMOXA32-PDMS72-PMOXA32 | 10.7 | 1.83 | 0.47 | OmpF | 7-ADCA. PGME | X | NA | V | PFR & dialysis | Cargo → Encapsulated enzyme activity → Bacterial death | [66] |
| PMOXA11-PDMS73-PMOXA11 | 7.2 | 1.70 | 0.22 | LamB | DNA | X | 1:390 | V | MOr, SI & SEC | Fluorescence-labelled cargo | [67] |
| PMOXA11-PDMS73-PMOXA11 | 7.2 | 1.70 | 0.22 | OmpF | Nucleosides | X | 1:10, 1:100 | V | PPFR & SEC | Cargo → Encapsulated enzyme activity → Color change | [68] |
| PMOXA11-PDMS73-PMOXA11 | 7.2 | 1.70 | 0.22 | TsX | Nucleosides | X | 1:10, 1:100 | V | PPFR & SEC | Cargo → Encapsulated enzyme activity → Color change | [68] |
| PMOXA11-PDMS73-PMOXA11 | 7.2 | 1.70 | 0.22 | LamB | DNA | X | NA | P | MAq | [67] | |
| Lipids | |||||||||||
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | Alamethicin | Calcium | X | 1:24 | V | MAq | Cargo precipitation inside vesicle | [69,70] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | FhuA | Sulphorhodamine B | X | 1:6,000,000 | V | MOr, SI & SEC | Cargo → Quenching inside vesicle → Color change | [71,72,73] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | FhuA | TMB | X | 1:4500. 1:3,600,000 | V | MAq/ & biobeads/MOr, SI & SEC | Cargo → Encapsulated enzyme activity → Color change | [71,72,74] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | FhuA | ND | 3000:1 | P | MAq | [72] | ||
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | FhuA | NAD | - | NA | V | MAq | Cargo → Encapsulated enzyme activity→ Absorbance change of cargo | [73] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | FhuA | DNA | - | NA | V | MOr, SI & SEC | Fluorescence-labelled cargo | [73] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | LamB | Sugar | X | NA | P | MAq | Current change at varying cargo concentration | [75] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | OmpF | e− | X | NA | P | MAq | Current change | [75] |
| PMOXA21-PDMS73-PMOXA21 | 9.0 | 1.70 | 0.36 | OmpF | Ampicillin | X | 1:1000 | V | MOr & SEC | Cargo → Hydrolysis inside vesicle → Color change | [12,76] |
| PMOXA20-PDMS75-PMOXA20 | 9.0 | 1.46 | 0.34 | AqpZ | H2O | X | 1:25, 1:50, 1:200 | V | PFR & biobeads | Vesicle size change | [77] |
| PMOXA11-PDMS76-PMOXA11 | 7.8 | 1.48 | 0.25 | BR | H+ | X | NA | V/Mc | MOr & SI | pH change | [78,79] |
| PMOXA11-PDMS76-PMOXA11 | 7.8 | 1.48 | 0.25 | BR & ATPase | H+ | X | 1:180 | V | MOr & dialysis | pH change & bioluminescence assay | [15] |
| PMOXA11-PDMS76-PMOXA11 | 7.8 | 1.48 | 0.25 | BR & ATPase | H+ | X | 1:20 | V | PBR & dialysis | pH change | [80,81,82] |
| PMOXA6-PDMS90-PMOXA6 | 9.5 | NA | 0.12 | OmpF | L-ascorbic acid, CO, Na2 S2 O4, ONOO− | X | 1:1300 | V | PFR, dialysis & SEC | Cargo → Absorbance change of encapsulated protein | [83] |
| PMOXA21-PDMS97-PMOXA21 | 9.0 | 1.70 | 0.30 | Hemaglutinin | X | 1:3800 | V | MAq & biobeads | MP → Fusion with fluorescence-labelled liposomes | [74] | |
| PMOXA9-PDMS106-PMOXA9 | 9.4 | 1.38 | 0.14 | NADH reductase | e− | X | 1:1400 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA13-PDMS110-PMOXA13 | 10.4 | 1.44 | 0.19 | NADH reductase | e− | X | 1:1200 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA14-PDMS110-PMOXA14 | 10.5 | 1.36 | 0.20 | NADH reductase | e− | X | 1:1200 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] |
| PMOXA15-PDMS110-PMOXA15 | 10.7 | 1.62 | 0.21 | AqpZ | H2O | X | 1:25–1:500 | V | PFR & SEC | Vesicle size change | [17,29] |
| PMOXA15-PDMS110-PMOXA15 | 10.7 | 1.62 | 0.21 | OmpF | ND | NA | P | MAq | [84] | ||
| PMOXA-PDMS-PMOXA | 20.0 | NA | FhuA | Calcein | X | 1:2,700,000 | V | MOr, SI & SEC | Cargo release → Color change | [85] | |
| PMOXA65-PDMS165-PMOXA65 | 23.3 | 1.63 | NADH reductase | e− | X | 1:550 | V | MAq, biobeads & SEC | Cargo → Reduction of MP → EPR signal | [49] | |
| PMOXA-PDMS-PMOXA | NA | NA | NA | BR | H+ | X | NA | P | MAq | pH change | [86,87] |
| PMOXA-PDMS-PMOXA | NA | NA | NA | BR & CcO | H+ & e− | X | NA | V | MOr, SI & SEC | Current & pH change | [87,88] |
| PMOXA-PDMS-PMOXA | NA | NA | NA | CcO | e− | X | NA | P | MOr, SI & SEC | Current change | [86,87] |
| PMOXA-PDMS-PMOXA | NA | NA | NA | OmpF | H+ | X | NA | P | MAq | Current change | [89] |
| PMOXA110-PDMS40-PEO25 | 13.4 | NA | 0.75 | AQP0 | H2O | ND | 1:200 | V | MOr, SI & SEC | [25] | |
| PMOXA45-PDMS40-PMOXA67 | 10.6 | NA | 0.68 | AQP0 | H2O | ND | 1:200 | V | MOr, SI & SEC | [25] | |
| MPEG-PVL | 6.5 | <1.2 | 0.00 | Polymyxin B | Calcein | X | 1:2 | V | MAq | Cargo release → Color change | [73] |
| P2VP-PEO | NA | NA | NA | FhuA | NAD | - | NA | V | MOr, SI & SEC | Cargo → Enzyme reaction inside vesicle → Absorbance change of cargo | [73] |
| PB12-PEO10 | 1.1 | 1.09 | 0.32 | AQP0 | H2O | X | 1:5–1:250 | V | MAq & dialysis | Vesicle size change | [18] |
| PB12-PEO10 | 1.1 | 1.09 | 0.32 | AQP0 | H2O | ND | 1:1.3 | Cr | MAq & dialysis | [18] | |
| PB12-PEO10 | 1.1 | 1.09 | 0.32 | AQP0 | H2O | ND | 1:1–1:10 | P | MAq & dialysis | [18] | |
| PB12-PEO10 | 1.1 | 1.09 | 0.3 | AqpZ | H2O | X | 1:50-1:1000 | V | MAq & dialysis | Vesicle size change | [90] |
| PB12-PEO10 | 1.1 | 1.09 | 0.32 | BR | H+ | X | 1:500 | V | MAq & biobeads | pH change | [91] |
| PB12-PEO10 | 1.1 | 1.09 | 0.3 | SoPIP2;1 | H2O | - | 1:200 | V | MAq & biobeads | Vesicle size change | [90] |
| PB12-PEO10 | 1.1 | NA | 0.34 | Hemolysin | Calcein | X | 1:33,000 | V | MAq & dialysis | Cargo release → Color change | [92] |
| PB22-PEO14 | 1.8 | 1.17 | 0.28 | AQP0 | H2O | ND | 2:1–1:300 | P | MAq & dialysis | [18] | |
| PB22-PEO23 | 2.2 | 1.09 | 0.39 | AqpZ | H2O | X | 1:15–1:200 | V | MAq & dialysis | Vesicle size change | [90] |
| PB22-PEO23 | 2.2 | 1.09 | 0.39 | SoPIP2;1 | H2O | - | 1:15, 1:200 | V | MAq & dialysis | Vesicle size change | [90] |
| PB29-PEO16 | 2.3 | 1.00 | 0.25 | AQP10 | H2O | - | 1:990 | V | PFR & SE | Vesicle size change | - |
| PB35-PEO14 | 2.5 | 1.09 | 0.19 | AqpZ | H2O | - | 1:15 | V | MAq & dialysis | Vesicle size change | [90] |
| PB35-PEO14 | 2.5 | 1.09 | 0.19 | SoPIP2;1 | H2O | - | 1:15 | V | MAq & dialysis | Vesicle size change | [90] |
| PB43-PEO32 | 3.7 | 1.03 | 0.31 | AQP10 | H2O | X | 1:600 | V | PFR & SE | Vesicle size change | [93] |
| PB46-PEO30 | 3.8 | 1.04 | 0.28 | AqpZ | H2O | - | 1:50,1:100,1:200 | V | MAq & dialysis | Vesicle size change | [90] |
| PB46-PEO32 | 3.9 | 1.00 | 0.30 | AQP10 | H2O | - | 1:580 | V | PFR & SE | Vesicle size change | - |
| PB52-PEO29 | 4.1 | <1.1 | 0.25 | Hemolysin | e− | X | NA | P | MAq | Current change | [94] |
| PB52-PEO29 | 4.1 | <1.1 | 0.25 | Polymyxin B | X | NA | P | MAq | Current change | [95] | |
| PB52-PEO29-LA | 4.1 | <1.1 | 0.25 | Hemolysin | e− | X | NA | P | MAq | Current change | [94] |
| PB52-PEO29-LA | 4.1 | <1.1 | 0.25 | Polymyxin B | X | NA | P | MAq | Current change | [95] | |
| PB92-PEO78 | 8.4 | 1.08 | 0.34 | AQP10 | H2O | - | 1:270 | V | PFR & SE | Vesicle size change | - |
| PB125-PEO80 | 8.9 | <1.1 | 0.28 | Alamethicin | Calcein | - | 1:2-1:8 | V | MAq | Cargo release → Color change | [96] |
| PHEMA25-PBMA25-PHEMA25 | 14.3 | 1.30 | 0.83 | AqpZ | - | NA | P | MAq & biobeads | Current change | [97] | |
| PHEMA25-PBMA25-PHEMA25 | 14.3 | 1.30 | 0.83 | Hemolysin | X | NA | P | MAq | Current change | [97] | |
| PHEMA25-PBMA25-PHEMA25 | 14.3 | 1.30 | 0.83 | OmpF | - | 1:70 | P | MAq & biobeads | Current change | [97] | |
| PEE37-PEO40 | 3.9 | <1.1 | 0.39 | Alamethicin | Calcein | X | 1:2–1:8 | V | MAq | Cargo release → Color change | [96] |
| PPO34-PGM14 | 6.5 | 1.30 | 0.66 | Strepatividin-BSA | ND | 1:5, 1:15, 1:50 | V | PPFR | [98] | ||
| PI93-PEO87 | 10.2 | 1.00 | 0.31 | FhuA | TMB | X | 1:6700. 1:5,300,000 | V | MOr, SI & SEC | Cargo → Encapsulated enzyme activity → Color change | [73] |
| PEO136-PIB18-PEO136 | 8.0 | 1.86 | 0.90 | Cecropin A | Calcein | X | 1:30 | V | MAq & SEC | Cargo release → Color change | [99] |
| P4MVP21-PS26-P4MVP21 | 13.1 | NA | 0.80 | PR | X | 1:10 | V | MAq & precipitation | Absorbance change in membrane protein | [100] | |
| P4MVP21-PS38-P4MVP21 | 14.3 | 1.19 | 0.74 | PR | X | 1:10 | V | MAq & precipitation | Absorbance change in membrane protein | [100] | |
| P4MVP29-PS42-P4MVP29 | 18.7 | NA | 0.78 | PR | X | 1:10 | V | MAq & precipitation | Absorbance change in membrane protein | [100] | |
| P4MVP22-PB28-P4MVP22 | 15.0 | NA | 0.92 | PR | ND | 1:10 | V | MAq & precipitation | [101] | ||
| P4MVP22-PB28-P4MVP22 | 15.0 | NA | 0.92 | RC | e− | X | 1:25 | V | MAq & precipitation | Cargo → Reduction of MP → EPR signal | [102] |
| P4VP22-PB28-P4VP22 | 7.1 | NA | 0.82 | PR | ND | 1:10 | V | MAq & precipitation | [101] | ||
| P4MVP29-PB56-P4MVP29 | 17.4 | 1.08 | 0.81 | RC | e− | X | 1:25 | V | MAq & precipitation | Cargo → Reduction of MP → EPR signal | [102] |
| P4MVP18-PB93-P4MVP18 | 13.9 | 1.06 | 0.62 | PR | ND | 1:10 | V | MAq & precipitation | [101] | ||
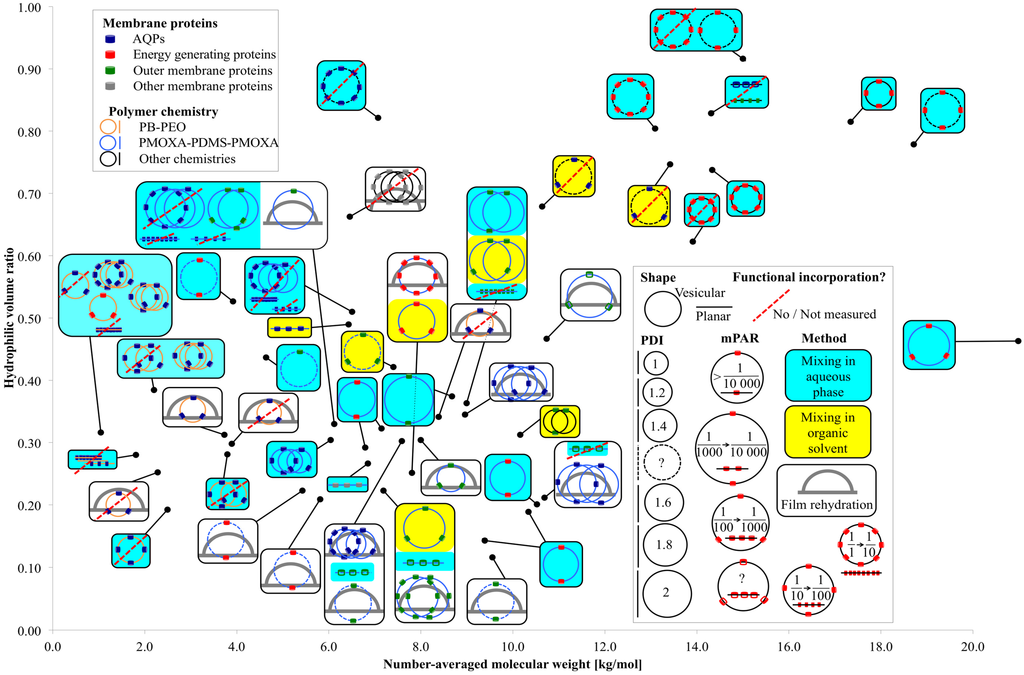
Figure 3.
Overview of relevant parameters for membrane protein incorporation into amphiphilic block copolymers.
Figure 3 presents an overview of membrane protein incorporation into polymers with a known Mn and f. Each black dot represents one polymer. The connected box shows the polymer chemistry, the incorporated membrane protein family, the self-assembled morphology (vesicular or planar), the incorporation method, the PDI of the polymer (not of the polymersomes), the mPAR and if the incorporation was functional or not, or respectively not measured. If there are several sketches in the box, several different experiments have been performed on the polymer. If there are two crossing circles and two close lines respectively, two different mPARs were investigated, where all other parameters remained the same. If there are three crossing circles, three or more mPARs were investigated. In the case of varying another parameter than mPAR (incorporation method, polymer chemistry, incorporated membrane protein etc.) a new sketch is drawn. Generally, polymers capable of functional incorporation have an f between 0.2 and 0.35 and Mn was in between 2 and 12 kg/mol. Compared with PB-PEO, PMOXA-PDMS-PMOXA has a far broader PDI [46], its bilayer is highly water impermeable [17] and they do not collapse in dried form [47]. PB-PEO is more lipid-like as far it collapses easier and has higher water permeability [18]. The polymers that did not achieve functional AQP incorporation were mainly PB-PEO polymers with small Mn and PDI. Energy generating (BR, CcO, NADH reductase, ATPase, RC, PR) and outer membrane proteins (OmpF, OmpG, FhuA, TsX) were incorporated mainly into PMOXA-PDMS-PMOXA polymers, but outer membrane proteins have also been incorporated in more exotic chemistries in an f range where one would not expect vesicular structures. The great majority of functional incorporation trials were performed with vesicular structures, where mixing was done in aqueous phase. Generally, at smaller PDI values, no functional membrane proteins can be incorporated, which is in agreement with the findings from Pata et al. [20]. A wide range of mPARs have been used with no optimal ratios detected. However, mPARs are based on the initial or nominal concentrations of membrane proteins and polymers and the final mPAR after incorporation may be different [48]. In the next section, we will discuss how to quantify membrane proteins (with focus on AQPs) after incorporation.
