Improved Antimicrobial Properties of White Wastewater Protein Hydrolysate Through Electrodialysis with an Ultrafiltration Membrane (EDUF)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Chemicals
2.1.2. White Wastewater Hydrolysate
2.1.3. Membrane Materials
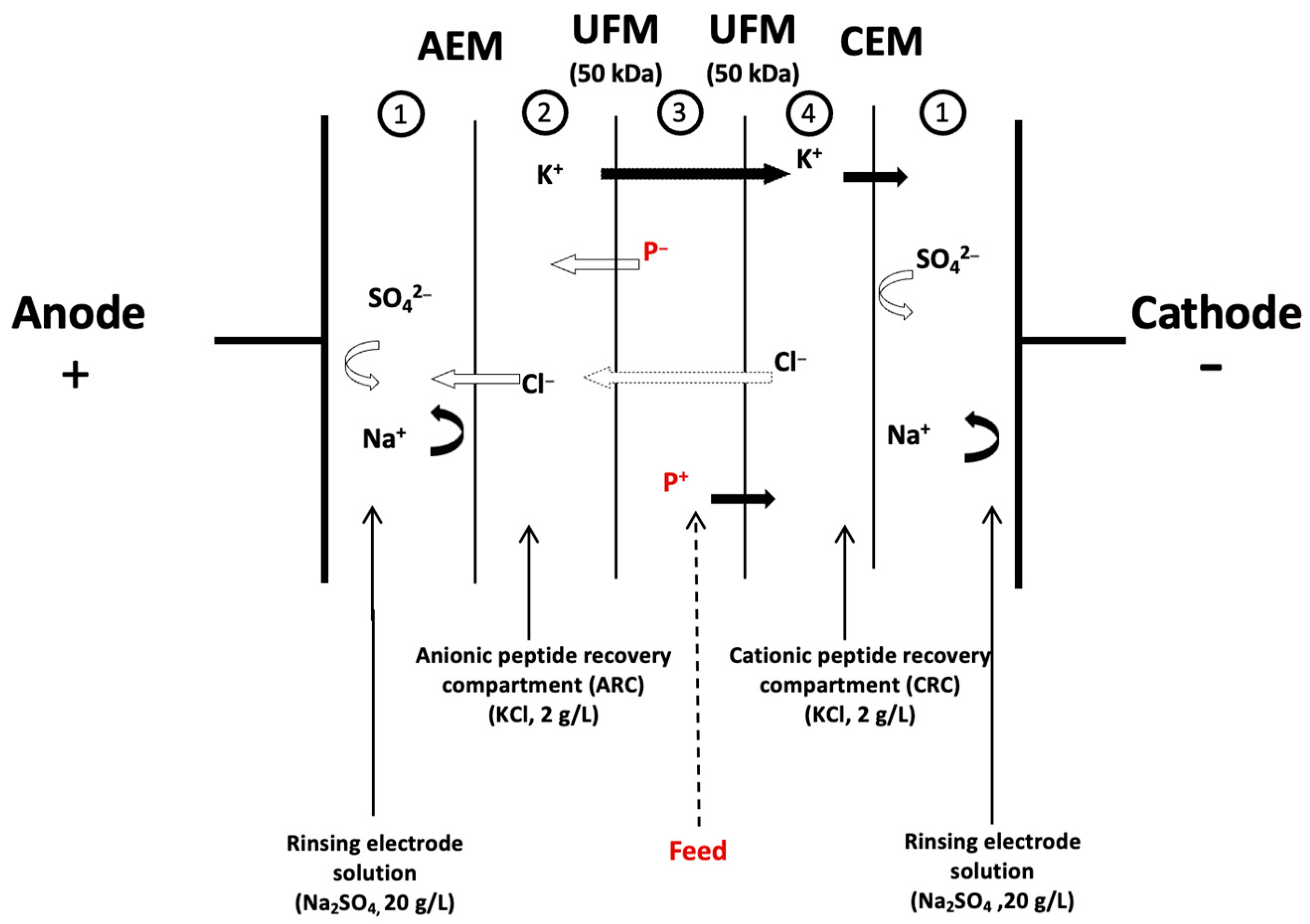
2.1.4. Electrodialysis Configuration
2.2. Protocol
2.3. Analyses
2.3.1. Conductivity and pH Measurements
2.3.2. Protein/Peptide Concentrations
2.3.3. Peptide Recovery Yield
2.3.4. Migration Rate
2.3.5. Relative Energy Consumption and Global Resistance
2.3.6. Thickness and Electrical Conductivity of Membranes
2.3.7. Peptides Characterization by UPLC-MS/MS Analysis
2.3.8. In Silico Bioinformatic Analyses
2.3.9. Antimicrobial Assays
2.3.10. Statistical Analyses
UPLC-MS/MS Data Treatment
Other Statistical Analyses
3. Results and Discussion
3.1. Electrodialysis: Process Performance Evaluation
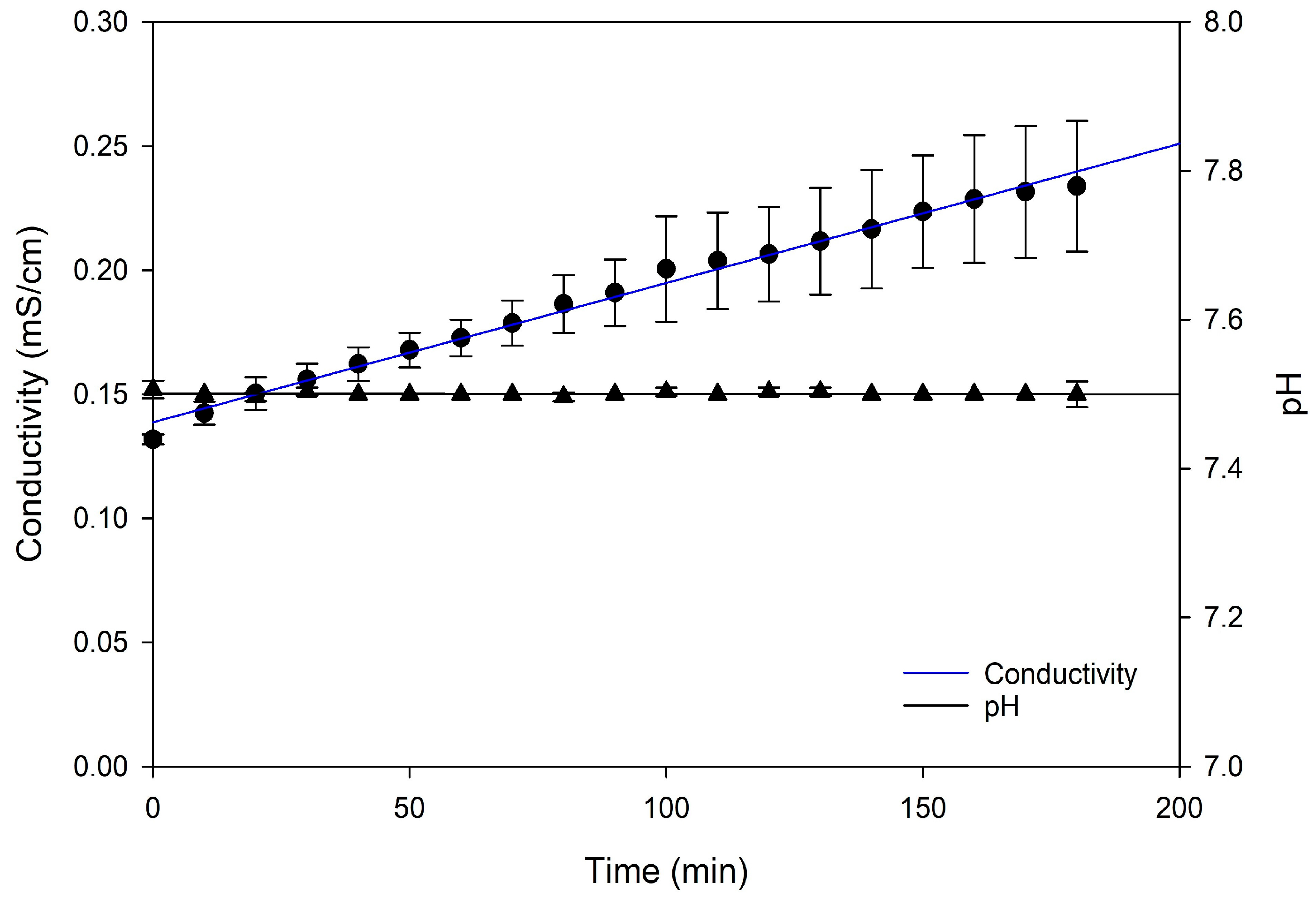
3.1.1. Progression of Conductivity and pH in ARC and CRC
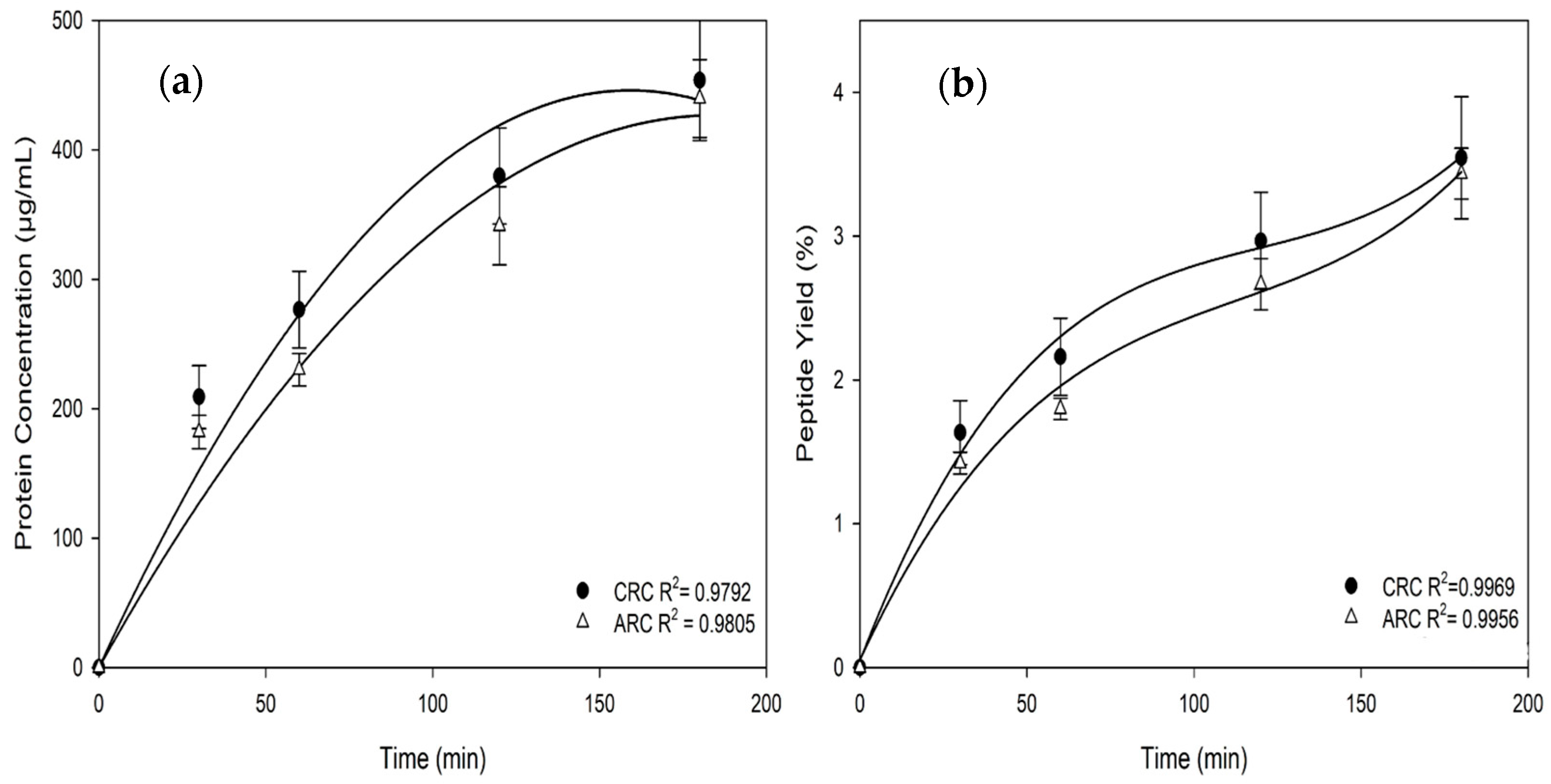
3.1.2. Protein/Peptide Concentrations, Yields, and Migration Rates in the Recovery Compartments
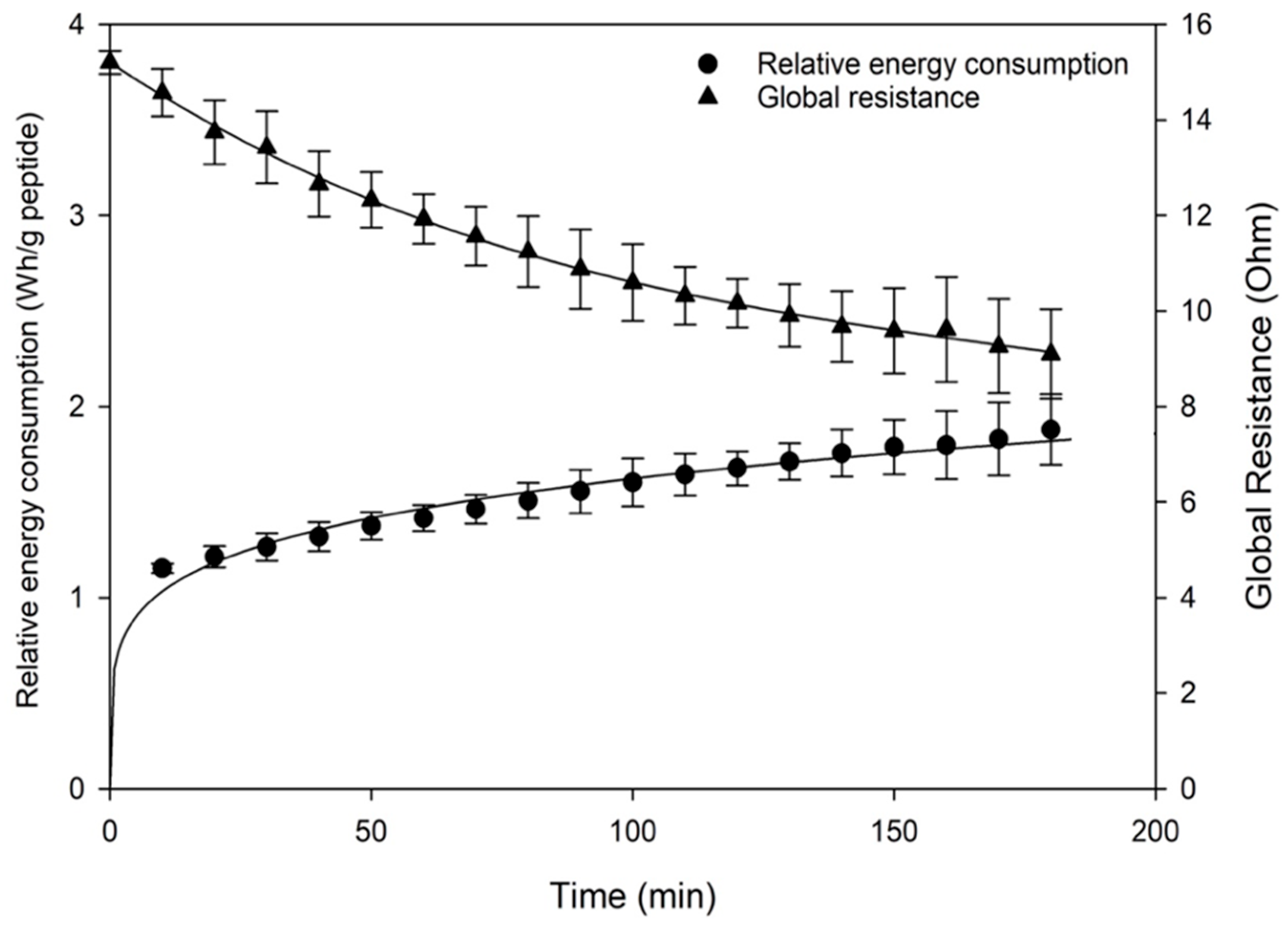
3.1.3. Global Resistance and REC
3.1.4. Membrane Characterization
3.2. Peptide Population Characterization
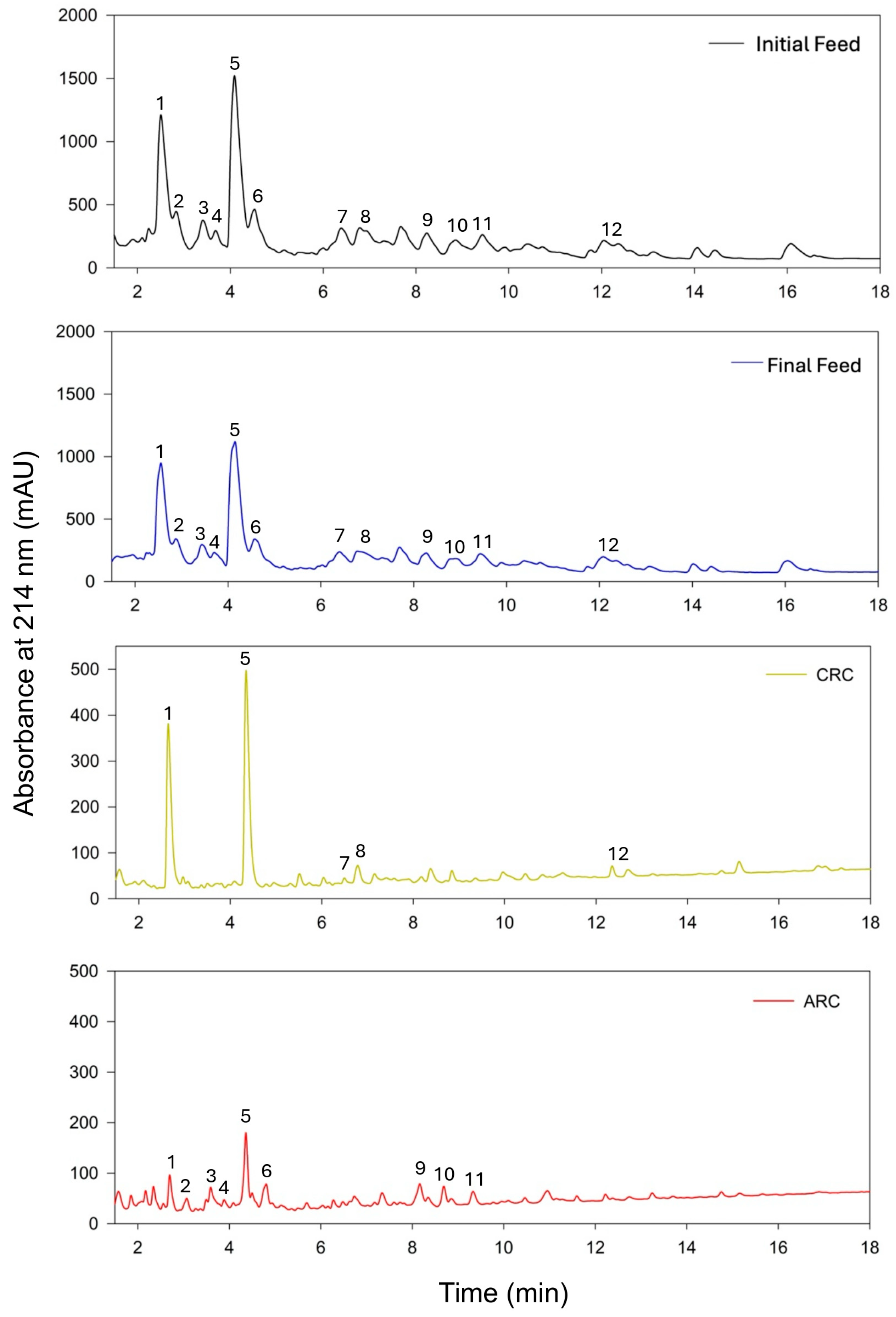
3.2.1. Analyses of UV Chromatograms
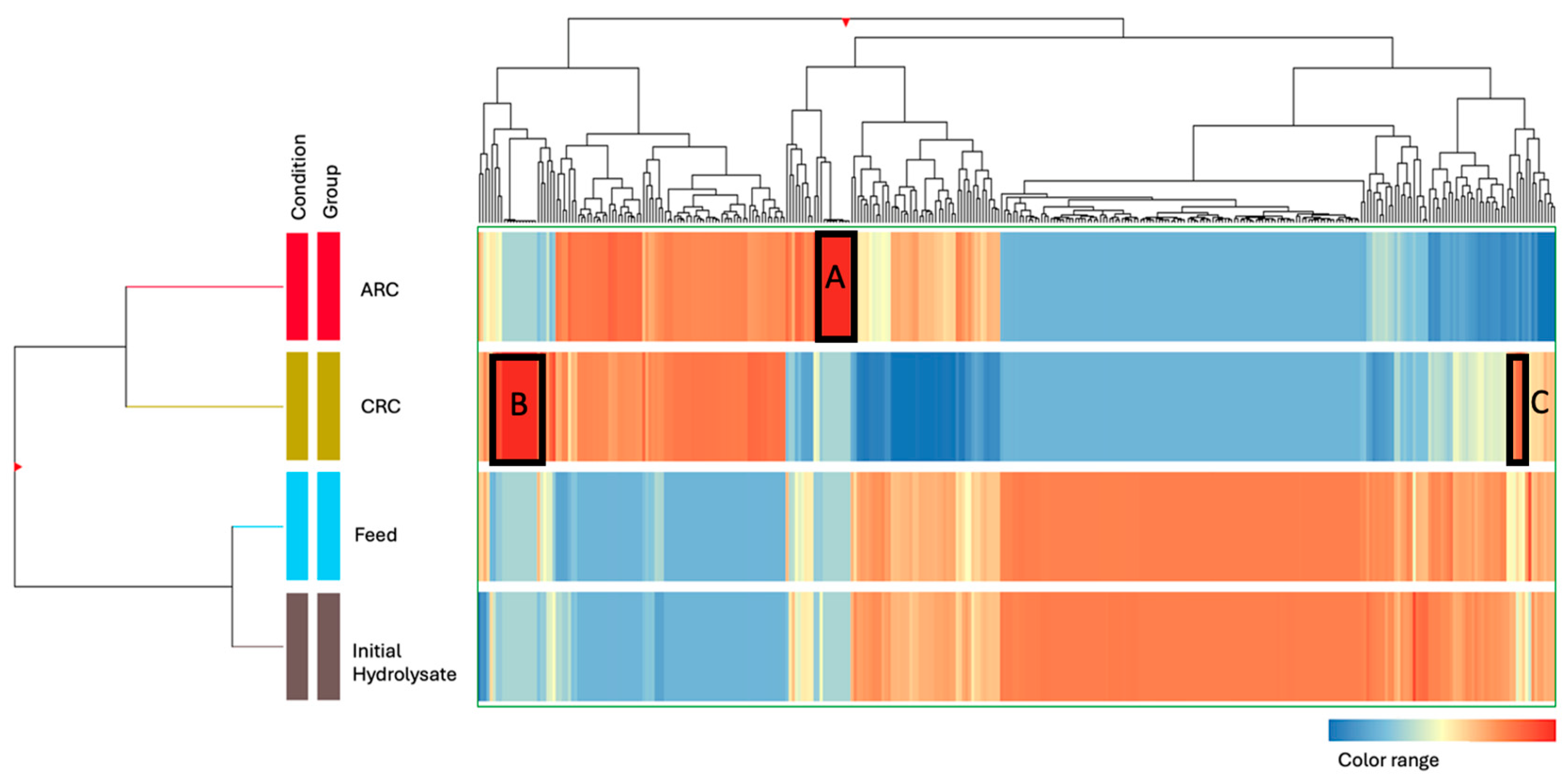
3.2.2. Hierarchical Clustering and Heatmap Visualization
3.2.3. Peptide Identification and Characterization
3.3. Antimicrobial Activities
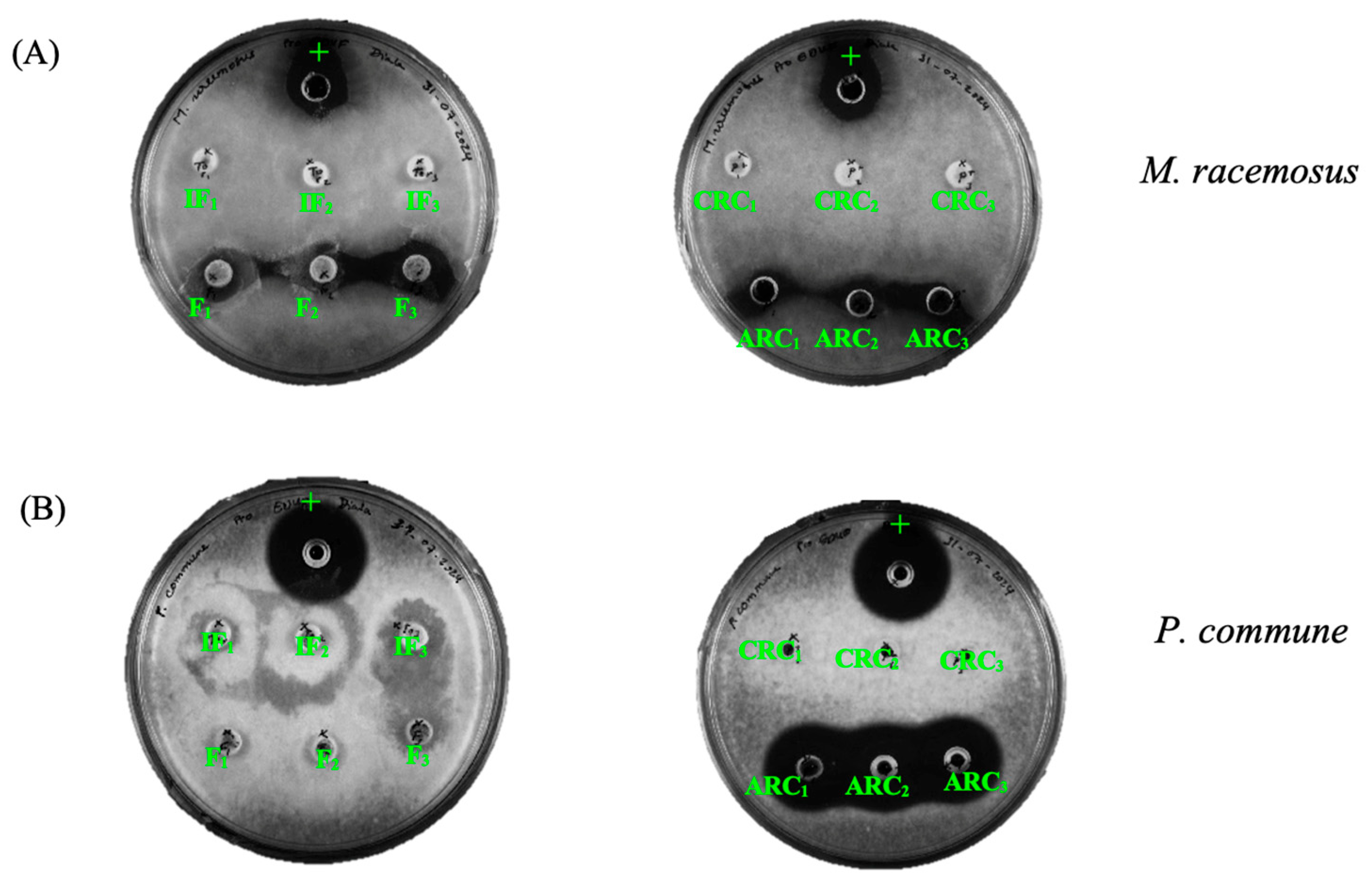
3.3.1. Antibacterial Activity of the Recovered Solutions
3.3.2. Antifungal Activity
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A.; Duche, R.T.; Wandhare, A.G.; Sian, J.K.; Singh, B.P.; Sihag, M.K.; Singh, K.S.; Sangwan, V.; Talan, S.; Panwar, H. Milk-Derived Antimicrobial Peptides: Overview, Applications, and Future Perspectives. Probiotics Antimicrob. Proteins 2023, 15, 44–62. [Google Scholar] [CrossRef]
- Cournoyer, A.; Bazinet, L. Electrodialysis Processes an Answer to Industrial Sustainability: Toward the Concept of Eco-Circular Economy?—A Review. Membranes 2023, 13, 205. [Google Scholar] [CrossRef]
