Evolution of the Concepts of Architecture and Supramolecular Dynamics of the Plasma Membrane
Abstract
1. Introduction
2. Evolution of the Concept of Plasma Membrane or Biomembrane
3. Concept of the Plasmatic Membrane and Transport
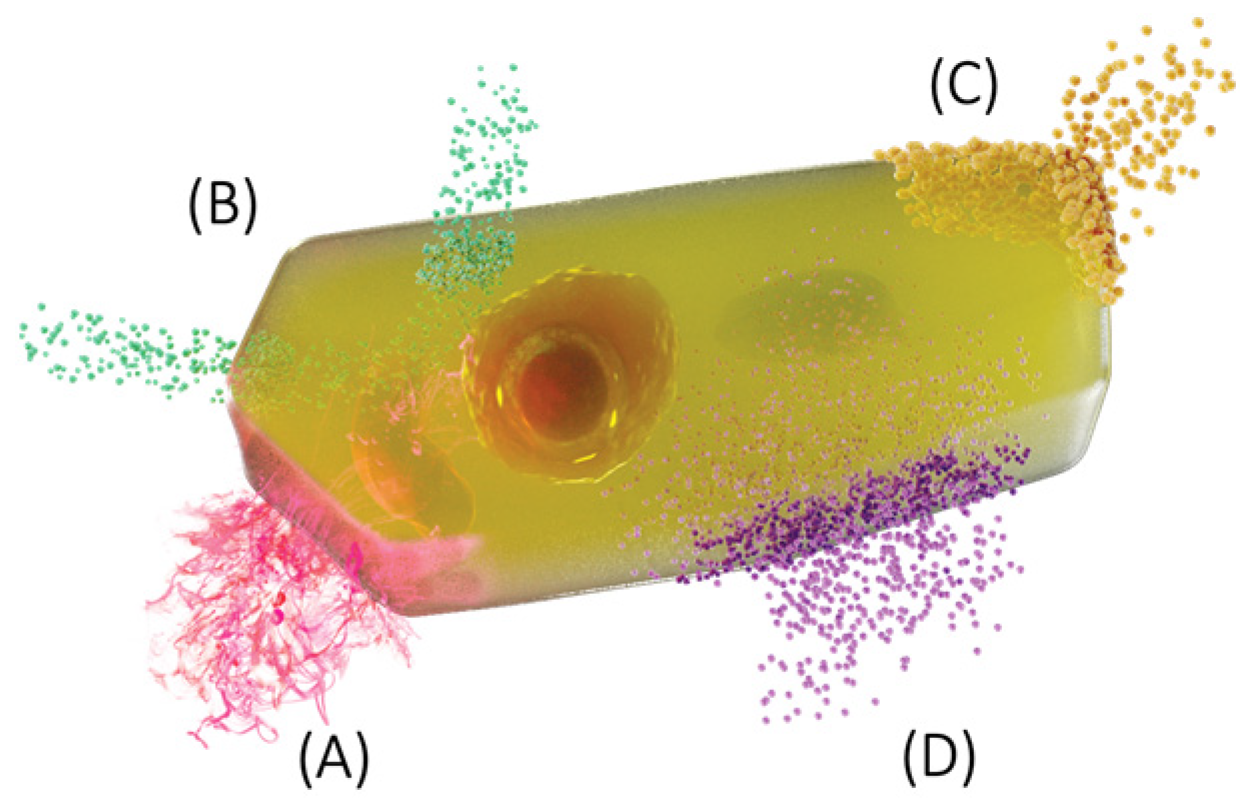
4. Membrane Models and Contributions to the Concept of the Membrane