3. Evaluation of AQP Incorporation Characterization Methods
Detecting functional incorporation of AQPs is challenging, as the permeating solute is neutral water molecules. Protein mediated transport of neutral molecules (in particular at the single protein level) is harder to measure than transport of charged molecules (ion or protons) or a specific chemical reaction (e.g., ATPase enzyme activity). Although deuterated water labeling has been proposed for measurements via Raman spectroscopy [103], these type of measurements is complicated by the fact that water transport rate in the AQP channel is different for deuterated water molecules compared to that for normal water molecules [104].
A popular method for measuring functional incorporation is SFLS. In SFLS, proteopolymersomes are rapidly mixed with an osmotically active agent (NaCl or sucrose) in a defined volume. In the case of a hyperosmotic shock, proteopolymersomes will shrink, which will give rise to an increase in light scattering. With an increasing amount of incorporated AQPs, the shrinking rate will increase as well. This method is however strongly affected by the quality (size distribution) of the polymersomes, of the AQP concentration in the polymersome and the concentration of the osmolytes [56].
In principle, direct visual quantification can be achieved by FF-TEM, although FF-TEM will not reveal any functional information. In FF-TEM, proteopolymersomes are captured in their original shape by quick-freezing. The frozen sample is fractured, where the fracture plane is along the proteopolymersome bilayer, which is the weakest point of the whole system. The sample with incorporated AQPs (or the cavities, where AQPs were embedded in the bilayer) is then exposed to carbon/metal coating. The forming replica is removed from the thawed sample and AQPs/cavities can be observed on the replica as distinct spots on the proteopolymersomes.
Another method is FCS of fluorescently labelled AQP. In FCS time-dependent fluctuations of fluorescence intensities in a microscopic space, the so-called confocal volume, are monitored and subjected to an autocorrelation function. Dependent from the different diffusion time of the particles diffusing through the confocal volume, one can obtain the number of particles in the confocal volume within a given time interval. When proteoliposomes or proteopolymersomes are monitored, then solubilized to micelles and monitored again, the proteins-per-vesicle-ratio (mean number of membrane proteins incorporated in the bilayer of one vesicle) can be obtained by dividing the latter number by the first. It is assumed that micelles contain only one AQP, thus the micelle-per-vesicle ratio is equal to the proteins-per-vesicle-ratio. Further details to the theory are given in [48]. Alternatively, one can obtain the proteins-per-vesicle-ratio by correlating the proteopolymersome solution with the AQP stock solution. Both correlations have advantages and challenges that are further described in the FCS subsection.
In characterizing biological material, SAXS is also a versatile tool because it gives structural information on particles in solution on a length-scale from 1 to 100 nm where data are presented as scattering intensity as a function of the magnitude of the scattering vector q. This quantity is independent of the particular geometry of the experimental set-up and directly related to the scattering angle 2θ as q = 4π sin(θ)/λ where λ is the wavelength of the X-ray beam. Two scattering points separated by a distance d within a particle gives rise to interference showing up as increased intensity in the scattering curve at q = 2π/d. This means that large features are probed at low q while smaller details are probed in the high-q region of the curves. The strength, with which a particle scatters, its contrast, is proportional to its excess electron density, i.e., the difference between the electron densities of the sample and the solvent. The downside is that SAXS requires access to elaborated synchrotron radiation sources.
Here, we will exemplify SFLS, FF-TEM, FCS and SAXS analyses with a series of diblock copolymers with optimal Mn and f range for functional membrane protein incorporation: PB29-PEO16, PB33-PEO18, PB45-PEO14, PB43-PEO32, PB46-PEO32 and PB92-PEO78. PB-PEO was chosen because it showed functional AQP incorporation as discussed before and the Mn and f range is easier to control compared to PMOXA-PDMS-PMOXA. For SFLS, FF-TEM and SAXS, AqpZ is used as the incorporated membrane protein, where GFP-tagged human aquaglyceroporin AQP10 is used for the FCS experiments. Details are provided in the supporting material.
3.1. Stopped-Flow Light Scattering
In order to exemplify an SFLS analysis, data for PB45-PEO14 and PB33-PEO18 diblock copolymer proteo- and polymersomes (meaning with and without AqpZ) is shown in Figure 4. For PB33-PEO18 the rate constant associated with the increase in light scattering intensity was slightly higher with AqpZ, for PB45-PEO14 it was even lower. This illustrates one of the major challenges in SFLS. The absence of a significant response to the change in extravesicular osmolarity could be due to an increase in the bilayer bending modulus induced by the presence of (non-functional or blocked) ApqZ. We observed similar problems in previous experiments with AqpZ and SoPIP2; 1, where only the smallest polymers (PB12-PEO10 and PB22-PEO23) showed a significant difference in SFLS between proteo- and polymersomes (results not shown). Another reason for the similar SFLS signal might be the blockage of the AqpZ channels by PEO chains. In this case, AqpZ would simply sit in the bilayer as an impermeable hydrophobic block, as suggested from Kumar et al. [18], because water permeation is blocked by the areas corresponding to the incorporated AqpZ, lower permeabilities of proteopolymersomes can be expected as compared to polymersomes. On the other hand the incorporated AqpZ could be fully functional, but the polymer matrix is resistant to changes in volume. This underscores the notion that SFLS is not a stand-alone technique.
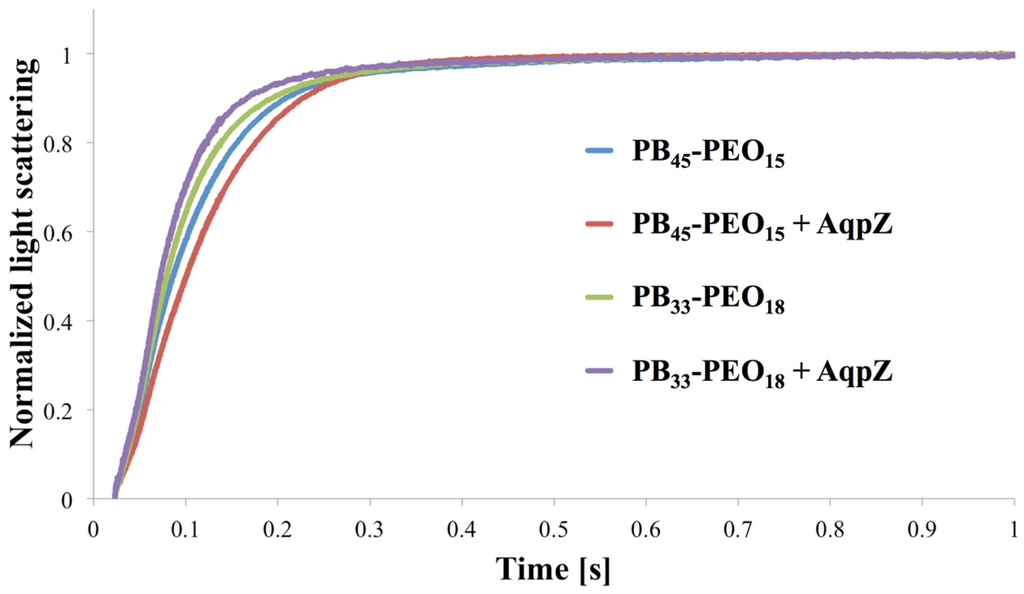
Figure 4.
Normalized light scattering vs. time for proteo- and polymersomes of PB45-PEO14 and PB33-PEO18 at an mPAR of 1:100. For PB45-PEO14 the apparent water permeability is slightly decreased for the proteopolymersomes versus polymersomes, whereas for PB33-PEO18 it is slightly increased.
3.2. Freeze Fracture Transmission Electron Microscopy
Results of FF-TEM for PB45-PEO14 proteopolymersomes are shown in Figure 5. Proteopolymersomes with an mPAR of 1:100 were produced using film rehydration (FR), frozen and fractured in a Leica MED20 station, where two planchets with frozen sample are separated, thus the fracture is more a “crack” than a “cut,” thereby minimizing smearing effects from usual FF procedures (for details see supporting information). All proteo- and polymersome had a pronounced raspberry-like surface, potentially due to collapsed PB chains. However, the “typical” spots that have been claimed to be associated with AQP in a study on proteoliposomes [105] were not observed. In Figure 5, the bubble-like spots are distributed equally among polymersomes (Figure 5a–d) and proteopolymersomes (b,c,e,f). The spots could be either PB chain accumulations (Figure 5a–c) or artifacts due to bad fracturing (d–f). Proteo- and polymersomes of other PB-PEO polymers at other Mn and f showed similar behavior. It thus seems that FF-TEM sample preparation plays a major role in false positive results. Occasionally, we observed dots all over the sample that were clearly not AqpZ but potentially polymer micelles. These dots could be eliminated by omitting an up concentration step and by carefully controlling temperature, sample and cutting handling or metal coating parameters (the optimized protocol is given in the supporting information). Even among the polymers with the shortest PB chains (PB32-PEO30 and PB45-PEO14), we could not ascertain the presence of AqpZ. However, from these experiments alone we cannot exclude the possibility that AqpZ tetramers could be present as the hydrophilic PEO chains are still large compared to lipid head groups. Thus the AqpZ could be concealed in the PB core.
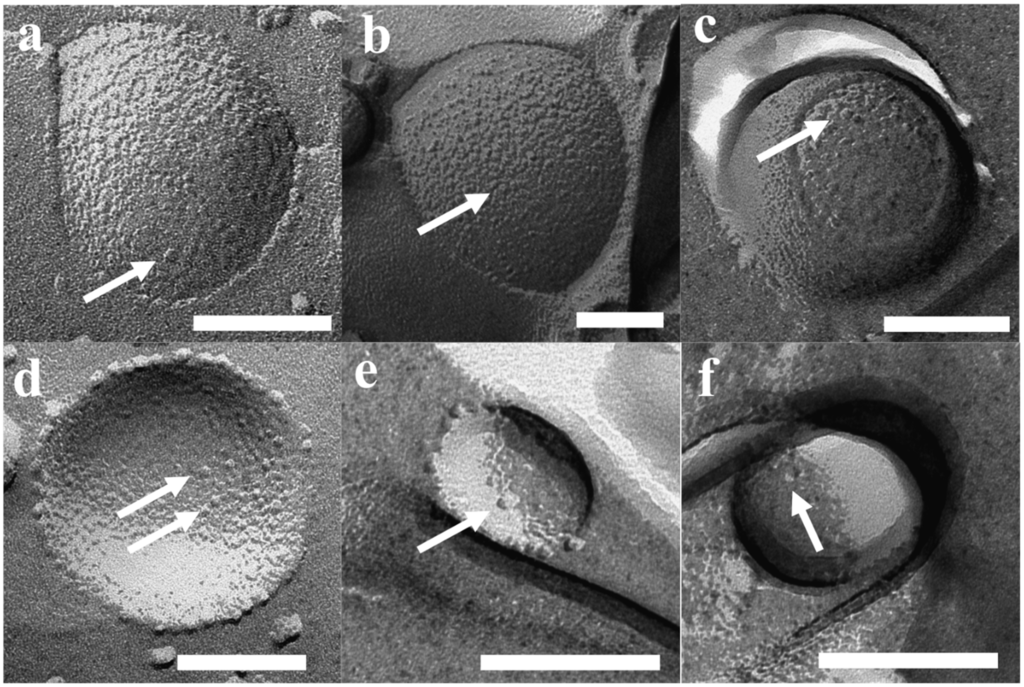
Figure 5.
FF-TEM images of PB45-PEO14 proteo—(b,c,e,f) and polymersomes (a,d). All vesicles revealed spots, potentially not from AqpZ but rather collapsed PB chains (a–c) or bad fracturing artifacts (d–f). Scale bar is 100 nm.
3.3. Fluorescence Correlation Spectroscopy
As both SFLS and FF-TEM present challenges as tools for evaluating protein incorporation into polymersomes, we also evaluated FCS as a novel method for getting quantitative information about AQP incorporation. This was inspired by a recent paper by Erbakan et al. describing various AqpZ isoforms, tagged with a fluorophore in proteoliposomes, where the protein-per-vesicle ratio was determined and further substantiated using SFLS [48]. Initially, we attempted to reproduce the proteoliposome experiments described in [48]. At an mPAR of 1:200, our measurements revealed a proteins-per-vesicle-ratio of 5.35, which was comparable to the ones obtained in Erbakan et al. (around 7.5). The difference could be due to the different AQP and tagged fluorophore used.
After having optimized the FCS instrument parameters for proteopolymersomes (for details please refer to the supporting information), we performed FCS on proteopolymersomes of PB45-PEO14 (mPAR 1:100) with AQP10-GFP and with OG-solubilized protein micelles. The results are shown in Figure 6. We obtained a higher species number in the proteopolymersomes sample than in the protein micelle sample. This could be due to the same OG-induced aggregation. We therefore decided to correlate the proteopolymersomes to the AQP10-GFP stock. Erbakan et al. could not do this, because the fluorophore used (mBanana fluorescent protein) exhibited a decreased fluorescence lifetime in pure OG environment (stock solution) compared to lipid/OG environment (solubilized protein micelles). GFP, however, did not seem to alter fluorescence lifetime significantly whether the AQP10-GFP is in OG (1.8 ns) or polymer/OG environment (1.97 ns, Figure 6b). They are comparable to fluorophore used by Erbakan et al. (4 ns [48]) and to standard GFP fluorescence lifetime (3 ns [106]). The difference between our GFP fluorescence lifetime and the standard one could be due to shielding of the attached AQP10 and the OG environment, as well as to the fitting algorithm of the instrument.
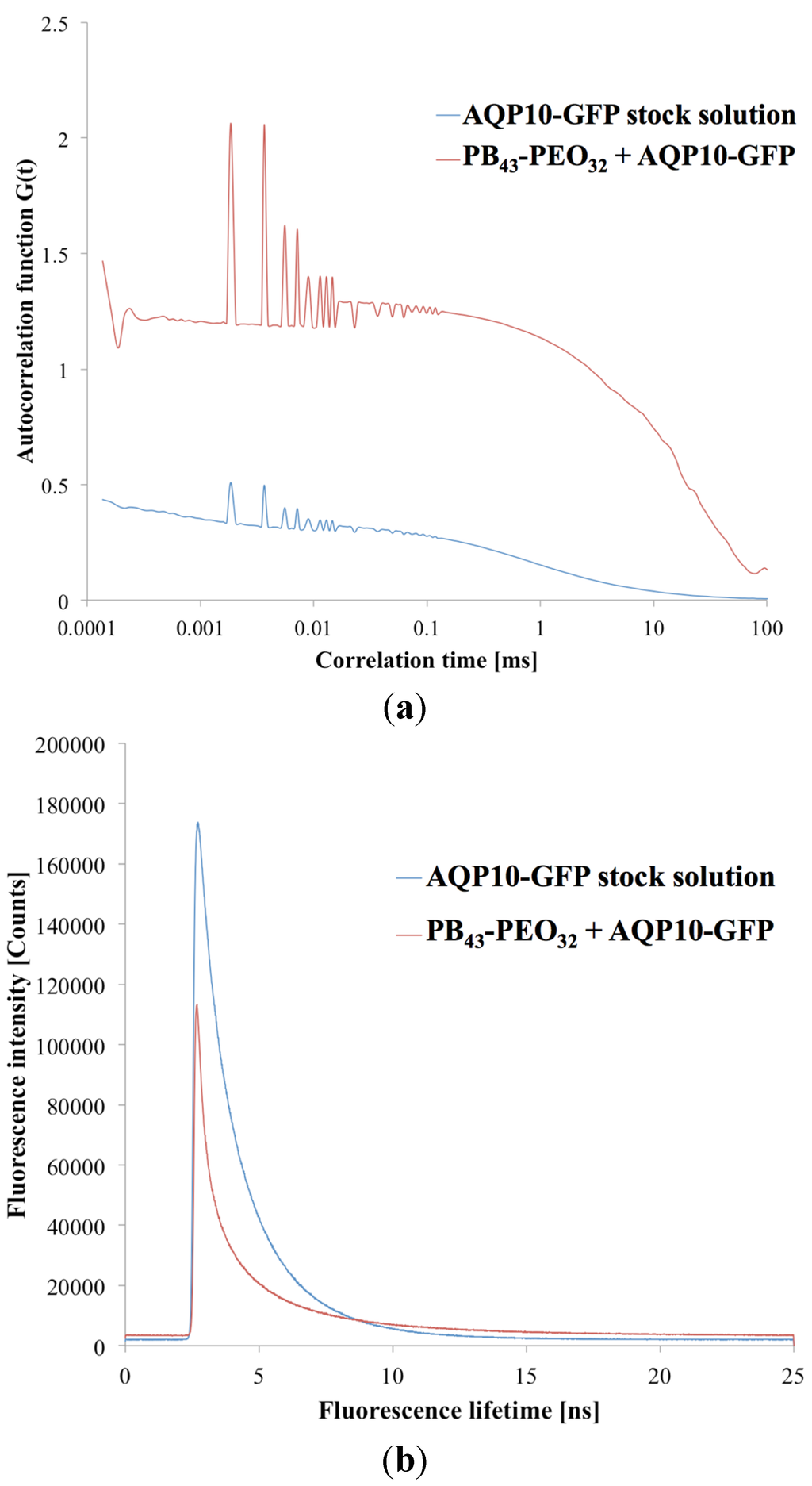
Figure 6.