- Sanchez-Reinoso, Z.; Todeschini, S.; Thibodeau, J.; Ben Said, L.; Fliss, I.; Bazinet, L.; Mikhaylin, S. Impact of Pulsed Electric Fields and pH on Enzyme Inactivation and Bioactivities of Peptic Hydrolysates Produced from Bovine and Porcine Hemoglobin. Foods 2022, 11, 3313. [Google Scholar] [CrossRef]
- Alalam, S.; Ben-Souilah, F.; Lessard, M.-H.; Chamberland, J.; Perreault, V.; Pouliot, Y.; Labrie, S.; Doyen, A. Characterization of Chemical and Bacterial Compositions of Dairy Wastewaters. Dairy 2021, 2, 179–190. [Google Scholar] [CrossRef]
- Damen, D.; Thibodeau, J.; Gaaloul, S.; Fliss, I.; Labrie, S.; Hamoudi, S.; Bazinet, L. Influence of Enzymatic Hydrolysis Conditions on Antimicrobial Activities and Peptide Profiles of Milk Protein-Derived Hydrolysates from White Wastewater. Clean. Waste Syst. 2024, 9, 100172. [Google Scholar] [CrossRef]
- Cournoyer, A.; Daigle, G.; Thibodeau, J.; Perreault, V.; Bazinet, L. Effects of applied current modes on the migration selectivity of peptides from a porcine cruor hydrolysate during electrodialysis with ultrafiltration membrane. Sep. Purif. Technol. 2024, 338, 126280. [Google Scholar] [CrossRef]
- Brião, V.B.; Mossmann, J.; Seguenka, B.; Graciola, S.; Piccin, J.S. Integrating Whey Processing: Ultrafiltration, Nanofiltration, and Water Reuse from Diafiltration. Membranes 2024, 14, 191. [Google Scholar] [CrossRef] [PubMed]
- Bargeman, G.; Houwing, J.; Recio, I.; Koops, G.; Van Der Horst, C. Electro-membrane filtration for the selective isolation of bioactive peptides from an α s2 -casein hydrolysate. Biotech Bioeng. 2002, 80, 599–609. [Google Scholar] [CrossRef]
- Sun, L.; Chen, Q.; Lu, H.; Wang, J.; Zhao, J.; Li, P. Electrodialysis with porous membrane for bioproduct separation: Technology, features, and progress. Food Res. Int. 2020, 137, 109343. [Google Scholar] [CrossRef]
- Kadel, S.; Pellerin, G.; Thibodeau, J.; Perreault, V.; Lainé, C.; Bazinet, L. How Molecular Weight Cut-Offs and Physicochemical Properties of Polyether Sulfone Membranes Affect Peptide Migration and Selectivity during Electrodialysis with Filtration Membranes. Membranes 2019, 9, 153. [Google Scholar] [CrossRef]