4.1. Lipid Structure
4.1.1. Lipid Monolayer
4.1.2. Lipid Bilayer
4.1.3. Cell Bilayer Thickness
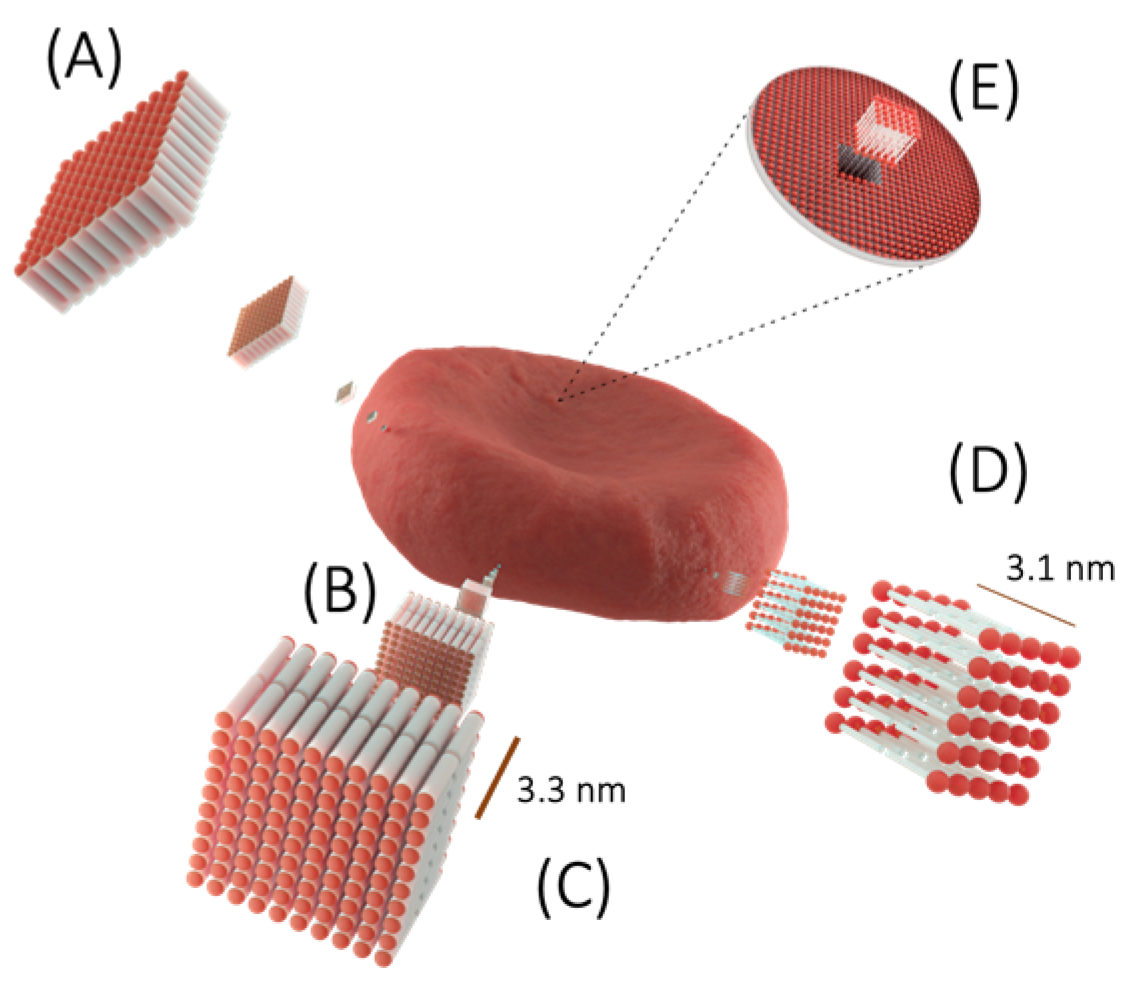
4.2. Lipid Bilayer and Membrane Proteins
4.2.1. The Lipid Bilayer Contains Proteins
4.2.2. Composition and Types of Lipids in the Plasmatic Membrane
4.2.3. Membrane Unit or Unitary Model