(a) Correlation diagram of proteopolymersomes and AQP10-GFP stock solution as a function of correlation time τ against autocorrelation function G(τ). The higher autocorrelation signal indicates a lower number of particles in the confocal volume, due to slower diffusion time. (b) Fluorescence lifetimes of the same samples as a function of lifetime against intensity signal. Where the intensities varied, the fluorescence lifetime was in a comparable range.
Accordingly, the sample correlation depend on the single components of the system. In the case of sensitive fluorophores, it is better to compare AQP vesicles and AQP micelles not to influence the fluorophore environment. In the case of polymers as the protein matrix, it is better to correlate the AQP-fluorophore stock solution as the polymeric AQP micelles aggregate easily. A disadvantage of correlating AQP-fluorophore stock with AQP vesicles is that the final concentration of AQP is not known, complicating a correlation with similar AQP concentration.
Calculating the species number of pure AQP10-GFP from the stock in the confocal volume and the one from the proteopolymersome solution (Figure 6a), we obtained a proteins-per-vesicle-ratio of 2.87. These results demonstrate that FCS can serve as a tool to quantify AQPs in proteopolymersomes. This opens possibility for conducting a systematic study in which f and Mn are varied in order to obtain quantitative information about which polymers can be used to achieve the highest proteins-per-vesicle-ratio.
3.4. Small-Angle X-Ray Scattering
Scattering curves for FR prepared proteo- and polymersomes of PB45-PEO14 and PB33-PEO18 are shown in Figure 7. The samples were extruded and centrifuged prior to measurements. At low q-values a typical linear slope is observed in the log-log plot, with the intensity following a power law of q−2. This behaviour is typical of flat laminar structures. The fact that the slope extends below the lowest detectable q-region indicates a low curvature (flat structure) even on the largest detectable length scale of q = 2π/0.1 nm ≈ 60 nm. At higher q, a characteristic oscillatory behaviour is observed. This is attributed to the complex interference between the negative contrast of PB and the positive contrast of PEO.
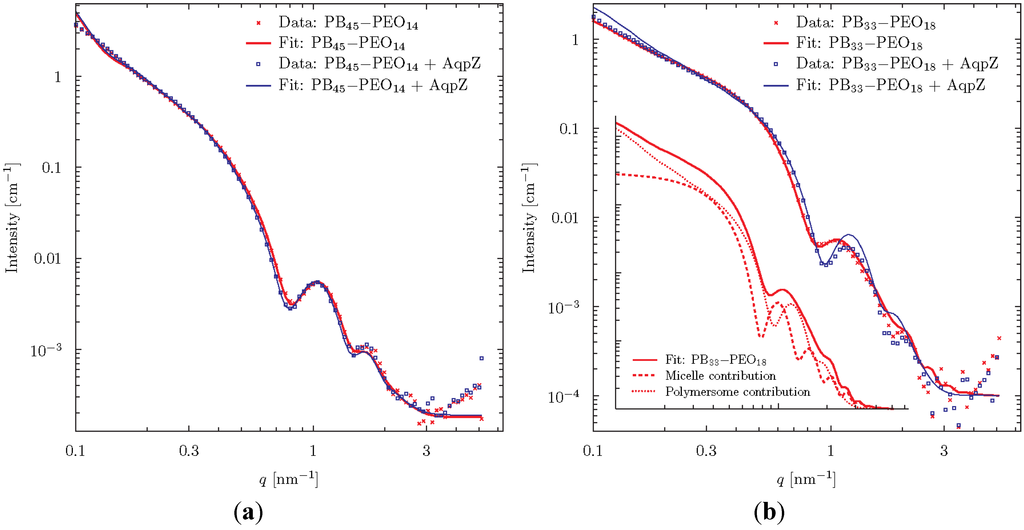
Figure 7.
SAXS data for proteo- and polymersomes of PB45-PEO14 (a) and PB33-PEO18 (b). The fits were obtained using a vesicle model consisting of three concentric spherical shells. To fit the polymersomes of PB33-PEO18, it was necessary to include an additional contribution from block-copolymer micelles as shown in the insert.
The theoretical scattering from various simple geometrical objects such as spheres, cylinders and ellipsoids of varying contrast can readily be calculated. These can be combined to form simplified models of the studied particles. We choose to analyze the data using a vesicle-model consisting of three concentric spherical shells of alternating contrast, corresponding to shells of PEO, PB, and PEO, respectively. The thickness of the individual shells was varied to give the best fit to data using a least squares fitting routine.
Excellent fits were obtained for the PB45-PEO14-system meaning that data are in good agreement with the assumption that the diblock copolymers form spherical vesicles. The fits were especially sensitive to changes in the parameter determining the thickness of the central hydrophobic bilayer constituted by the PB-groups. These were fitted to 9.10 ± 0.1 nm and 8.94 ± 0.07 nm in the presence or absence of AqpZ respectively. Concerning the overall vesicle diameter, we can conclude from the model that it is larger than 60 nm, which is not surprising given the initial analysis above. It is evident from the data and the fit parameter values that well defined bilayer vesicles are formed and that the incorporation of AqpZ introduces only minor differences to the structure of the vesicles.
For the PB33-PEO18 proteopolymersomes, reasonably good fits were obtained with the vesicle model with a hydrophobic bilayer thickness of 7.66 ± 0.05 nm. However, for the polymersomes, no fit to the data gave reasonable physical parameters. The data fit required the assumption that a population of block copolymer micelles co-exists with the vesicles. The combined model fit showed that 76 wt% of the population consisted of proteopolymersomes and 24 wt% were micelles with a hydrophobic core of diameter 11.7 ± 0.3 nm gives a good fit with the data. The insert of Figure 7 shows the separate vesicle and micelle contributions.
In conclusion, the SAXS analysis reveals that, for PB45-PEO14, vesicles are formed both with and without AQP where AQP incorporation leads to a minor differences in average hydrophobic vesicle wall thickness, which could indicate a dimpling or puckering of polymers close to the incorporated AQPs. In the case of PB33-PEO18, some micelle-formation is observed, but this tendency is reduced when AQP is incorporated.
To summarize this chapter, the characterization methods for functional incorporation of AQPs in PB-PEO diblock copolymers were investigated. SFLS and FF-TEM are in principle powerful tools but, for polymer systems, the analysis can give ambiguous results. On the other hand, FCS and SAXS can provide detailed information, but the latter requires access to large-scale facilities in the form of synchrotron radiation sources.
4. Recent Developments in AQP Membrane Designs
Provided that the performance of AqpZ proteopolymersome described by Kumar et al. [17] could be scaled up, they could create a water separation membrane that reaches fluxes of 11,000 L m−2 h−1, a value that is several orders of magnitude higher than conventional industrial membranes. In highly packed 2D AqpZ crystal arrays, fluxes of up to 16,000 L m−2 h−1 could be achieved in principle [107]. However, these values will very probably never be achieved due to upscaling issues—but they show the huge potential of biomimetic membranes. The development is rapid: in 2011, ABPMs were regarded as the most revolutionary membrane advances but also the ones most farthest away from a potential commercial viability [108]. Now, four years later, ABPM membranes are commercially available with areas of tens of m2 [109]. It will still take time before the technology is widespread, but it has definitely moved outside the fundamental research domain. In the next sections, we will highlight the AQP biomimetic membrane technology development in detail.
4.1. Membrane Designs Based on Planar Biomimetic Structures
The first industrial approaches are made from two Danish companies, Aquaporin A/S, and AquaZ (now Applied Biomimetic). Together with the Danish Technical University (DTU), the University of Southern Denmark (SDU), DHI, Lund University, Sweden, Ben-Gurion University of the Negev, Israel, Malaga University, Spain, Vilnius University, Lithuania and Veolia Water, France, Aquaporin A/S joined the EU funded MEMBAQ project 2006–2010 which aimed at utilizing AQPs for industrial applications [110]. At the same time, AquaZ started on membrane development based on a patent from Carlo Montemagno where he described conceptually how AQPs, embedded in polymeric or lipid bilayers, could function as a biomimetic membrane, although without any concrete design of such a membrane [111].
The first membrane design from Aquaporin A/S was based on an ethylene tetrafluoroethylene (ETFE) scaffold with holes of 300 µm, produced by laser-ablation, which are inspired by painting/folding lipid chambers from the 70s [112,113]. A freestanding lipid-bilayer film is established by “painting” a two-phase solution over the hole, where the lipids move from the organic solvent to the aqueous phase, accumulate around the holes and establish a bridging layer. Several membrane proteins and peptides were incorporated in the freestanding layer including porins [36]. In addition, freestanding PMOXA-PDMS-PMOXA polymer membranes with incorporated gramicidin A channels were developed [37] and characterized [114]. In subsequent designs, the membrane is supported by PEO-dimethacrylate (PEO-DMA) based hydrogels [115] or stabilized using surface plasma polymerization [116]. Moreover, a strategy was explored to form interface lipid bilayer between lipid-coated water drops in a continuous oil phase [117]. A later liquid membrane approach investigated SoPIP2;1 proteoliposomes in a sandwich between NF membranes that could prove an AQP fingerprint for the first time, however at modest water flux [118]. These designs [118,119,120,121,122] later paved the way for developing membrane-based biosensor designs [45]. The hydrogel approach from Aquaporin A/S was adapted in 2010, when Montemagno and AquaZ claimed an ABLM design with internally UV cross linked and PA-interconnected proteoliposomes that are immobilized on a lipid-coated PA layer and supported with a PEO hydrogel [123].
In 2009, Aquaporin A/S and DHI Singapore initiated collaborative research with the SMTC on biomimetic membranes. At the same time, the Chung lab from NUS started biomimetic research in collaboration with Wolfgang Meier and coworkers. NUS followed up on Aquaporin’s hydrogel approach and tried to achieve a planar proteobilayer, starting with AqpZ proteoliposome fusion on pure and PEO coated porous alumina and found an increasing stability with increasing mPAR [124]. In 2012, they described an approach based on a Langmuir-Blodgett-film with Nickel-chelated lipids that bind to His-tagged AqpZ, similar to the approach from Kumar [29] but using lipids with subsequent Langmuir-Schäffer deposition-mediated transfer on a mica surface [125]. This was followed by Kaufman et al. who incoporated spinach AQP (SoPIP2;1) in positively charged bolalipid micelles which were then fused on a negatively charged silica surface [126]. Chuyang Tang et al. investigated on fusion behavior of proteoliposoes on pure and polymer-coated silica via quartz crystal microbalance with dissipation (QCM-D) [127]. They found increasing robustness and fusion resistance with increasing mPAR, and further proteoliposome stabilization with polyelectrolyte layers at the highest mPAR (1:25) in 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) liposomes [127].
The SMTC group also investigated ABLMs, following Kaufman’s approach of liposome fusion on nanofiltration (NF) membranes [128,129] and fused AqpZ proteoliposomes on NF PA-polysulfone (PSf) membranes that were precoated with positively charged lipids via spin-coating [130]. The proteoliposomes were placed on the NF membrane and slightly pressurized with 0.5 bar. They found a linear relationship between the roughness of the ABLM surface and mPAR indicating AqpZ incorporation, but no effect from AqpZ on the water flux Jv and the reverse salt flux Js could be observed [130].
4.2. Membrane Designs Based on Vesicular Biomimetic Structures
A different approach was initiated jointly by SMTC and Aquaporin A/S in which AqpZ proteoliposomes were embedded in the standard PA layer made from interfacial polymerization of m-phenyl diamine (MPD) and trimesoyl chloride (TMC) on a PSf support structure [1,131,132]. ABLMs were tested with functional AqpZ proteoliposomes, proteoliposomes with an inactive AqpZ mutant and PA-PSf membranes without proteoliposomes. ABLMs were further benchmarked against commercially available membranes with cross-flow RO tests on 42 cm2 effective coupon area. The ABLMs with AqpZ proteoliposomes had a significantly higher Jv than the ABLM with inactive AqpZ and the PA-PSf membrane while Js values were similar in all cases. Furthermore the ABLMs were able to withstand 10 bar pressure making them well-suited for low pressure RO applications. Jv of the AqpZ ABLM was ~40% higher compared to the commercial brackish water RO membrane (BW30) and an order of magnitude higher compared to a seawater RO membrane (SW30HR).
This was followed up by a systematic study, which revealed that 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)-based proteoliposomes and proteoliposomes of mPAR of 1:200 gave optimal water flux as judged by SFLS and that cholesterol addition could seal defects on the proteoliposomes [133].
To achieve higher loading and better sealing, the SMTC group coated proteoliposomes with polydopamine (PDA) and immobilized them on a 28 cm2 NF polyamide imide (PAI) membrane by embedding them in branched polyethyleneimine (PEI), cross linked per PA bond at elevated temperature [134]. The SFLS data showed, that the elevated temperature had a higher negative influence on the permeability of the proteoliposomes than the PDA coating itself. Even so, AqpZ function was demonstrated with an optimal performance mPAR of 1:200 when reconstituted and integrated into the PAI-PEI layer. In contrast, the best SFLS response was achieved at an mPAR of 1:100 [133]. This discrepancy could be due to AqpZ being affected by the PDA coating or the PEI branches. Still, the Jv was measured to be 36 L m−2 h−1 bar−1) making it the highest among all biomimetic membranes so far [134].
In addition, proteopolymersomes can be functionalized to get bound chemically to a counterpart functionalized membrane. Functionalization of both liposomes and polymersomes has been studied extensively since decades [135,136,137].
ABPMs with functionalized proteopolymersomes are first mentioned in a patent of Montemagno in 2011, where he claimed a concept of proteopolymersomes made of polyethyloxazoline-polydimethylsiloxane-polyethyloxazoline (PEOXA-PDMS-PEOXA) triblock copolymers, where the methacrylate-functionalized PEOXA block is immobilizing the proteopolymersomes on a methacrylate functionalized cellulosic membrane [138].
The first experimental results on this approach were presented by the NUS group [53]. They made proteopolymersomes containing AqpZ in methacrylate-functionalized PMOXA-PDMS-PMOXA and tested them with SFLS. In contrast to Kumar [17], no significant difference SFLS signals with varying mPAR was observed—likely a reflection of the issues with SFLS on rigid structures mentioned in Section 3. Proteopolymersomes were deposited onto acrylate-functionalized polycarbonate track-etched (PCTE) membranes and immobilized by UV-crosslinking of the acrylate groups with the methacrylate of the PMOXA, as claimed from Montemagno et al. [138]. Afterwards, the proteopolymersomes were further immobilized by pressure-assisted adsorption and possibly ruptured by “smooth extrusion”. AQP resulted in an increasing Jv with increasing mPAR, whereas there was no Jv with polymersomes alone; however, AFM and field emission-scanning electron microscopy (FE-SEM) revealed that the layer had some defects [53]. In a subsequent study, they followed the same approach using an acrylate-functionalized cellulose acetate membrane [54]. Here, they found an increase in Jv and decrease in NaCl rejection with proteopolymersomes of higher mPAR. The increase in Jv could indicate AQP activity, but the NaCl rejection was however still quite low (33%) and the measured membrane area was only 7 mm2 [54].
In another approach, gold-disulphide binding to immobilize disulphide functionalized PMOXA-PDMS-PMOXA AqpZ proteopolymersomes on gold-coated porous alumina and silicon surfaces has been described [77]. Here, FE-SEM revealed that full coverage of the pores was achieved at pore diameter of 55 nm, where larger (100nm diameter) pores remained open. Again, an effect of incorporating AQP was observed but NaCl rejection was modest [77]. To obtain a better sealing, cysteamine was added with PDA and histidine coatings after the proteopolymersome immobilization on gold-coated PCTE [3]. Jv increased and Js decreased with increasing amount of PDA-His-layers; however, the best sealing was obtained without proteopolymersomes. Pressure retarded osmosis (PRO) mode testing (AL to the water receiving draw side) resulted in significantly higher Js than forward osmosis (FO) mode testing (AL to feed side) [3]. Mathematical simulations on this ABPM indicated that in PRO mode, Jv is determined by vesicle size and permeability. In FO mode, hydrostatic pressure is determined by the vesicle interior solute concentration [52].
Another slightly different design has been experimentally realized afterwards with AqpZ and methacrylate- and carboxyl-functionalized PMOXA-PDMS-PMOXA on amine-functionalized CA [63]. Here, proteopolymersomes are first covalently attached to the CA, where the carboxyl-groups of PMOXA and the amine groups on CA formed a PA bond. Then, a methacrylic cross linking polymerization is performed by dipping the membrane into a mixture of methyl methacrylate, ethylene glycol dimethacrylate and initiator. Jv is linearly increasing and NaCl rejection decreasing with polymerization time. An increase in Jv and decrease in NaCl rejection of ABPMs compared to only methacrylated CA and polymersome coated CA in both FO and NF mode evidenced the presence of AQP. However the NaCl rejection (61%) still indicated significant defects [63].