- Persico, M.; Dhulster, P.; Bazinet, L. Redundancy analysis for determination of the main physicochemical characteristics of filtration membranes explaining their fouling by peptides. J. Membr. Sci. 2018, 563, 708–717. [Google Scholar] [CrossRef]
- Cowan, D.A.; Brown, J.H. Effect of Turbulence on Limiting Current in Electrodialysis Cells. Ind. Eng. Chem. 1959, 51, 1445–1448. [Google Scholar] [CrossRef]
- Kadel, S.; Daigle, G.; Thibodeau, J.; Perreault, V.; Pellerin, G.; Lainé, C.; Bazinet, L. How physicochemical properties of filtration membranes impact peptide migration and selectivity during electrodialysis with filtration membranes: Development of predictive statistical models and understanding of mechanisms involved. J. Membr. Sci. 2021, 619, 118175. [Google Scholar] [CrossRef]
- Suwal, S.; Roblet, C.; Amiot, J.; Doyen, A.; Beaulieu, L.; Legault, J.; Bazinet, L. Recovery of valuable peptides from marine protein hydrolysate by electrodialysis with ultrafiltration membrane: Impact of ionic strength. Food Res. Int. 2014, 65, 407–415. [Google Scholar] [CrossRef]
- Pranata, J. Measurement of Casein Content in Milk and Milk-Based Products Using Kjeldahl, Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis, and Mid Infrared Spectroscopy. 2020. Available online: https://hdl.handle.net/1813/103493 (accessed on 9 September 2024).
- Lteif, R.; Dammak, L.; Larchet, C.; Auclair, B. Conductivitéélectrique membranaire: Étude de l’effet de la concentration, de la nature de l’électrolyte et de la structure membranaire. Eur. Polym. J. 1999, 35, 1187–1195. [Google Scholar] [CrossRef]
- Lebrun, L.; Da Silva, E.; Pourcelly, G.; Métayer, M. Elaboration and characterisation of ion-exchange films used in the fabrication of bipolar membranes. J. Membr. Sci. 2003, 227, 95–111. [Google Scholar] [CrossRef]
- Martin, N.; Torres-Frenzel, P.; Wiedmann, M. Invited review: Controlling dairy product spoilage to reduce food loss and waste. J. Dairy Sci. 2021, 104, 1251–1261. [Google Scholar] [CrossRef]
- CLSI M100-S27; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; CLSI 2016/2017.
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin. Microbiol. Infect. 2000, 6, 509–515. [CrossRef]
- Ostertagová, E.; Ostertag, O.; Kováč, J. Methodology and Application of the Kruskal-Wallis Test. Appl. Mech. Mater. 2014, 611, 115–120. [Google Scholar] [CrossRef]