4.2.4. Peripheral Proteins
4.2.5. Cell Surface
4.2.6. Models of the 1960s: Membrane Proteins
Protein Subunit Model
The Lipoprotein-Repeating Unit’s Model and the Protein-Crystal Model
Fracture of Frozen Membranes
Concept of Fluidity
Mosaic Model
Micellar Model
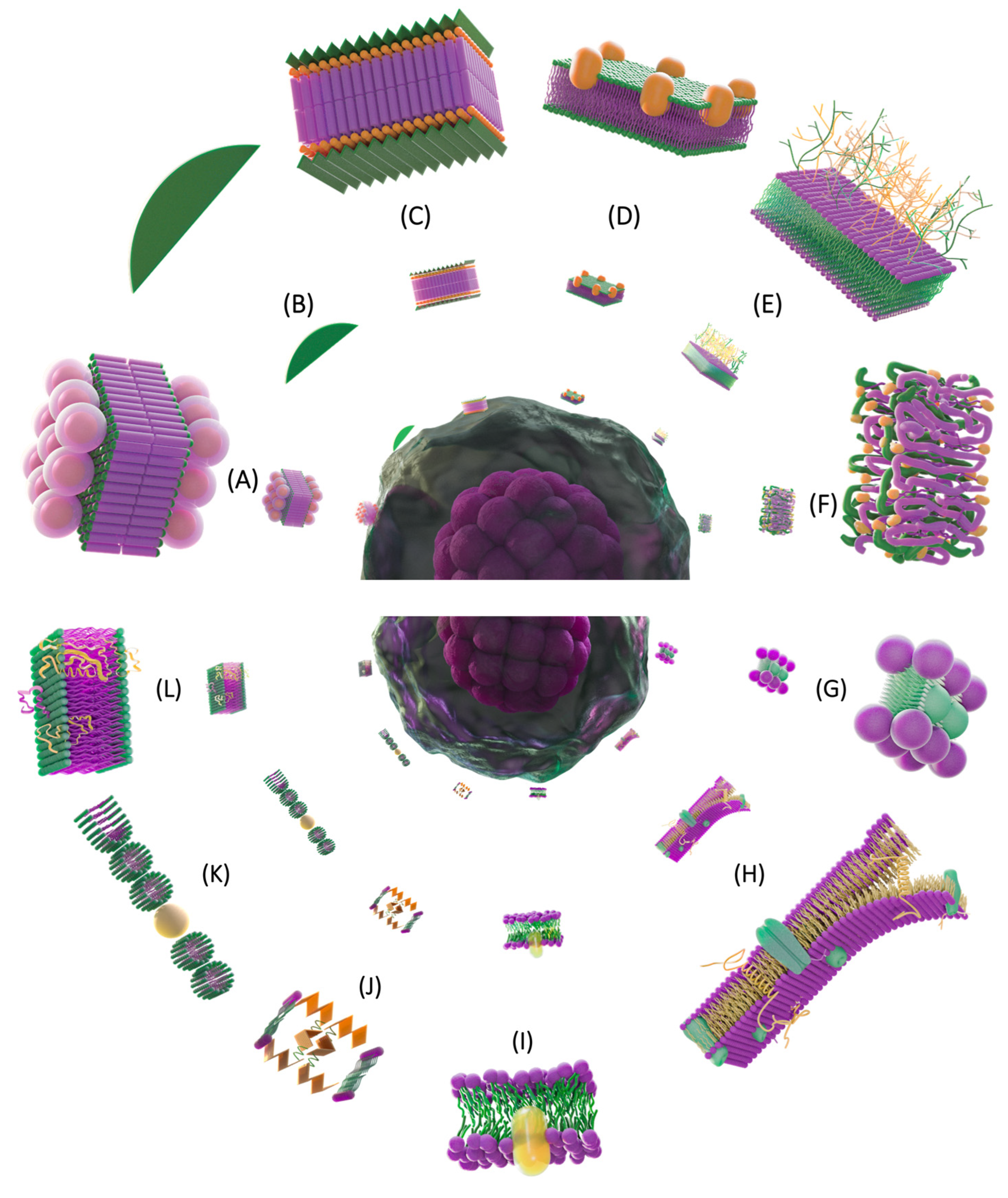
4.3. Dynamics of the Plasma Membrane