Another example of methacrylate cross-linking involves amine-functionalized AqpZ proteoliposomes on a PDA precoated ultrafiltration (UF) polyacrylonitrile (PAN) membrane [139]. Here, proteoliposomes are internally cross linked via methacrylate and gently pressurized onto the PDA-PAN support, allowing the amines of PDA and functionalized lipids to react. Further stabilization is achieved via glutaraldehyde. The internal cross linking of the proteoliposomes has a positive effect on stability. Jv and NaCl rejection between liposome-coated membranes and ABLMs showed some effects of AqpZ presence; however, FE-SEM images and low NaCl rejections revealed that defects in the ABLM played a strong role in the membrane performance [139].
Instead of chemical bonding, proteoliposomes or -polymersomes can be bound by electrostatic forces. Using this approach, Kaufman et al. attempted to fuse positively charged bolamphiphilic proteoliposomes onto negatively charged NF PA and sulfonate PSf (PSS) membranes [140]. The proteoliposome loading was enhanced with the more negatively charged PSS membrane. However, proteoliposome loading also led to a decrease in Jv together with an increase in NaCl rejection, probably due to induced defects in the bolamphiphilic bilayer by SoPIP2;1 [140].
Another electrostatic-binding-based approach employed the embedment of positively charged poly-L-lysine covered AqpZ proteoliposomes in the anionic part of a layer-by-layer (LbL) sandwich on an UF PAN membrane [2]. The anionic part is made of polyacrylic acid (PAA) and PSS, where the cationic counterpart was polyallylamine hydrochloride (PAH). Here, a clear AqpZ effect could be observed as Jv increased by 30%–50% after addition of proteoliposomes, where the effect was stronger when there was a higher amount of negatively charged lipids present. The MgCl2 rejection was comparable to the work of Zhao et al. [1]; however, no NaCl rejection was presented [2]. This work was extended by encapsulating magnetic nanoparticles to force more proteoliposomes magnetically to adsorb on the polyanionic film. In FO mode, they measured an increase in both, Jv and Js with increasing mPAR, which speaks for remaining defects despite the efforts to load more vesicles onto the supporting substrate [141].
Wang and coworkers from Ocean University of China followed up on that approach and immobilized AqpZ proteoliposomes with positively charged lipids on top of a negatively charged PSS layer, followed by PEI on an UF PAN membrane [142]. Modest NaCl rejection and Jv decrease indicated a highly defective membrane. An increase in Jv between liposomes and proteoliposomes as well as further increase in Jv with higher mPAR could indicate the presence of AQP. NaCl rejection remained however unchanged between all membranes. They further showed that membrane performance was compromised after detergent treatment [142].
All designs are summarized in Figure 8, and based on the results obtained so far, we conclude that the embedment of proteopolymersomes or -liposomes in a layer results in more efficient membranes than layer-based immobilization. A great advantage of the PA-embedment technique is that no precoating/functionalization is needed which otherwise severely limits any upscaling [1]. All reported performances are however still modest compared to theoretical predictions and clearly more development is required. The major dilemma seems to be that with increasing mPAR, Jv increases, but the matrix layer becomes weaker and more prone to salt leakage. Introduction of sealing and stabilizing polymer networks could improve rejection but may also compromise Jv [107].
Next to ABPMs and ABLMs, there are two main directions aiming to achieve biomimetic membranes by AQP mimicking artificial channels: carbon nanotubes (CNTs) and organic building block nano channels [143]. CNT is more prominent because fast water permeation is proven in theory [144] and experimentally [145]. With regard to organic nano channels, there are five promising structures found to compete with ABPMs, ABLMs and CNTs: zinc and N,N-diacetic acid imidazolium bromide based zwitterionic coordination polymers [146], helical pores of dendritic dipeptides [147], imidazole compounds with urea ribbons [148], hydrazine-appended pillar[5]arenes, macrocycles of m-phenylene ethynylene [149]. Their great advantage is their smaller size with comparable channel diameter (3–10 Å) [143].
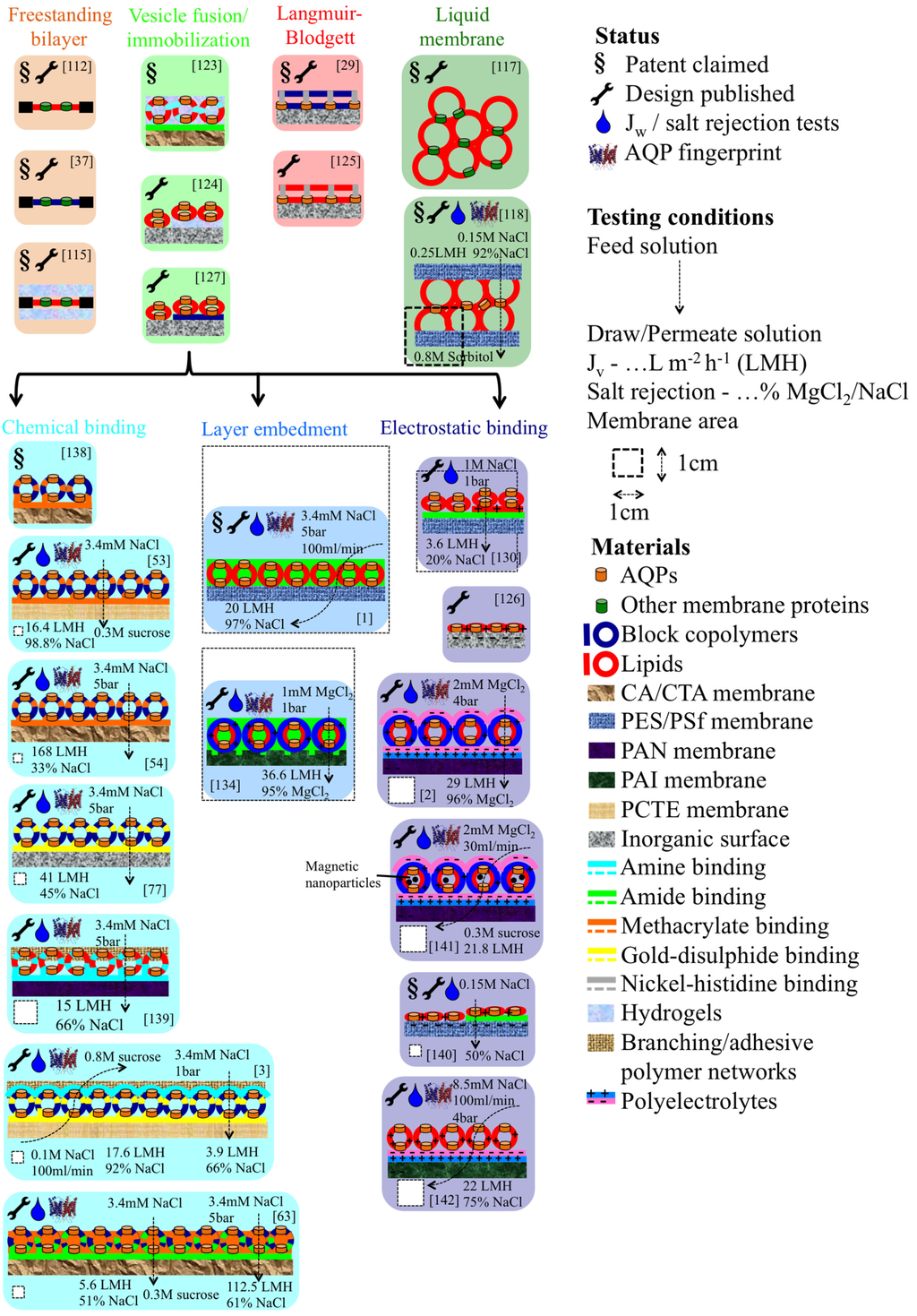
Figure 8.
Schematic overview of all published designs for ABPMs and ABLMs. Pioneer work is mainly done by Kumar and Aquaporin AIS. The most experimental designs has been done by NUS, where NTU published the most promising layer embedment ABLMs. The main recent work is on LbL-based electrostatic binding, for example binding of proteoliposomes on a polyelectrolyte layer [142].
4.3. POSS—A Novel Element in Interfacial Polymerization
Nearly all RO and FO membranes are PA-based, often referred to as thin film composite (TFC) membranes due to their superior performance compared to other membrane designs. A PA-layer is generally generated by a reaction between an amine and an acyl chloride [150]. This reaction can be prepared by dissolving the amine group in an aqueous phase, and the acyl chloride group in an organic phase [151]. Typically, a membrane is wetted with the aqueous phase, containing the amine group and dried a little to remove visible liquid while keeping the surface moist. Then the organic phase with the acyl chloride group is added on top. The reaction growth is believed to be directed into the organic phase [152], due to a preferential solubility of the amine group in the organic phase compared to the solubility of the acyl chloride in the aqueous phase. This results in the well-known ridge and valley form of PA layer. The standard amine-acyl chloride combination is MPD and TMC, and typically these are supplemented with additives (molecules with similar chemistries) in low concentrations to improve flux, rejection or chlorine resistance [150].
The ideal AL of a PA membrane for water separation has to be highly water permeable, while rejecting all other solutes and being resistant against cleaning. An ideal AL of an ABPM could even be water impermeable if it enables sufficient integration of proteopolymersomes in such a way that water only passes the incorporated proteins. Therefore, novel AL components have to be explored. An AL with homogenous thickness could facilitate such proteopolymersome integration.
We have used POSS (amine linker) and TMC (acyl chloride linker) for their potential use for the integration of proteopolymersomes in ABPMs. In a recent study POSS has been introduced as an AL layer components and POSS-TMC-layer exhibited a well-defined layer without ridges and valleys but with high mechanical stability on PAN membranes [153]. This may be a better platform for the integration of proteopolymersomes compared to the ridge-and-valley MPD-TMC network. The reaction is schematically depicted in Figure 9.
Here, we prepared PA layers (hereinafter referred to as AL) of POSS+TMC containing polymersomes of PB29-PEO16 in the aqueous phase. The influence of vesicles on the AL properties were determined, in order to provide a basis for subsequent addition of AQPs. We selected PB29-PEO16 due to its ability to form large amounts of stable polymersomes in aqueous phase compared to other PB-PEO polymersomes [154]. For the microfluidic approach, we used proteopolymersomes (AqpZ, PB33-PEO18, mPAR 1:100). PB33-PEO18 forms large amounts of stable polymersomes in aqueous phase as well and showed successful AqpZ incorporation as evidenced by SAXS. We used MilliQ water as the aqueous phase and hexane as organic phase and in order to achieve the lowest possible polydispersity, polymersomes were sonicated resulting in 95% of the polymersomes with a diameter of 196±83 nm as determined by dynamic light scattering (DLS).
We produced a non-supported AL by simply adding both phases after another in a beaker and an AL supported by microfiltration (MF) polyethersulfone (PES) layers using different coating procedures (for details see supplementary information). Characterization of the non-supported AL was done using Fourier-transformed infrared spectroscopy (FTIR), SEM and a novel, recently published microfluidic approach that allows for direct monitoring of the polymerization process [10]. Characterization of the supported AL was also achieved via FTIR and SEM, where it was also tested for functionality using standard flux and rejection test in FO mode and methylviolet staining.
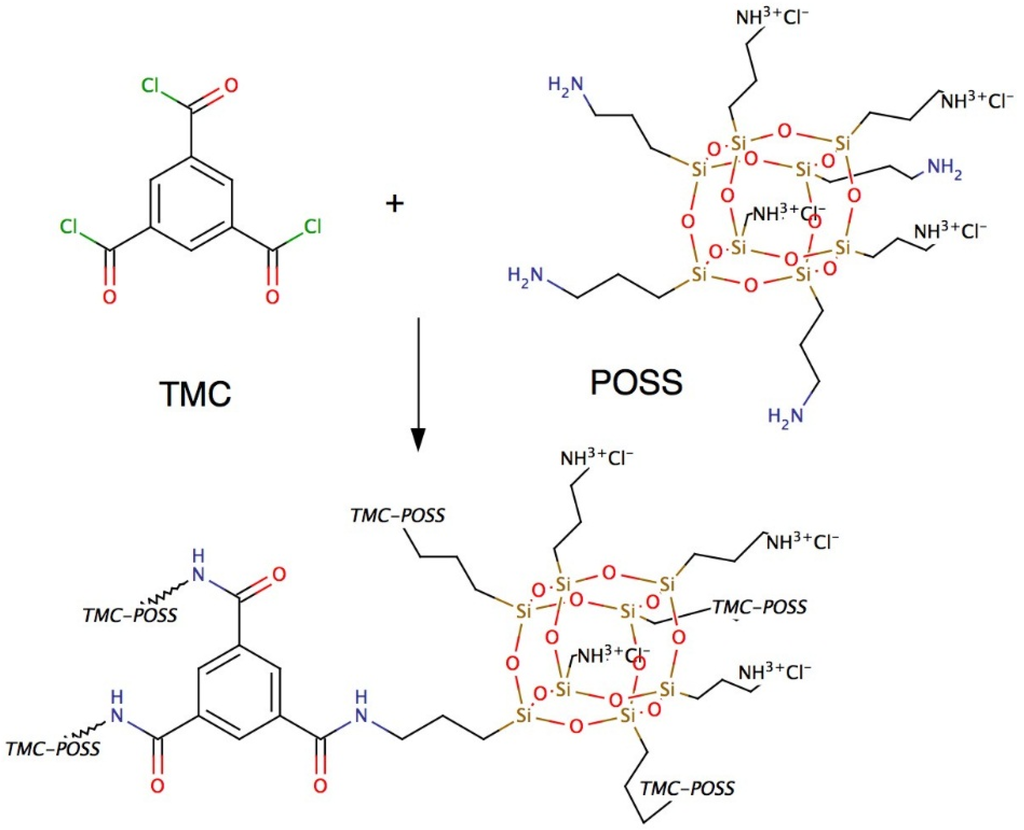
Figure 9.
Chemical structure of POSS and TMC and the resulting AL. POSS as the amine linker generate a highly stable and well-defined AL with TMC.
After adding both phases, pieces of the formed (non-supported) AL were air dried then vacuum-dried where they crumbled to flake-like structures. FTIR analysis of POSS+TMC with addition of polymersomes revealed the presence of block copolymers in the AL, see Figure 10. AL with polymersomes had an absorption peak around 3000 cm−1 (C-H stretch), which can also be found in spectra of PB and PEO [155,156]. The polymersome-free AL exhibited a broad peak at that wavelength range but not a distinct maximum as for the polymersome-containing AL. This could indicate a successful polymersome integration in the AL. Polymersomes furthermore did not seem to block PA formation, because the characteristic peaks of a PA bond, the C=O stretch at 1636 cm−1, as well as the N-H stretch at 1545 cm−1 [153] were clearly visible in the AL with polymersomes. Finally, partial hydrolysis of the POSS leading to the AL formation is not substantially affected by the presence of the polymerosomes as far as the characteristic peaks for the POSS-cage and ladder (1125 cm−1 and 1040 cm−1 [153]) were present in both AL. There was however an apparent influence of the polymersomes on TMC reactivity. Originally, Dalwani et al. used 2g/L TMC for their non-supported and supported AL [153]. In our case we could not form a non-supported AL with 2 g/L but with 0.5 g/L TMC. Potentially the TMC-POSS-stochiometry was artificially increased by the presence of another species in the aqueous phase. An excess of TMC could hinder network structure formation, because TMC will not connect POSS cages, resulting only in low molecular weight networks. We used 0.5 g/L TMC for the non-supported POSS+TMC AL and POSS/polymersomes+TMC AL.
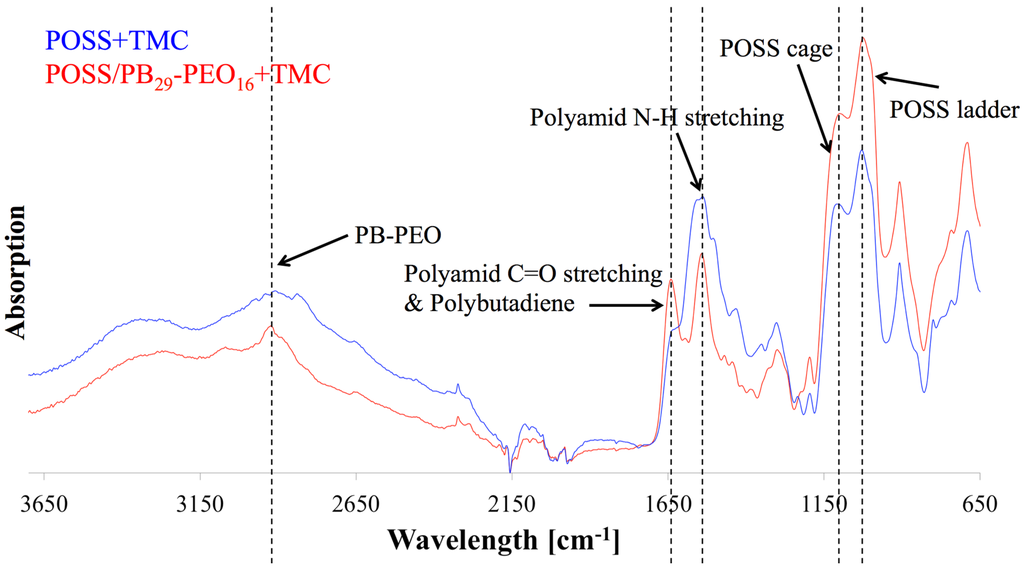
Figure 10.