- Shuken, S.R.; McNerney, M.W. Costs and Benefits of Popular P-Value Correction Methods in Three Models of Quantitative Omic Experiments. Anal. Chem. 2023, 95, 2732–2740. [Google Scholar] [CrossRef]
- Jaccard, J.; Becker, M.A.; Wood, G. Pairwise multiple comparison procedures: A review. Psychol. Bull. 1984, 96, 589–596. [Google Scholar] [CrossRef]
- Geoffroy, T.; Bernier, M.; Thibodeau, J.; Francezon, N.; Beaulieu, L.; Mikhaylin, S.; Langevin, M.; Lutin, F.; Bazinet, L. Semi-industrial scale-up of EDUF technology for the electroseparation of bioactive cationic peptides: Impact of process parameters and cell configurations on eco-efficiency. J. Membr. Sci. 2022, 641, 119856. [Google Scholar] [CrossRef]
- Sanchez-Reinoso, Z.; Bazinet, M.; Leblanc, B.; Clément, J.-P.; Germain, P.; Bazinet, L. Application of machine learning tools to study the synergistic impact of physicochemical properties of peptides and filtration membranes on peptide migration during electrodialysis with filtration membranes. Sep. Purif. Technol. 2025, 360, 130877. [Google Scholar] [CrossRef]
- an Kan, E.J.M.; Demel, R.A.; Breukink, E.; van der Bent, A.; de Kruijff, B. Clavanin Permeabilizes Target Mem-branes via Two Distinctly Different pH-Dependent Mechanisms. Biochemistry 2002, 41, 7529–7539. [Google Scholar] [CrossRef]
- Ndiaye, N.; Pouliot, Y.; Saucier, L.; Beaulieu, L.; Bazinet, L. Electroseparation of bovine lactoferrin from model and whey solutions. Sep. Purif. Technol. 2010, 74, 93–99. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, G.Q.; Kentish, S.E. Isolation of lactoferrin and immunoglobulins from dairy whey by an electrodialysis with filtration membrane process. Sep. Purif. Technol. 2020, 233, 115987. [Google Scholar] [CrossRef]
- Henaux, L.; Thibodeau, J.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. How Charge and Triple Size-Selective Membrane Separation of Peptides from Salmon Protein Hydrolysate Orientate their Biological Response on Glucose Uptake. Int. J. Mol. Sci. 2019, 20, 1939. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Sun, L.; Shou, Q.; Liang, X.; Liu, H. Electrodialysis Deacidification of Acid Hydrolysate in Hemicellulose Saccharification Process: Membrane Fouling Identification and Mechanisms. Membranes 2023, 13, 256. [Google Scholar] [CrossRef] [PubMed]