FTIR diagram of POSS/polymersomes+TMC AL (labelled red) and POSS+TMC control AL (labelled blue) as a function of wavelength against absorption. The AL with polymersomes had an absorption peak around 3000 cm−1, that responds to PB and PEO, indicating their presence in the AL, where the characteristic absorption peaks for PA bonds and POSS were present as well.
The FTIR results were complemented with SEM analysis of the same samples, see in Figure 11. The POSS+TMC AL appeared smooth and well-defined, in agreement with previous work [153], see Figure 11a,b. When polymersomes were added (Figure 11c–e) a clear distinction can be made between the side towards the organic phase, that does not reveal presence of polymersomes (Figure 11c) and the side that faced the aqueous layer, which is well-covered with polymersomes (Figure 11e).
Most of the polymersomes seemed to sit loosely on top of the AL, whereas some polymersomes seemed to be covered to a certain extent by the AL, their shape less sharp than the others (indicated by the dotted circles in image Figure 11d). A few polymersomes were directly embedded inside the AL, visible from its cracked profile (arrows in Figure 11d). This could indicate that the POSS approach can be used to embed polymersomes in such a way that they would be suitable for membrane fabrication.
Recently, a novel approach from the microfluidic field was published [10] that allows visual study of the evolution of the location of interfacial polymerisation reactions. This involves a chip containing a hydrophobized micro-chamber that is separated in two compartments by an array of micro-pillars each with a diameter of 30 µm and a height of 50 µm. The aqueous phase with amine linker was introduced via micro capillary connections into one compartment and formed a water-air-interface between the pillars. Then the organic phase with acyl chloride linker was introduced into the other compartment. AL formation at the interface between the solutions was observed using an optical microscope. Depending on the linkers, the resulting AL will have a different morphology and formation time. POSS+TMC forms well-defined AL with a formation time within 4 s. In contrast, for instance the apparent growth of a film from Jeffamine+TMC is not finalized after 15 min and the film reveals the ridge and valley structures that are typical for AL formed by interfacial polymerization [10].
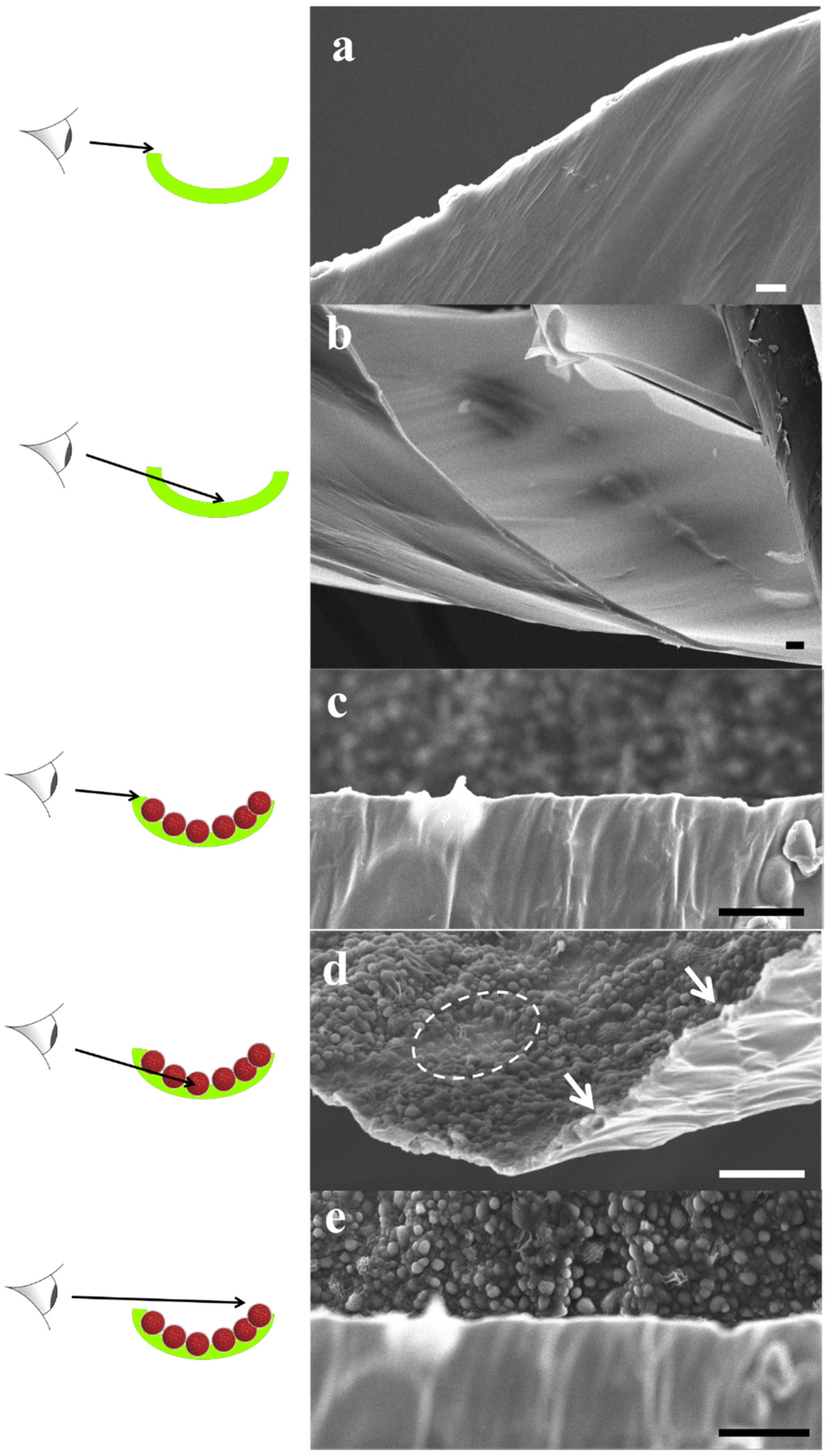
Figure 11.
SEM images of POSS+TMC AL (a,b) and of POSS/polymersomes+TMC AL (c–e) with schematic sketches, which part of the layer is being captured. Images were taken from different parts of the flakes (labelled green in the sketch) of the AL, that were generated during the SEM preparation. The AL without polymersomes was smooth and well-defined, which remained on the organic side when polymersomes were added. The aqueous side was covered with loosely attached and half-covered polymersomes (dotted circle in (d)). A few could be observed inside the AL (arrows in (d)). Scale bar is 3 µm.
We used this approach to monitor POSS/proteopolymersomes+TMC AL (AqpZ & PB33-PEO18, mPAR 1:100), see Figure 12. The chip that was used was not hydrophobized optimally, which resulted in partial infusion of the aqueous phase into the channel with the organic phase. The hydrophobization was still sufficiently efficient to hinder the aqueous phase from passing entirely to the other compartment. Other reasons for the shift of the interface from the pillar structures to the organic phase could be overpressure from the aqueous phase, which is hard to control since the offered pressure is in the range of 104 Pa. When the organic phase containing TMC was introduced, the typical sharp AL was formed at the aqueous-organic interface (Figure 12b dotted line 1). After that, the reaction was continued by the diffused amine into the organic phase and the formed AL that connected the initial interfaces, exhibited a new aqueous-organic interface (Figure 12b dotted line 2). Such observation demonstrates a less denser AL formed by POSS/proteopolymersomes-TMC compared to that formed by POSS-TMC reaction. The formation time was on the order of seconds. The film remained in the same shape and no further growth was observed in the following 12 h.
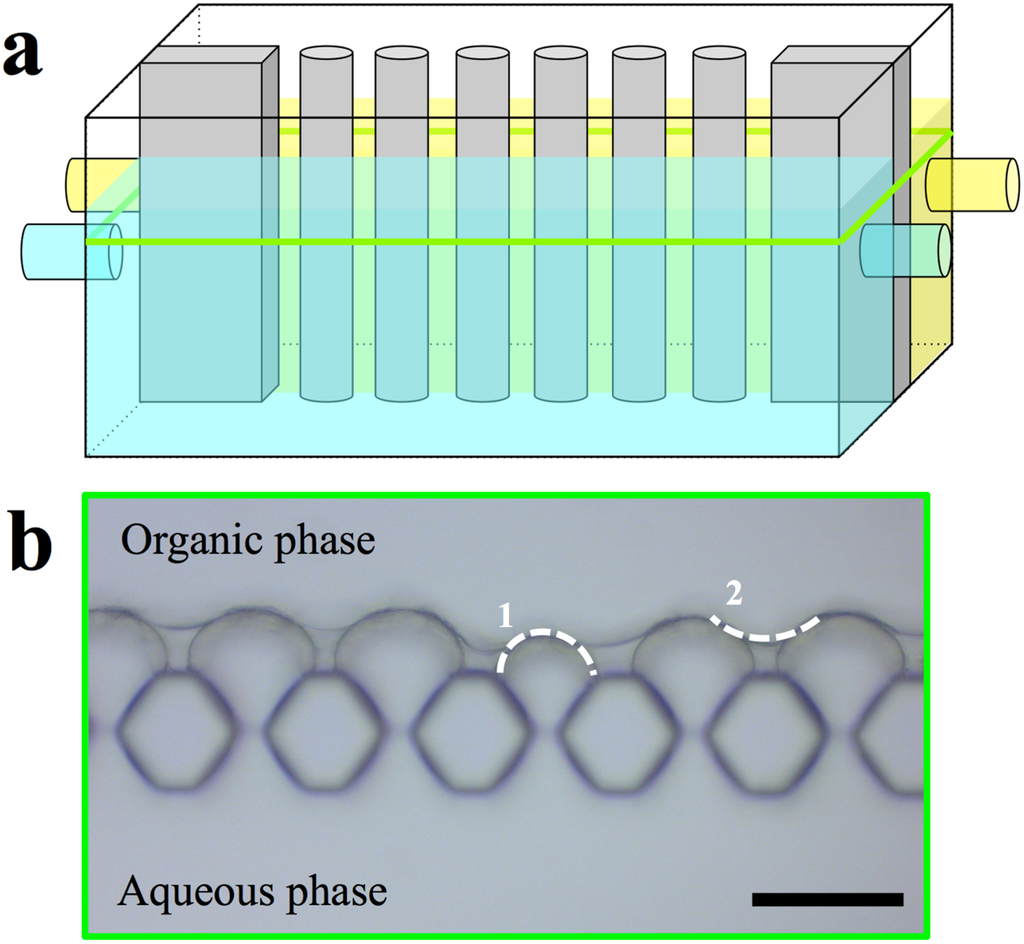
Figure 12.
(a) Schematic sketch of the microfluidic chamber and micrographs of POSS/proteopolymersomes+TMC AL and (b) micrograph of the compartment. The aqueous phase reached into the other compartment. After introducing the organic phase, a well-defined AL formed. Scale bar is 50 µm.
We then investigated POSS+TMC on MF PES support material coated following the procedures in Dalwani et al. [153], which is further described in the supporting information. The MF PES itself was supported by a nonwoven. FTIR spectroscopy revealed the presence of polymersomes in the supported AL, however, the PA formation is significantly reduced compared to the non-supported POSS/polymersomes+TMC AL, see Figure 13. A main challenge of analyzing supported AL with FTIR is the potential absorption of the PES support, which has a strong absorption especially in the region between 700 and 2000 cm−1. Especially, the POSS absorption peaks interfered strongly with PES peaks. In the supported POSS/polymersomes+TMC AL the PB-PEO signal at 3000 cm−1 was present as well as another small peak around 1700 cm−1 that also appeared in the FTIR spectra of PB [155]. Interestingly, it could not be found in the non-supported AL. Potentially, it was overlayed from the background signal in the region between 1600–3650 cm−1 that was more significant at the FTIR analysis spectra from the non-supported AL. Both PA bonds were present in the supported POSS+TMC AL but strongly reduced in the one with polymersomes. The large peak (at 1580 cm−1) close to the N-H stretching peak, is associated with PES. The N-H stretching peak (1545 cm−1) was only present in the supported POSS+TMC AL. The broad peak of this AL from 3150–3650 cm−1 is likely associated to water and/or unreacted amine groups.
The reason for the suppression of the PA-signal in the supported POSS/polymersomes+TMC AL is not clear. It may be related to the TMC reactivity as discussed before. We used 2 g/L TMC for the supported POSS+TMC AL and POSS/polymersomes+TMC AL, because there was no AL formation at 0.5 g/L. Another TMC concentration may be more optimal for the supported POSS/polymersomes+TMC AL. The potential blockage of PA formation induced by polymersomes should have suppressed the PA formation in the non-supported AL as well, which it did not. However, AL formation was significantly decreased with supported POSS+TMC AL when changing from 2 g/L to 0.5 g/L. Another hypothesis could be that POSS+TMC do not form easily on MF PES. To our knowledge, no former POSS+TMC AL formation on MF PES has been reported. MF PES has significantly bigger pore sizes than PAN. This could hamper the formation of a smooth layer.
In contrast to the FTIR analysis, SEM analysis showed a completely covered POSS/polymersomes+TMC AL on the MF PES (Figure 14). In addition, POSS and TMC alone seemed to cover the microporous PES structure completely with the smooth layer, although less defined than for PAN substrates [153]. This could be due to the different pore size as mentioned before. When polymersomes were added, the AL exhibited sub-micron sized bumps. They are 1.5–2 µm in length and 0.5–1 µm in height. Considering a covering AL of 100 nm thickness [153] (Figure 14 sketch in bottom left corner) there would be groups of 6–9 polymersomes in a row in 1–3 layers. In contrast to the non-supported ALs, we can only observe the side facing the organic phase. In the case of the supported POSS/polymersomes+TMC AL, the polymersomes influence the shape of the AL side facing the organic phase to a far higher extent than in the non-supported form. This is most probably due to the different preparations, with regard to POSS/polymersomes being in solution at the non-supported AL formation and being at the water-air-interface or even dried on the MF PES at the supported AL formation. Thus, the chances of polymersomes being integrated in the AL is higher for the supported AL than for the non-supported one.
In conclusion, SEM analysis revealed a successful embedment of polymersomes in a supported AL, whereas FTIR data were less informative. A limitation of FTIR and SEM analysis of supported AL is that only a small fraction of the whole membrane is observed. Another aspect is that the AL could become brittle during drying, and delaminate, or break off when exposed to liquid nitrogen that is used for SEM sample preparation.
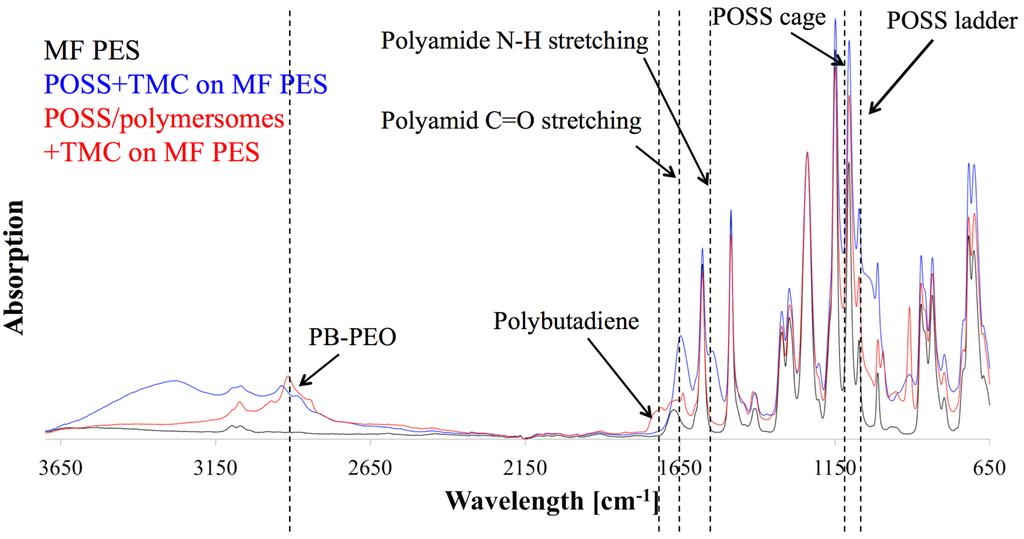
Figure 13.
FTIR analysis of supported POSS/polymersomes+TMC AL (red) and POSS+TMC control AL (blue) on MF PES and pure MF PES (black). The PES supporting material had high absorption and interfered with many absorption peaks. A subtraction from the absorption spectra of pure PES resulted in negative peaks. We therefore only normalized the spectra. PB-PEO was present in the AL with polymersomes; however, the PA formation was strongly suppressed.
We also attempted to test the POSS/polymersomes+TMC AL on MF PES in terms of flux and rejection measurements in FO mode. However, we did not see any FO performance. Approximately one third of the membranes tested were impermeable to salt as evidenced by the low conductivity change in the feed solution within 2 h. The rest of the membranes were leaky as evidenced by an immediate increase in conductivity. The fraction of sealed and leaky membranes of POSS+TMC and POSS/polymersomes+TMC were comparable. In the sealed membranes, the pores are likely clogged by several accumulated layers of POSS+TMC AL. However, after staining with methyl blue no pinholes or scratches were detected on the surface, suggesting that the supporting PES was covered with the AL. As mentioned before, MF PES may not be a suited support for POSS+TMC ALs in general. PAN support did not show any flux without hydraulic pressure, due to the small pore size (5–30 nm) [157]. It may be suited for POSS+TMC for a NF membrane, but not for FO. A compromise would be to use UF PES membranes as used by Lee et al. [158].
To conclude this subchapter, we obtained insights in non-supported and supported AL containing POSS with polymersomes in the aqueous phase and TMC in the organic phase. The non-supported POSS/polymersomes+TMC AL was formed successfully with high amounts of polymersomes covered and some of them even integrated inside the AL. The supported POSS/polymersomes+TMC showed a different characteristics. FTIR data indicated a high suppression of the AL formation at polymersome addition, whereas SEM images showed a completely covered and significantly different AL upon polymersome addition. None of the membranes produced, containing POSS and TMC had any reasonable performance, probably due to incomplete coverage of the AL. Still, it was interesting to get an insight into how POSS, TMC and proteopolymersomes are interacting. Further challenges will be to create a functional water separation membrane from these components.
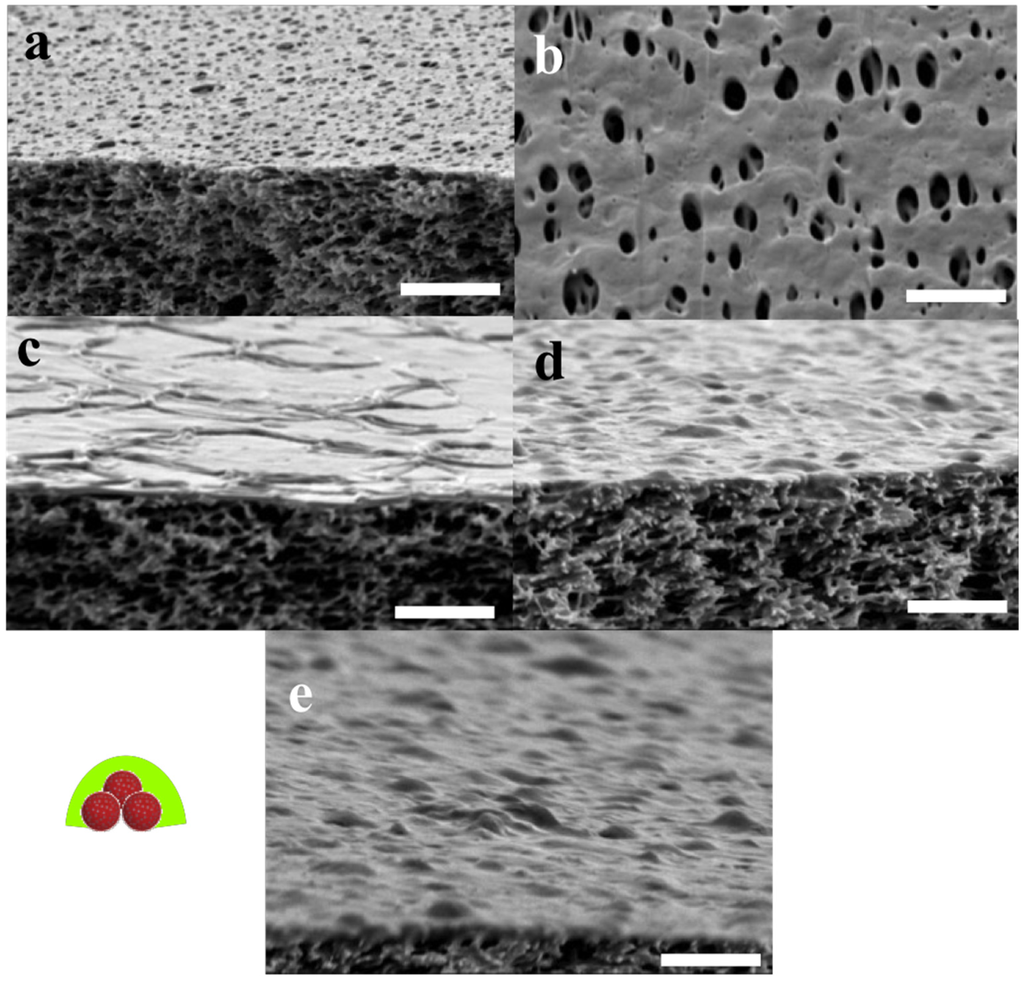
Figure 14.
SEM images of MF PES (a,b) supported POSS+TMC AL on MF PES (c) and supported POSS/polymersomes+TMC on MF PES (d,e). Schematic sketch of polymersome coverage left to (e). Micropores of the MF PES were covered completely by the POSS+TMC AL. After addition of polymersomes, small bumps with dimensions similar to the polymersomes were observed on the organic faced side of the AL. Greater bumps may be attributed to accumulations of covered polymersomes. Scale bar is 3 µm.
5. Perspectives
As mentioned in other reviews [4], ABPMs are rapidly evolving and coming of age. Future challenges will be the upscaling production of both AQPs and block copolymers. Another relevant economic issue is the use of AQP-solubilizing detergents that have a broad price range. ABPMs need to be comparable to established membrane technologies in terms of cost and scale. As seen in Figure 8, all published studies about ABPMs are tested in small-scale laboratory experiments. Even though there are commercially available ABPMs in m2-scale, more development will be needed. The lessons learnt from nature are not completely transferred yet.
Supplementary Files
Supplementary File 1Acknowledgments
We thank Michiel Raaijmakers, Yali Zhang, Evelien Maaskant and Nieck Benes, University of Twente, especially Michiel Raaijmakers and Yali Zhang for helpful discussion and assistance in the lab for all experiments with POSS+TMC membranes; Daniela Pirner, University of Bayreuth, Germany, and Sokol Ndoni, DTU Nanotech, for helpful discussions and Simon Levinsen, DTU Nanotech for assistance in polymer synthesis; Fabian Itel and Mohamed Chami, both University of Basel, Switzerland, and Manish Kumar, Pennsylvania State University, USA, for useful discussions about polymersomes in general and freeze fracture; Fadoua Sbai, Aquaporin A/S for assistance with polymersome preparation; Per Amstrup Pedersen for providing AQP10-GFP; Klaus Qvortrup and Ramon Liebrechts, University of Copenhagen, for providing assistance with freeze fracture and access to the Leica freeze fracture instruments, Krzysztof Trzaskus, Joris de Grooth and Harmen Zwijnenberg for providing assistance with the membrane flux and rejection tests. We thank NXUS for the collection and analysis of the SAXS-data. NXUS is a pilot project enabling industrial use of large scale research facilities in collaboration with university experts in the field of neutron and X-ray science. NXUS is funded by the University of Copenhagen and the capital region of Denmark. JH was supported by an industrial PhD grant from Innovation Fund Denmark. CHN was supported by the IBISS—Industrial Biomimetic Sensing and separation platform (http://www.ibiss.dtu.dk) funded by the Danish Innovation Fund Grant No. 097-2012-4.
Author Contributions
Conceived and designed the experiments: Joachim Habel, Anayo Ogbonna. Performed the experiments: Joachim Habel, Michael Hansen, Nanna Larsen, Søren Kynde, Søren Roi Midtgaard. Analyzed the data: Joachim Habel, Michael Hansen, Søren Kynde, Søren Roi Midtgaard, Grethe Vestergaard Jensen. Contributed reagents/materials/analysis tools: Joachim Habel, Michael Hansen, Søren Kynde, Søren Roi Midtgaard, Grethe Vestergaard Jensen, Julie Bomholt, Anayo Ogbonna, Kristoffer Almdal, Alexander Schulz, Claus Hélix-Nielsen. Wrote the paper: Joachim Habel, Søren Kynde, Claus Hélix-Nielsen.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations/Nomenclature
| ABLM | Aquaporin-based lipidic biomimetic membrane |
| ABM | Aquaporin-based biomimetic membrane |
| ABPM | Aquaporin-based polymeric biomimetic membrane |
| 7-ADCA | 7-aminodesacetoxycephalosporanic acid |
| AFM | Atomic force microscopy |
| AL | Active layer |
| AQP | Aquaporin |
| AqpZ | Aquaporin Z |
| BR | Bacteriorhodopsin |
| BSA | Bovine serum albumin |
| CA | Cellulose acetate |
| CcO | Cytochrome c oxidase |
| CNT | Carbon nanotube |
| Cr | Planar shape with protein crystals |
| DLS | Dynamic light scattering |
| DMA | Dimethacrylate |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| DPhPC | 1,2-diphytanoyl-sn-glycero-3-phosphocholine |
| DTU | Danish Technical University |
| EFTE | Ethylene tetrafluoroethylene |
| ELF | Enzyme-labelled fluorescence |
| EPR | Electron paramagnetic resonance |
| f | Hydrophilic volume ratio |
| FCS | Fluorescence correlation spectroscopy |
| FE-SEM | Field-emission scanning electron microscopy |
| FF-TEM | Freeze-fracture transmission electron microscopy |
| FhuA | Ferric hydroxamate uptake protein |
| FI | Functional incorporation |
| FO | Forward osmosis |
| FTIR | Fourier-transformed infrared spectroscopy |
| FR | Film rehydration |
| Js | Membrane reverse salt flux |
| Jv | Membrane water flux |
| LA | Lipoic acid |
| LamB | Phage lambda receptor |
| LbL | Layer-by-layer |
| Mn | Number-averaged molecular weight |
| Mw | Weight-averaged molecular weight |
| MAq | Mixing in aqueous phase |
| Mc | Micellar shape |
| MF | Microfiltration |
| MloK1 | Potassium channel from Mesorhizobium loti |
| MPD | m-phenyl diamine |
| MPEG | Methyl polyethyleneglycol |
| mPAR | Molecular amphiphile-to-protein-ratio |
| MOr | Mixing in organic phase |
| PVL | Polyvalerolactone |
| NA | Not announced |
| NAD | β-nicotinamide adenine dinucleotide |
| NADH | Hydrogenated β-nicotinamide adenine dinucleotide |
| ND | Not determined |
| NF | Nanofiltration |
| NtAQP1 | Tobacco plasma membrane intrinsic protein 1 |
| NtPIP2;1 | Tobacco plasma membrane intrinsic protein 2 |
| NUS | National University of Singapore |
| OG | n-Octyl-β-D-Glucopyranoside |
| OmpF | Outer membrane protein F |
| OmpG | Outer membrane protein G |
| P | Planar shape |
| P2VP | Poly-2-vinyl pyridine |
| P4MVP | Poly-4-vinyl methylpyridine iodide |
| P4VP | Poly-4-vinyl pyridine iodide |
| PA | Polyamide |
| PAA | Polyacrylic acid |
| PAI | Polyamide imide |
| PAH | Polyallylamine hydrochloride |
| PAN | Polyacrylonitrile |
| PB | Polybutadiene |
| PBR | Polymer bulk rehydration |
| PBMA | Polybutyl methacrylate |
| PCTE | Polycarbonate track-etched |
| PDA | Polydopamine |
| PDA | Polydopamine |
| P DI | Polydispersity index |
| PDMS | Polydimethylsiloxane |
| PEE | Polyethylethylene |
| PEOXA | Polyethylene oxazoline |
| PEO | Polyethylene oxide |
| PES | Polyethersulfone |
| PFR | Polymer film rehydration |
| PGM | Polyglycerol monomethacrylate |
| PGME | Phenylglycine methyl ester |
| PHEMA | Polyhydroxyethyl methacrylate |
| PI | Polyisoprene |
| PIB | Polyisobutylene |
| PMOXA | Polymethyloxazoline |
| POSS | Polyhedral oligomeric silsesquioxane |
| PPFR | Protein/polymer film rehydration |
| PPO | Polypropylene oxide |
| PR | Proteorhodopsin |
| PRO | Pressure retarded osmosis |
| PS | Polystyrene |
| PSf | Polysulfone |
| PSS | Sulfonate polysulfone |
| QCM-D | Quartz crystal microbalance with dissipation |
| RC | Reaction centre |
| RO | Reverse osmosis |
| S | Shape |
| SFLS | Stopped-flow light scattering |
| SAXS | Small-angle X-ray scattering |
| SDU | Southern Danish University |
| SE | Solvent evaporation |
| SEC | Size exclusion chromatography |
| SEM | Scanning electron microscopy |
| SI | Solvent injection |
| SMTC | Singapore Membrane technology Centre |
| SoPIP2;1 | Spinach plasma membrane intrinsic protein 2;1 |
| TFC | Thin film composite |
| TMB | 3,3,5,5-tetramethyl-benzidine |
| TMC | Trimesoyl chloride |
| UF | Ultrafiltration |
| V | Vesicular shape |
References
- Zhao, Y.; Qiu, C.; Li, X.; Vararattanavech, A.; Shen, W.; Torres, J.; Hélix-Nielsen, C.; Wang, R.; Hu, X.; Fane, A.G.; et al. Synthesis of robust and high-performance aquaporin-based biomimetic membranes by interfacial polymerization-membrane preparation and RO performance characterization. J. Membr. Sci. 2012, 423–424, 422–428. [Google Scholar] [CrossRef]
- Sun, G.; Chung, T.S.; Jeyaseelan, K.; Armugam, A. A layer-by-layer self-assembly approach to developing an aquaporin-embedded mixed matrix membrane. RSC Adv. 2013, 3, 473. [Google Scholar] [CrossRef]
- Wang, H.L.; Chung, T.S.; Tong, Y.W.; Jeyaseelan, K.; Armugam, A.; Duong, H.H.P.; Fu, F.; Seah, H.; Yang, J.; Hong, M. Mechanically robust and highly permeable AquaporinZ biomimetic membranes. J. Membr. Sci. 2013, 434, 130–136. [Google Scholar] [CrossRef]
- Tang, C.Y.; Wang, Z.; Petrinic´, I.; Fane, A.G.; Hélix-Nielsen, C. Biomimetic aquaporin membranes coming of age. Desalination 2015, 368, 89–105. [Google Scholar] [CrossRef]
- Tang, C.Y.; Zhao, Y.; Wang, R.; Hélix-Nielsen, C.; Fane, A.G. Desalination by biomimetic aquaporin membranes: Review of status and prospects. Desalination 2013, 308, 34–40. [Google Scholar] [CrossRef]
- Hélix-Nielsen, C. Biomimetic Membranes for Sensor and Separation Applications; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Kita-Tokarczyk, K.; Meier, W. Biomimetic block copolymer membranes. CHIMIA 2008, 62, 820–825. [Google Scholar] [CrossRef]
- Raaijmakers, M.J.T.; Hempenius, M.A.; Schön, P.M.; Vancso, G.J.; Nijmeijer, A.; Wessling, M.; Benes, N.E. Sieving of Hot Gases by Hyper-Cross-Linked Nanoscale-Hybrid Membranes. J. Am. Chem. Soc. 2014, 136, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Ajit Walter, P.; Muthukumar, T.; Reddy, B. Assessment of antifouling efficacy of polyhedral oligomeric silsesquioxane based poly (urea-urethane-imide) hybrid membranes. Lett. Appl. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Benes, N.E.; Lammertink, R.G.H. Visualization and characterization of interfacial polymerization layer formation. Lab Chip 2015, 15, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, Y.; Racker, E. Partial Resolution of the Enzymes Catalyzing Oxidative Phosphorylation. J. Biol. Chem. 1971, 246, 5477–5487. [Google Scholar]
- Nardin, C.; Thoeni, S.; Widmer, J.; Winterhalter, M.; Meier, W. Nanoreactors based on (polymerized) ABA-triblock copolymer vesicles. Chem. Commun. 2000, 1433–1434. [Google Scholar] [CrossRef]
- Zhang, X.; Tanner, P.; Graff, A.; Palivan, C.G.; Meier, W. Mimicking the cell membrane with block copolymer membranes. J. Polym. Sci. Part A: Polym. Chem. 2012, 50, 2293–2318. [Google Scholar] [CrossRef]
- Tanner, P.; Baumann, P.; Enea, R.; Onaca, O.; Palivan, C.; Meier, W. Polymeric Vesicles: From Drug Carriers to Nanoreactors and Artificial Organelles. Acc. Chem. Res. 2011, 44, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Montemagno, C. Artificial organelle: ATP synthesis from cellular mimetic polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef] [PubMed]
- Vriezema, D.M.; Garcia, P.M.L.; Sancho Oltra, N.; Hatzakis, N.S.; Kuiper, S.M.; Nolte, R.J.M.; Rowan, A.E.; van Hest, J.C.M. Positional Assembly of Enzymes in Polymersome Nanoreactors for Cascade Reactions. Angew. Chem. Int. Ed. 2007, 46, 7378–7382. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Grzelakowski, M.; Zilles, J.; Meier, W.P. Highly permeable polymeric membranes based on the incorporation of the functional water channel protein Aquaporin Z. Proc. Natl. Acad. Sci. USA 2007, 104, 20719–20724. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Habel, J.; Shen, Y.x.; Meier, W.; Walz, T. High-Density Reconstitution of Functional Water Channels into Vesicular and Planar Block Copolymer Membranes. J. Am. Chem. Soc. 2012, 134, 18631–18637. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O. Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Pata, V.; Dan, N. The Effect of Chain Length on Protein Solubilization in Polymer-Based Vesicles (Polymersomes). Biophys. J. 2003, 85, 2111–2118. [Google Scholar] [CrossRef]
- Srinivas, G.; Discher, D.; Klein, M. Key Roles for Chain Flexibility in Block Copolymer Membranes that Contain Pores or Make Tubes. Nano Lett. 2005, 5, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Ortiz, V.; Srinivas, G.; Klein, M.L.; Kim, Y.; Christian, D.; Cai, S.; Photos, P.; Ahmed, F. Emerging applications of polymersomes in delivery: From molecular dynamics to shrinkage of tumors. Prog. Polym. Sci. 2007, 32, 838–857. [Google Scholar] [CrossRef] [PubMed]
- </b>Aponte-Santamaría, C.; Briones, R.; Schenk, A.; Walz, T.; de Groot, B. Molecular driving forces defining lipid positions around aquaporin-0. Proc. Natl. Acad. Sci. USA 2012, 109, 9887–9892. [Google Scholar]
- Hansen, J.S.; Vararattanavech, A.; Plasencia, I.; Greisen, P.J.; Bomholt, J.; Torres, J.; Emnéus, J.; Hélix-Nielsen, C. Interaction between sodium dodecyl sulfate and membrane reconstituted aquaporins: A comparative study of spinach SoPIP2;1 and E. coli AqpZ. Biochim. Biophys. Acta (BBA)—Biomembr. 2011, 1808, 2600–2607. [Google Scholar] [CrossRef] [PubMed]
- Stoenescu, R.; Graff, A.; Meier, W. Asymmetric ABC-Triblock Copolymer Membranes Induce a Directed Insertion of Membrane Proteins. Macromol. Biosci. 2004, 4, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Hite, R.K.; Li, Z.; Walz, T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010, 29, 1652–1658. [Google Scholar] [CrossRef] [PubMed]
- Nehring, R.; Palivan, C.G.; Casse, O.; Tanner, P.; Tüxen, J.; Meier, W. Amphiphilic Diblock Copolymers for Molecular Recognition: Metal-Nitrilotriacetic Acid Functionalized Vesicles. Langmuir 2009, 25, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; Abeyrathne, P.; Dukovski, D. The Affinity Grid: A pre-fabricated EM grid for monolayer purification. J. Mol. Boil. 2008, 382, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M. Biomimetic Membranes as New Materials for Applications in Environmental engineering and Biology. Ph.D. Thesis, University Illinois, Champaign, IL, USA, 2010. [Google Scholar]
- Kumar, M.; Meier, W. Highly Permeable Polymeric Membranes. Patent WO 2009/078174, 18 June 2009. [Google Scholar]
- Gonen, T.; Sliz, P.; Kistler, J.; Cheng, Y.; Walz, T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 2004, 429, 193–197. [Google Scholar] [CrossRef] [PubMed]
- </b>Chandy, G.; Zampighi, G.; Kreman, M.; Hall, J. Comparison of the Water Transporting Properties of MIP and AQP1. J. Membr. Biol. 1997, 159, 29–39. [Google Scholar]