- Dufton, G.; Mikhaylin, S.; Gaaloul, S.; Bazinet, L. Positive Impact of Pulsed Electric Field on Lactic Acid Removal, Demineralization and Membrane Scaling during Acid Whey Electrodialysis. Int. J. Mol. Sci. 2019, 20, 797. [Google Scholar] [CrossRef]
- Saini, S.; Kumar, R.; Chawla, J.; Kaur, I. Adsorption of bivalent lead ions from an aqueous phase system: Equilibrium, thermodynamic, kinetics, and optimization studies. Water Environ. Res. 2019, 91, 1692–1704. [Google Scholar] [CrossRef]
- Firdaous, L.; Malériat, J.; Schlumpf, J.; Quéméneur, F. Transfer of Monovalent and Divalent Cations in Salt Solutions by Electrodialysis. Sep. Sci. Technol. 2007, 42, 931–948. [Google Scholar] [CrossRef]
- Galizia, M.; Benedetti, F.M.; Paul, D.R.; Freeman, B.D. Monovalent and divalent ion sorption in a cation exchange membrane based on cross-linked poly (p-styrene sulfonate-co-divinylbenzene). J. Membr. Sci. 2017, 535, 132–142. [Google Scholar] [CrossRef]
- Kim, Y.; Walker, W.S.; Lawler, D.F. Competitive separation of di- vs. mono-valent cations in electrodialysis: Effects of the boundary layer properties. Water Res. 2012, 46, 2042–2056. [Google Scholar] [CrossRef]
- Xu, X.; He, Q.; Ma, G.; Wang, H.; Nirmalakhandan, N.; Xu, P. Selective separation of mono- and di-valent cations in electrodialysis during brackish water desalination: Bench and pilot-scale studies. Desalination 2018, 428, 146–160. [Google Scholar] [CrossRef]
- Persico, M.; Mikhaylin, S.; Doyen, A.; Firdaous, L.; Hammami, R.; Chevalier, M.; Flahaut, C.; Dhulster, P.; Bazinet, L. Formation of peptide layers and adsorption mechanisms on a negatively charged cation-exchange mem-brane. J. Colloid Interface Sci. 2017, 508, 488–499. [Google Scholar] [CrossRef]
- Huang, T.; Qian, Y.; Fu, X.; Huang, S.; Li, Y.; Zhou, C. De Novo Design of Triblock Amphiphilic Short Antimicrobial Peptides. ACS Appl. Polym. Mater. 2020, 2, 3988–3992. [Google Scholar] [CrossRef]
- Chen, X.; Fan, R.; Wang, Y.; Munir, M.; Li, C.; Wang, C.; Hou, Z.; Zhang, G.; Liu, L.; He, J. Bovine milk β-casein: Structure, properties, isolation, and targeted application of isolated products. Com-Prehensive Rev. Food Sci. Food Saf. 2024, 23, e13311. [Google Scholar] [CrossRef] [PubMed]