- Kumar, M.; Walz, T. High Density Membrane Protein Membranes. Patent WO 2014/028923, 20 February 2014. [Google Scholar]
- Habel, J. Structural and Functional Characterization of Aquaporin 0 Incorporated in Block Copolymers and Their Resulting Aggregate Morphologies. Master Thesis, Universität Basel, Basel, Switzerland, 2011. [Google Scholar]
- Hélix-Nielsen, C. Biomimetic membranes for sensor and separation applications. Anal. Bioanal. Chem. 2009, 395, 697–718. [Google Scholar] [CrossRef] [PubMed]
- Pszon-Bartosz, K.; Hansen, J.S.; Stibius, K.B.; Groth, J.S.; Emnéus, J.; Geschke, O.; Hélix-Nielsen, C. Assessing the efficacy of vesicle fusion with planar membrane arrays using a mitochondrial porin as reporter. Biochem. Biophys. Res. Commun. 2011, 406, 96–100. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, A.; Stibius, K.; Vissing, T. Biomimetic triblock copolymer membrane arrays: A stable template for functional membrane proteins. Langmuir 2009, 25, 10447–10450. [Google Scholar] [CrossRef] [PubMed]
- Andreasson-Ochsner, M.; Fu, Z.; May, S.; Ying Xiu, L.; Nallani, M.; Sinner, E.K. Selective Deposition and Self-Assembly of Triblock Copolymers into Matrix Arrays for Membrane Protein Production. Langmuir 2012, 28, 2044–2048. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Jamshad, M.; Knowles, T.J.; Morrison, K.A.; Downing, R.; Cant, N.; Collins, R.; Koenderink, J.B.; Ford, R.C.; Overduin, M.; et al. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014, 461, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.P.; Dafforn, T.; Overduin, M. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Scott, A.; Rotem, D.; Mehta, K.K.; Bayley, H.; Dekker, C. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat. Nanotechnol. 2010, 5, 874–877. [Google Scholar] [CrossRef] [PubMed]
- Balme, S.; Janot, J.M.; Berardo, L.; Henn, F.; Bonhenry, D.; Kraszewski, S.; Picaud, F.; Ramseyer, C. New Bioinspired Membrane Made of a Biological Ion Channel Confined into the Cylindrical Nanopore of a Solid-State Polymer. Nano Lett. 2011, 11, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Bodor, S.; Zook, J.M.; Lindner, E.; Tóth, K.; Gyurcsányi, R.E. Electrochemical methods for the determination of the diffusion coefficient of ionophores and ionophore-ion complexes in plasticized PVC membranes. Analyst 2008, 133, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, S. Supporting and stabilizing biomimetic membranes. Ph.D. Thesis, Danish Technical University, Kgs. Lyngby, Denmark, 2011. [Google Scholar]
- Mech-Dorosz, A.; Heiskanen, A.; Bäckström, S.; Perry, M.; Muhammad, H.B.; Hélix-Nielsen, C.; Emnéus, J. A reusable device for electrochemical applications of hydrogel supported black lipid membranes. Biomed. Microdevices 2015, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E. Polymer Vesicles. Science 2002, 297, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Nardin, C.; Hirt, T.; Leukel, J.; Meier, W. Polymerized ABA Triblock Copolymer Vesicles. Langmuir 2000, 16, 1035–1041. [Google Scholar] [CrossRef]
- Erbakan, M.; Shen, Y.X.; Grzelakowski, M.; Butler, P.J.; Kumar, M.; Curtis, W.R. Molecular Cloning, Overexpression and Characterization of a Novel Water Channel Protein from Rhodobacter sphaeroides. PLoS ONE 2014, 9, e86830. [Google Scholar] [CrossRef] [PubMed]
- Graff, A. Amphiphilic Copolymer Membranes Promote NADH: Ubiquinone Oxidoreductase Activity: Towards an Electron-Transfer Nanodevice. Macromol. Chem. Phys. 2010, 211, 229–238. [Google Scholar] [CrossRef]
- Wong, D.; Jeon, T.J.; Schmidt, J. Single molecule measurements of channel proteins incorporated into biomimetic polymer membranes. Nanotechnology 2006, 17, 3710–3717. [Google Scholar] [CrossRef]
- Uehlein, N.; Otto, B.; Eilingsfeld, A.; Itel, F.; Meier, W.; Kaldenhoff, R. Gas-tight triblock-copolymer membranes are converted to CO2 permeable by insertion of plant aquaporins. Sci. Rep. 2012, 2, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chung, T.S.; Tong, Y.W. Study on water transport through a mechanically robust Aquaporin Z biomimetic membrane. J. Membr. Sci. 2013, 445, 47–52. [Google Scholar] [CrossRef]
- Wang, H.; Chung, T.S.; Tong, Y.W.; Jeyaseelan, K.; Armugam, A.; Chen, Z.; Hong, M.; Meier, W. Highly Permeable and Selective Pore-Spanning Biomimetic Membrane Embedded with Aquaporin Z. Small 2012, 8, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.S.; Chung, T.S.; Jeyaseelan, K.; Armugam, A. Aquaporin-embedded biomimetic membranes for nanofiltration. J. Membr. Sci. 2012, 407–408, 27–33. [Google Scholar] [CrossRef]
- De Vocht, C.; Ranquin, A.; Willaert, R.; Van Ginderachter, J.A.; Vanhaecke, T.; Rogiers, V.; Versées, W.; Van Gelder, P.; Steyaert, J. Assessment of stability, toxicity and immunogenicity of new polymeric nanoreactors for use in enzyme replacement therapy of MNGIE. J. Control. Release 2009, 137, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Grzelakowski, M.; Cherenet, M.F.; Shen, Y.x.; Kumar, M. A framework for accurate evaluation of the promise of aquaporin based biomimetic membranes. J. Membr. Sci. 2015, 1–32. [Google Scholar] [CrossRef]
- Grzelakowski, M.; Onaca, O.; Rigler, P.; Kumar, M.; Meier, W. Immobilized Protein-Polymer Nanoreactors. Small 2009, 5, 2545–2548. [Google Scholar] [CrossRef] [PubMed]
- Ihle, S.; Onaca, O.; Rigler, P.; Hauer, B.; Rodríguez-Ropero, F.; Fioroni, M.; Schwaneberg, U. Nanocompartments with a pH release system based on an engineered OmpF channel protein. Soft Matter 2011, 7, 532–539. [Google Scholar] [CrossRef]
- Tanner, P.; Balasubramanian, V.; Palivan, C.G. Aiding Nature’s Organelles: Artificial Peroxisomes Play Their Role. Nano Lett. 2013, 13, 2875–2883. [Google Scholar] [CrossRef] [PubMed]
- Tanner, P.; Onaca, O.; Balasubramanian, V.; Meier, W.; Palivan, C.G. Enzymatic Cascade Reactions inside Polymeric Nanocontainers: A Means to Combat Oxidative Stress. Chem. Eur. J. 2011, 17, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.Ł.; Kowal, J.K.; Wu, D.; Stahlberg, H.; Palivan, C.G.; Meier, W.P. Functional surface engineering by nucleotide-modulated potassium channel insertion into polymer membranes attached to solid supports. Biomaterials 2014, 35, 7286–7294. [Google Scholar] [CrossRef] [PubMed]
- Winterhalter, M.; Hilty, C.; Bezrukov, S.M.; Nardin, C.; Meier, W.; Fournier, D. Controlling membrane permeability with bacterial porins: Application to encapsulated enzymes. Talanta 2001, 55, 965–971. [Google Scholar] [CrossRef]
- Xie, W.; He, F.; Wang, B.; Chung, T.S.; Jeyaseelan, K.; Armugam, A.; Tong, Y.W. An aquaporin-based vesicle-embedded polymeric membrane for low energy water filtration. J. Mater. Chem. A 2013, 1, 7592–7600. [Google Scholar] [CrossRef]
- Heinisch, T.; Langowska, K.; Tanner, P.; Reymond, J.L.; Meier, W.; Palivan, C.; Ward, T.R. Fluorescence-Based Assay for the Optimization of the Activity of Artificial Transfer Hydrogenase within a Biocompatible Compartment. ChemCatChem 2013, 5, 720–723. [Google Scholar] [CrossRef]
- Broz, P.; Driamov, S.; Ziegler, J.; Ben-Haim, N.; Marsch, S.; Meier, W.; Hunziker, P. Toward Intelligent Nanosize Bioreactors: A pH-Switchable, Channel-Equipped, Functional Polymer Nanocontainer. Nano Lett. 2006, 6, 2349–2353. [Google Scholar] [CrossRef] [PubMed]
- Langowska, K.; Palivan, C.G.; Meier, W. Polymer nanoreactors shown to produce and release antibiotics locally. Chem. Commun. 2013, 49, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Graff, A.; Sauer, M.; Meier, W. Virus-assisted loading of polymer nanocontainer. Proc. Natl. Acad. Sci. USA 2002, 99, 5064–5068. [Google Scholar] [CrossRef] [PubMed]
- Ranquin, A.; Versées, W.; Meier, W.; Steyaert, J.; Van Gelder, P. Therapeutic Nanoreactors: Combining Chemistry and Biology in a Novel Triblock Copolymer Drug Delivery System. Nano Lett. 2005, 5, 2220–2224. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Haefele, T.; Graff, A.; Nardin, C.; Meier, W. Ion-carrier controlled precipitation of calcium phosphate in giant ABA triblock copolymer vesicles. Chem. Commun. 2001, 7, 2452–2453. [Google Scholar] [CrossRef]
- Haefele, T.; Kita-Tokarczyk, K.; Meier, W. Phase Behavior of Mixed Langmuir Monolayers from Amphiphilic Block Copolymers and an Antimicrobial Peptide. Langmuir 2006, 22, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Nallani, M.; Benito, S.; Onaca, O.; Graff, A.; Lindemann, M.; Winterhalter, M.; Meier, W.; Schwaneberg, U. A nanocompartment system (Synthosome) designed for biotechnological applications. J. Biotechnol. 2006, 123, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Nallani, M. A Novel Nanocompartment System Named Synthosome for Biotechnological Applications. Ph.D. Thesis, Universität Bremen, Bremen, Germany, 2005. [Google Scholar]
- Onaca, O. Functionalized polymer vesicles and interactions with Polymyxin B and derivates. Ph.D. Thesis, Universität Bremen, Bremen, Germany, 2007. [Google Scholar]
- Graff, A. Insertion of Membrane Proteins in Artificial Polymer Membranes. Ph.D. Thesis, Universität Basel, Basel, Switzerland, 2006. [Google Scholar]
- Meier, W.; Nardin, C.; Winterhalter, M. Reconstitution of channel proteins in (polymerized) ABA triblock copolymer membranes. Angew. Chem. Int. Ed. 2000, 39, 4599–4602. [Google Scholar] [CrossRef]
- Nardin, C.; Widmer, J.; Winterhalter, M.; Meier, W. Amphiphilic block copolymer nanocontainers as bioreactors. Eur. Phys. J. E 2001, 4, 403–410. [Google Scholar] [CrossRef]
- Duong, P.H.H.; Chung, T.S.; Jeyaseelan, K.; Armugam, A.; Chen, Z.; Yang, J.; Hong, M. Planar biomimetic aquaporin-incorporated triblock copolymer membranes on porous alumina supports for nanofiltration. J. Membr. Sci. 2012, 409-410, 34–43. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, H.; Montemagno, C.D. Toward hybrid proteo-polymeric vesicles generating a photoinduced proton gradient for biofuel cells. Nanotechnology 2005, 16, 1589–1597. [Google Scholar] [CrossRef]
- Lee, H.; Ho, D.; Kuo, K.; Montemagno, C.D. Vectorial insertion of bacteriorhodopsin for directed orientation assays in various polymeric biomembranes. Polymer 2006, 47, 2935–2941. [Google Scholar] [CrossRef]
- Choi, H.J.; Germain, J. Effects of different reconstitution procedures on membrane protein activities in proteopolymersomes. Nanotechnology 2006, 17, 1825–1830. [Google Scholar] [CrossRef]
- Choi, H.J.; Montemagno, C.D. Biosynthesis within a bubble architecture. Nanotechnology 2006, 17, 2198–2202. [Google Scholar] [CrossRef]
- Choi, H.J.; Montemagno, C. Light-Driven Hybrid Bioreactor Based on Protein-Incorporated Polymer Vesicles. IEEE Trans. Nanotechnol. 2007, 6, 171–176. [Google Scholar] [CrossRef]
- Dobrunz, D.; Toma, A.C.; Tanner, P.; Pfohl, T.; Palivan, C.G. Polymer Nanoreactors with Dual Functionality: Simultaneous Detoxification of Peroxynitrite and Oxygen Transport. Langmuir 2012, 28, 15889–15899. [Google Scholar] [CrossRef] [PubMed]
- Thoma, J.; Belegrinou, S.; Rossbach, P.; Grzelakowski, M.; Kita-Tokarczyk, K.; Meier, W. Membrane protein distribution in composite polymer-lipid thin films. Chem. Commun. 2012, 48, 8811–8813. [Google Scholar] [CrossRef] [PubMed]
- Onaca, O.; Sarkar, P.; Roccatano, D.; Friedrich, T.; Hauer, B.; Grzelakowski, M.; Güven, A.; Fioroni, M.; Schwaneberg, U. Functionalized Nanocompartments (Synthosomes) with a Reduction-Triggered Release System. Angew. Chem. Int. Ed. 2008, 47, 7029–7031. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Chu, B.; Lee, H.; Montemagno, C. Protein-driven energy transduction across polymeric biomembranes. Nanotechnology 2004. [Google Scholar] [CrossRef]
- Ho, D.; Chu, B.; Lee, H.; Brooks, E.K.; Kuo, K.; Montemagno, C.D. Fabrication of biomolecule–copolymer hybrid nanovesicles as energy conversion systems. Nanotechnology 2005, 16, 3120–3132. [Google Scholar] [CrossRef]
- Xi, J.Z.; Ho, D.; Chu, B.; Montemagno, C.D. Lessons Learned from Engineering Biologically Active Hybrid Nano/Micro Devices. Adv. Funct. Mater. 2005, 15, 1233–1240. [Google Scholar] [CrossRef]
- Ho, D.; Chu, B.; Schmidt, J.J.; Brooks, E.K.; Montemagno, C.D. Hybrid Protein-Polymer Biomimetic Membranes. IEEE Trans. Nanotechnol. 2004, 3, 256–263. [Google Scholar] [CrossRef]
- Habel, J. Functional and Chemical Characterization of Vesicular Diblock Copolymer Bilayers with Aquaporin Included; Technical Report; Aquaporin A/S: Copenhagen, Denmark, 2011. [Google Scholar]
- Espina, M. Barrier Properties of Biomimetic Membranes. Master Thesis, Danish Technical University (DTU), Kgs. Lyngby, Denmark, 2012. [Google Scholar]
- Nallani, M.; Andreasson-Ochsner, M.; Tan, C.W.D.; Sinner, E.K.; Wisantoso, Y.; Geifman-Shochat, S.; Hunziker, W. Proteopolymersomes: In vitro production of a membrane protein in polymersome membranes. Biointerfaces 2011, 6, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Bomholt, J. Human Aquaporins—From in vivo detection to industrial scale production. Ph.D. Thesis, University Copenhagen, Copenhagen, Denmark, 2014. [Google Scholar]
- Zhang, X.; Fu, W.; Palivan, C.G.; Meier, W. Natural channel protein inserts and functions in a completely artificial, solid-supported bilayer membrane. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Dorn, J.; Belegrinou, S.; Kreiter, M.; Sinner, E.K.; Meier, W. Planar Block Copolymer Membranes by Vesicle Spreading. Macromol. Biosci. 2011, 11, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, K.; Discher, D.E.; Lal, J.; Janmey, P.; Goulian, M. Interactions of Membrane-Active Peptides with Thick, Neutral, Nonzwitterionic Bilayers. J. Phys. Chem. B 2005, 109, 14356–14364. [Google Scholar] [CrossRef] [PubMed]
- Toughrai, S. Functional Surfaces through Biomimetic block Copolymer Membranes. Ph.D. Thesis, Universität Basel, Basel, Switzerland, 2014. [Google Scholar]
- Amado, E.; Schöps, R.; Brandt, W.; Kressler, J. Spontaneous Formation of Giant Bioactive Protein-Block Copolymer Vesicles in Water. ACS Macro Lett. 2012, 1, 1016–1019. [Google Scholar] [CrossRef]
- Noor, M.; Dworeck, T.; Schenk, A.; Shinde, P.; Fioroni, M.; Schwaneberg, U. Polymersome surface decoration by an EGFP fusion protein employing Cecropin A as peptide “anchor”. J. Biotechnol. 2012, 157, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Fernandes, D.A.; O’Halloran, M.; Zheng, W.; Jiang, Y.; Ladizhansky, V.; Brown, L.S.; Liang, H. “Frozen” Block Copolymer Nanomembranes with Light-Driven Proton Pumping Performance. ACS Nano 2014, 8, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Hua, D.; Kuang, L.; Liang, H. Self-Directed Reconstitution of Proteorhodopsin with Amphiphilic Block Copolymers Induces the Formation of Hierarchically Ordered Proteopolymer Membrane Arrays. J. Am. Chem. Soc. 2011, 133, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Olson, T.L.; Lin, S.; Flores, M.; Jiang, Y.; Zheng, W.; Williams, J.C.; Allen, J.P.; Liang, H. Interface for Light-Driven Electron Transfer by Photosynthetic Complexes across Block Copolymer Membranes. J. Phys. Chem. Lett. 2014, 5, 787–791. [Google Scholar] [CrossRef]
- Ibata, K.; Takimoto, S.; Morisaku, T.; Miyawaki, A.; Yasui, M. Analysis of Aquaporin-Mediated Diffusional Water Permeability byCoherent Anti-Stokes Raman Scattering Microscopy. Biophys. J. 2011, 101, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Mamonov, A.B.; Coalson, R.D.; Zeidel, M.L.; Mathai, J.C. Water and Deuterium Oxide Permeability through Aquaporin 1: MD Predictions and Experimental Verification. J. Gen. Physiol. 2007, 130, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Itel, F.; Al-Samir, S.; Oberg, F.; Chami, M.; Kumar, M.; Supuran, C.T.; Deen, P.M.T.; Meier, W.; Hedfalk, K.; Gros, G.; Endeward, V. CO2 permeability of cell membranes is regulated by membrane cholesterol and protein gas channels. FASEB J. 2012, 26, 5182–5191. [Google Scholar] [CrossRef] [PubMed]
- GFP-Lifetime. Available online: http://www.iss.com/ (accessed on 7 July 2015).