- Sah, B.N.P.; Vasiljevic, T.; McKechnie, S.; Donkor, O.N. Antioxidative and antibacterial peptides derived from bovine milk proteins. Crit. Rev. Food Sci. Nutr. 2018, 58, 726–740. [Google Scholar] [CrossRef]
- Koprivnjak, T.; Peschel, A. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci. 2011, 68, 2243–2254. [Google Scholar] [CrossRef]
- Nigenda, E.S.; Postma, T.M.; Hezwani, M.; Pirvan, A.; Gannon, S.; Smith, C.-A.; Riehle, M.; Liskamp, R.M.J. Synthesis and cellular penetration properties of new phosphonium based cationic amphiphilic peptides. MedChemComm 2018, 9, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Berger, I.; Loewy, Z.G. Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics. Bacteria 2024, 3, 171–182. [Google Scholar] [CrossRef]
- Khan, F. Multifaceted strategies for alleviating Pseudomonas aeruginosa infection by targeting protease activity: Natural and synthetic molecules. Int. J. Biol. Macromol. 2024, 278, 134533. [Google Scholar] [CrossRef] [PubMed]
- Sowa-Jasiłek, A.; Zdybicka-Barabas, A.; Stączek, S.; Pawlikowska-Pawlęga, B.; Grygorczuk-Płaneta, K.; Skrzypiec, K.; Gruszecki, W.I.; Mak, P.; Cytryńska, M. Antifungal Activity of Anionic Defense Peptides: Insight into the Action of Galleria mellonella Anionic Peptide 2. Int. J. Mol. Sci. 2020, 21, 1912. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef]
- Albadr, A.A.; Coulter, S.M.; Porter, S.L.; Thakur, R.R.S.; Laverty, G. Ultrashort Self-Assembling Peptide Hydrogel for the Treatment of Fungal Infections. Gels 2018, 4, 48. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Statistical Test | F or t | df | p-Value |
|---|---|---|---|---|
| Peptide concentration (CRC) | Linear regression | F = 95.62 | 1, 13 | <0.0001 |
| Peptide concentration (ARC) | Linear regression | F = 94.01 | 1, 13 | <0.0001 |
| Peptide yield (CRC) | Cubic regression | F = 52.69 | 3, 1 | 0.1009 |
| Peptide yield (ARC) | Cubic regression | F = 37.22 | 3, 1 | 0.1198 |
| Global resistance over time | Quadratic regression | F = 2236.69 | 2, 16 | <0.0001 |
| Peptide migration rate | Quadratic regression | F = 54.45 | 2, 2 | 0.0180 |