- Choi, H.J.; Montemagno, C. Recent Progress in Advanced Nanobiological Materials for Energy and Environmental Applications. Materials 2013, 6, 5821–5856. [Google Scholar] [CrossRef]
- Pendergast, M.M.; Hoek, E.M.V. A review of water treatment membrane nanotechnologies. Energy Environ. Sci. 2011, 4, 1946–1971. [Google Scholar] [CrossRef]
- Aquaporin Home Page. Available online: www.aquaporin.dk (accessed on 6 July 2015).
- MEMBAQ Home Page. Available online: www.membaq.eu (accessed on 6 July 2015).
- Montemagno, C.; Schmidt, J.; Tozzi, S. Biomimetic Membranes. Patent WO 2004/011600, 5 February 2004. [Google Scholar]
- Hansen, J.S.; Perry, M.; Vogel, J.; Vissing, T.; Hansen, C.R.; Geschke, O.; Emnéus, J.; Hélix-Nielsen, C. Development of an automation technique for the establishment of functional lipid bilayer arrays. J. Micromech. Microeng. 2009, 19. [Google Scholar] [CrossRef]
- Hansen, J.S. Development of supported biomimetic membranes for insertion of aquaporin protein water channels for novel water filtration applications. Ph.D. Thesis, Danish Technical University, Kgs. Lyngby, Denmark, 2010. [Google Scholar]
- Rein, C.; Pszon-Bartosz, K.; Stibius, K.B.; Bjørnholm, T.; Hélix-Nielsen, C. Free-Standing Biomimetic Polymer Membrane Imaged with Atomic Force Microscopy. Langmuir 2011, 27, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, S.; Stibius, K.; Szewczykowski, P.; Perry, M.; Bohr, H.; Hélix-Nielsen, C. Hydrogels for in situ encapsulation of biomimetic membrane arrays. Polym. Adv. Technol. 2010, 23, 182–189. [Google Scholar] [CrossRef]
- Perry, M.; Hansenz, J.; Stibius, K. Surface Modifications of Support Partitions for Stabilizing Biomimetic Membrane Arrays. J. Membr. Sci. Technol. 2011. [Google Scholar] [CrossRef]
- Rein, C. Stabilization and characterization of 2D and 3D biomimetic membranes. Ph.D. thesis, University of Copenhagen, Copenhagen, Denmark, 2011. [Google Scholar]
- Aquaporin A/S. Biomimetic Membranes and Uses Thereof. Patent WO 2010/146365, 23 December 2010. [Google Scholar]
- Aquaporin A/S. Membrane for Filtering of Water. Patent WO 2006/122566, 23 November 2006. [Google Scholar]
- Aquaporin A/S. Biomimetic Water Membrane Comprising Aquaporins Used in the Production of Salinity Power. Patent WO 2007/033675, 29 March 2007. [Google Scholar]
- Aquaporin A/S. Scaffold for Composite Biomimetic Membrane. Patent WO 2009/074155, 18 June 2009. [Google Scholar]
- Aquaporin A/S. Assays Relating to Biomimetic Membranes and Their Uses. Patent WO 2010/146366, 23 December 2010. [Google Scholar]
- Montemagno, C.; Aquaz; Danfoss. Nanofabricated Membrane Using Polymerized Proteoliposomes. Patent WO 2010/091078, 12 August 2010. [Google Scholar]
- Wang, H.; Chung, T.S.; Tong, Y.W.; Meier, W.; Chen, Z.; Hong, M.; Jeyaseelan, K.; Armugam, A. Preparation and characterization of pore-suspending biomimetic membranes embedded with Aquaporin Z on carboxylated polyethylene glycol polymer cushion. Soft Matter 2011, 7, 7274. [Google Scholar] [CrossRef]
- Sun, G.; Zhou, H.; Li, Y.; Jeyaseelan, K.; Armugam, A.; Chung, T.S. A novel method of AquaporinZ incorporation via binary-lipid Langmuir monolayers. Colloids Surf. B: Biointerfaces 2012, 89, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, Y.; Grinberg, S.; Linder, C.; Heldman, E.; Gilron, J.; Freger, V. Fusion of Bolaamphiphile Micelles: A Method to Prepare Stable Supported Biomimetic Membranes. Langmuir 2013, 29, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, R.; Wicaksana, F.; Zhao, Y.; Tang, C.Y.; Torres, J.; Fane, A.G. Fusion behaviour of aquaporin Z incorporated proteoliposomes investigated by quartz crystal microbalance with dissipation (QCM-D). Colloids Surf. B: Biointerfaces 2013, 111, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, Y.; Berman, A. Supported lipid bilayer membranes for water purification by reverse osmosis. Langmuir 2010, 26, 7388–7395. [Google Scholar] [CrossRef] [PubMed]
- Freger, V.; Kaufman, Y. Biomimetic Membranes, Their Production and Uses Thereof in Water Purification. Patent US 2011/0084026, 14 April 2011. [Google Scholar]
- Li, X.; Wang, R.; Tang, C.Y.; Vararattanavech, A.; Zhao, Y.; Torres, J.; Fane, A.G. Preparation of supported lipid membranes for aquaporin Z incorporation. Colloids Surf. B: Biointerfaces 2012, 94, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Aquaporin AS.; NTU. Aquaporin Based Thin Film Composite Membranes. Patent WO 2013/043118, 28 March 2013. [Google Scholar]
- Aquaporin AS. Systems for Water Extraction. Patent WO 2014/128293, 28 August 2014. [Google Scholar]
- Zhao, Y.; Vararattanavech, A.; Li, X.; Hélix-Nielsen, C.; Vissing, T.; Torres, J.; Wang, R.; Fane, A.G.; Tang, C.Y. Effects of Proteoliposome Composition and Draw Solution Types on Separation Performance of Aquaporin-Based Proteoliposomes: Implications for Seawater Desalination Using Aquaporin-Based Biomimetic Membranes. Environ. Sci. Technol. 2013, 5, 1496–1503. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, R.; Wicaksana, F.; Tang, C.Y.; Torres, J.; Fane, A.G. Preparation of high performance nanofiltration (NF) membranes incorporated with aquaporin Z. J. Membr. Sci. 2014, 450, 181–188. [Google Scholar] [CrossRef]
- Tanner, P. Design and Development of Protein-Polymer Assemblies to Engineer Artificial organelles. Ph.D. Thesis, Universität Basel, Basel, Switzerland, 2013. [Google Scholar]
- Pawar, P.V.; Gohil, S.V.; Jain, J.P.; Kumar, N. Functionalized polymersomes for biomedical applications. Polym. Chem. 2013, 4, 3160–3176. [Google Scholar] [CrossRef]
- Jesorka, A.; Orwar, O. Liposomes: Technologies and Analytical Applications. Annu. Rev. Anal. Chem. 2008, 1, 801–832. [Google Scholar] [CrossRef] [PubMed]
- Montemagno, C. Biomimetic membrane formed from a vesicle-thread conjugate. Patent WO 2010/040353, 15 April 2010. [Google Scholar]
- Sun, G.; Chung, T.S.; Jeyaseelan, K.; Armugam, A. Stabilization and immobilization of aquaporin reconstituted lipid vesicles for water purification. Colloids Surf. B: Biointerfaces 2012, 102, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, Y.; Grinberg, S.; Linder, C.; Heldman, E.; Gilron, J.; Shen, Y.X.; Kumar, M.; Lammertink, R.G.H.; Freger, V. Towards supported bolaamphiphile membranes for water filtration: Roles of lipid and substrate. J. Membr. Sci. 2014, 457, 50–61. [Google Scholar] [CrossRef]
- Sun, G.; Chung, T.S.; Chen, N.; Lu, X.; Zhao, Q. Highly permeable aquaporin-embedded biomimetic membranes featuring a magnetic-aided approach. RSC Adv. 2013, 3, 9178–9184. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Wang, X.; Wang, S.; Ding, W.; Gao, C. Layer-by-Layer Assembly of Aquaporin Z-Incorporated Biomimetic Membranes for Water Purification. Environ. Sci. Technol. 2015, 49, 3761–3768. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.x.; Saboe, P.O.; Sines, I.T.; Erbakan, M.; Kumar, M. Biomimetic membranes: A review. J. Membr. Sci. 2014, 454, 359–381. [Google Scholar] [CrossRef]
- Thomas, J.A.; McGaughey, A.J.H. Reassessing Fast Water Transport through Carbon Nanotubes. Nano Lett. 2008, 8, 2788–2793. [Google Scholar] [CrossRef] [PubMed]
- Holt, J.; Park, H.; Wang, Y.; Stadermann, M.; Artyukhin, A.; Grigoropoulos, C.; Noy, A.; Bakajin, O. Fast Mass Transport Through Sub-2-Nanometer Carbon Nanotubes. Science 2006, 312, 1034–1037. [Google Scholar] [CrossRef] [PubMed]
- Fei, Z.; Zhao, D.; Geldbach, T.J.; Scopelliti, R.; Dyson, P.J.; Antonijevic, S.; Bodenhausen, G. A Synthetic Zwitterionic Water Channel: Characterization in the Solid State by X-ray Crystallography and NMR Spectroscopy. Angew. Chem. Int. Ed. 2005, 44, 5720–5725. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Dulcey, A.E.; Balagurusamy, V.S.K.; Miura, Y.; Smidrkal, J.; Peterca, M.; Nummelin, S.; Edlund, U.; Hudson, S.D.; Heiney, P.A.; Duan, H.; Magonov, S.N.; Vinogradov, S.A. Self-assembly of amphiphilic dendritic dipeptides into helical pores. Nature 2004, 430, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Barboiu, M. Artificial Water Channels. Angew. Chem. Int. Ed. 2012, 51, 11674–11676. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, G.; Yamato, K.; Shen, Y.; Cheng, R.; Wei, X.; Bai, W.; Gao, Y.; Li, H.; Liu, Y.; et al. Self-assembling subnanometer pores with unusual mass-transport properties. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Alsvik, I.; Hägg, M.B. Pressure Retarded Osmosis and Forward Osmosis Membranes: Materials and Methods. Polymers 2013, 5, 303–327. [Google Scholar] [CrossRef]
- Wittbecker, E.L.; Morgan, P.W. Interfacial polycondensation. I. J. Polym. Sci. 1959, 40, 289–297. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Jeong, B.H.; Huang, X.; Hoek, E.M.V. Impacts of reaction and curing conditions on polyamide composite reverse osmosis membrane properties. J. Membr. Sci. 2008, 311, 34–45. [Google Scholar] [CrossRef]
- Dalwani, M.; Zheng, J.; Hempenius, M.; Raaijmakers, M.J.T.; Doherty, C.M.; Hill, A.J.; Wessling, M.; Benes, N.E. Ultra-thin hybrid polyhedral silsesquioxane-polyamide films with potentially unlimited 2D dimensions. J. Mater. Chem. 2012, 22, 14835–14838. [Google Scholar] [CrossRef]
- Habel, J.; Ogbonna, A.; Skaarup, N.; Schulte, L.; Almdal, K.; Hélix-Nielsen, C. How molecular internal-geometric parameters affect PB-PEO polymersome size in aqueous solution. Phys. Chem. Chem. Phys. 2015. To be submitted. [Google Scholar]
- PB-FTIR-Spectrum. Available online: http://webbook.nist.gov/cgi/cbook.cgi?ID=B6002924Mask=80 (accessed on 6 July 2015).
- PEO-FTIR-Spectrum. Available online: http://webbook.nist.gov/cgi/cbook.cgi?ID=B6002924Mask=80 (accessed on 6 July 2015).
- Germic, L.; Ebert, K.; Bouma, R.H.B.; Borneman, Z.; Mulder, M.H.V.; Strathmann, H. Characterization of polyacrylonitrile ultrafiltration membranes. J. Membr. Sci. 1997, 132, 131–145. [Google Scholar] [CrossRef]
- Lee, K.P.; Zheng, J.; Bargeman, G.; Kemperman, A.J.B.; Benes, N.E. pH stable thin film composite polyamine nanofiltration membranes by interfacial polymerisation. J. Membr. Sci. 2015, 478, 75–84. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).