| Protein concentration (CRC) | Quadratic regression | F = 47.18 | 2, 2 | 0.0208 |
| Protein concentration (ARC) | Quadratic regression | F = 50.39 | 2, 2 | 0.0195 |
| Membrane conductivity (AEM) | Paired t-test | t = 5.23 | 2 | <0.05 |
| Membrane conductivity (CEM) | Paired t-test | t = 4.87 | 2 | <0.05 |
| UFM thickness (UFM1 vs. UFM2) | One-way ANOVA | F = 1.22 | 2, 6 | >0.05 |
| Thickness (mm) | Conductivity (mS/cm) | |||
|---|---|---|---|---|
| Membrane | Before | After | Before | After |
| AEM | 0.138 ± 0.001 a* | 0.140 ± 0.002 a | 5.91 ± 0.12 a | 4.83 ± 0.12 b |
| CEM | 0.143 ± 0.002 a | 0.136 ± 0.004 a | 8.65 ± 0.27 a | 7.09 ± 0.32 b |
| UFM 1 | 0.184 ± 0.001 aA | 0.184 ± 0.001 aA | 9.74 ± 0.31 aA | 9.49 ± 0.26 aA |
| UFM 2 | 0.191 ± 0.002 aA | 0.185 ± 0.005 aA | 10.01 ± 0.27 aA | 9.52 ± 0.22 aA |
| Peak # | Sequence Name | Molecular Mass (Da) | Source | Isoelectric Point | Gravy Score | Net Charge at pH 7 |
|---|---|---|---|---|---|---|
| CRC | ||||||
| 1 | SRYP | 521.26 | α Lactalbumin | 8.46 | 1.00 | 0.99 |
| 1 | EALG | 388.20 | α Lactalbumin | 4.05 | −1.10 | −1.01 |
| 1 | QKP | 371.22 | κ casein (45–47) | 8.75 | −3.00 | 0.99 |
| 1 | VYP | 377.19 | α Lactalbumin | 5.97 | −1.27 | 0.00 |
| 1 | KPAA | 385.23 | κ casein (63–66) | 8.75 | −0.48 | 0.99 |
| 5 | TPV | 319.16 | Bsa (472–474) | 5.18 | 0.63 | 0.00 |
| 5 | MKEGIHAQQ | 1040.51 | Casein α s1 (123–131) | 6.50 | −1.09 | 0.09 |
| 5 | QRF | 449.24 | α Lactalbumin | 11.06 | 0.23 | 1.00 |
| 5 | VDPVN | 542.34 | α Lactalbumin | 3.09 | 0.04 | −1.00 |
| 5 | AHK | 354.19 | α Lactalbumin | 10.13 | 0.67 | 1.09 |
| 5 | KPDP | 455.23 | BSA (122–125) | 5.84 | −2.65 | −0.01 |
| 7 | YPEL | 520.25 | Casein α s1 (146–149) | 4.05 | −0.65 | −1.01 |
| 7 | VPQK | 470.29 | Casein β (173–136) | 8.72 | −1.20 | 1.00 |
| 7 | YVE | 409.27 | α Lactalbumin | 3.27 | −0.27 | −1.00 |
| 8 | PQLE | 485.25 | Casein α (107–110) | 4.60 | −1.20 | −1.00 |
| 8 | KIPA | 427.28 | Β lactoglobulin (77–80) | 8.75 | 0.20 | 0.99 |
| 8 | QPEV | 471.23 | α Lactalbumin | 3.27 | 0.43 | −1.00 |
| 12 | GPVRGPFP | 825.45 | Casein β (199–206) | 9.75 | −0.39 | 1.00 |
| 12 | LPVP | 424.22 | Casein β (171–174) | 5.53 | 1.20 | 0.00 |
| 12 | LPLP | 434.21 | Casein β (135–138) | 5.53 | 1.10 | 0.00 |
| ARC | ||||||
| 1 | VPQ | 342.18 | BSA (420–422) | 5.49 | −0.30 | 0.00 |
| 2 | KVPQ | 476.21 | BSA (419–422) | 8.75 | −1.20 | 0.99 |
| 4 | DEL | 375.17 | Casein β (43–45) | 4.05 | −1.07 | −2.00 |
| 4 | DEALEK | 703.34 | Β lactoglobulin (130–135) | 4.14 | −1.47 | −2.00 |
| 5 | MKEGIHAQQ | 1040.51 | Casein α s1 (123–131) | 6.50 | −1.09 | 0.09 |
| 6 | PQYLKT | 748.36 | α Lactalbumin | 9.67 | −0.22 | 1.00 |
| 9 | GDLEI | 545.27 | Β lactoglobulin (52–56) | 4.05 | 0.18 | −2.00 |
| 10 | ELEEL | 631.31 | Casein β (2–6) | 4.05 | −0.58 | −3.00 |
| 11 | EMPF | 522.22 | α Lactalbumin | 4.05 | −0.10 | −1.00 |
| Strains | Strain No. | Initial Hydrolysate | Recovered Feed | Cationic Solution | Anionic Solution | |
|---|---|---|---|---|---|---|
| Bacteria | C. tyrobutyricum | ATCC25755 | − | − | − | − |
| P. aeruginosa | ATCC15442 | − | − | − | − | |
| Molds | M. racemosus | LMA722 | − | + | − | + |
| P. commune | LMA72 | − | − | − | + | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damen, D.; Thibodeau, J.; Gaaloul, S.; Labrie, S.; Hamoudi, S.; Bazinet, L. Improved Antimicrobial Properties of White Wastewater Protein Hydrolysate Through Electrodialysis with an Ultrafiltration Membrane (EDUF). Membranes 2025, 15, 238. https://doi.org/10.3390/membranes15080238
Damen D, Thibodeau J, Gaaloul S, Labrie S, Hamoudi S, Bazinet L. Improved Antimicrobial Properties of White Wastewater Protein Hydrolysate Through Electrodialysis with an Ultrafiltration Membrane (EDUF). Membranes. 2025; 15(8):238. https://doi.org/10.3390/membranes15080238
Chicago/Turabian StyleDamen, Diala, Jacinthe Thibodeau, Sami Gaaloul, Steve Labrie, Safia Hamoudi, and Laurent Bazinet. 2025. "Improved Antimicrobial Properties of White Wastewater Protein Hydrolysate Through Electrodialysis with an Ultrafiltration Membrane (EDUF)" Membranes 15, no. 8: 238. https://doi.org/10.3390/membranes15080238
APA StyleDamen, D., Thibodeau, J., Gaaloul, S., Labrie, S., Hamoudi, S., & Bazinet, L. (2025). Improved Antimicrobial Properties of White Wastewater Protein Hydrolysate Through Electrodialysis with an Ultrafiltration Membrane (EDUF). Membranes, 15(8), 238. https://doi.org/10.3390/membranes15080238