4.3.1. Lateral Diffusion of Proteins
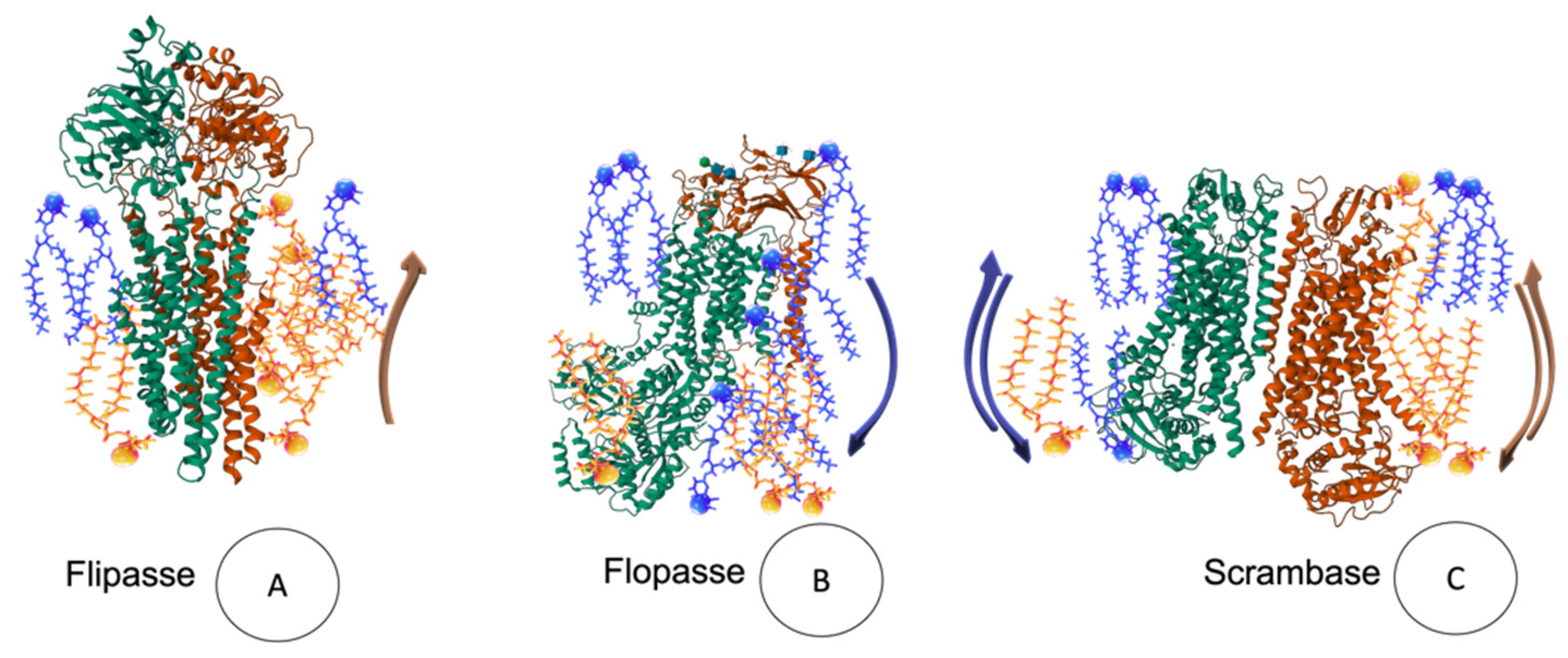
4.3.2. Flip-Flop
4.3.3. Fluid Mosaic Model
4.3.4. Membrane Plasticity
4.3.5. Asymmetric Lipid Bilayer
4.3.6. Hydrodynamic Model of Membrane Flow
4.3.7. Secondary Alpha Helix Proteins
4.3.8. Plate Model
4.3.9. Geometry of Molecules
4.3.10. Mattress Model
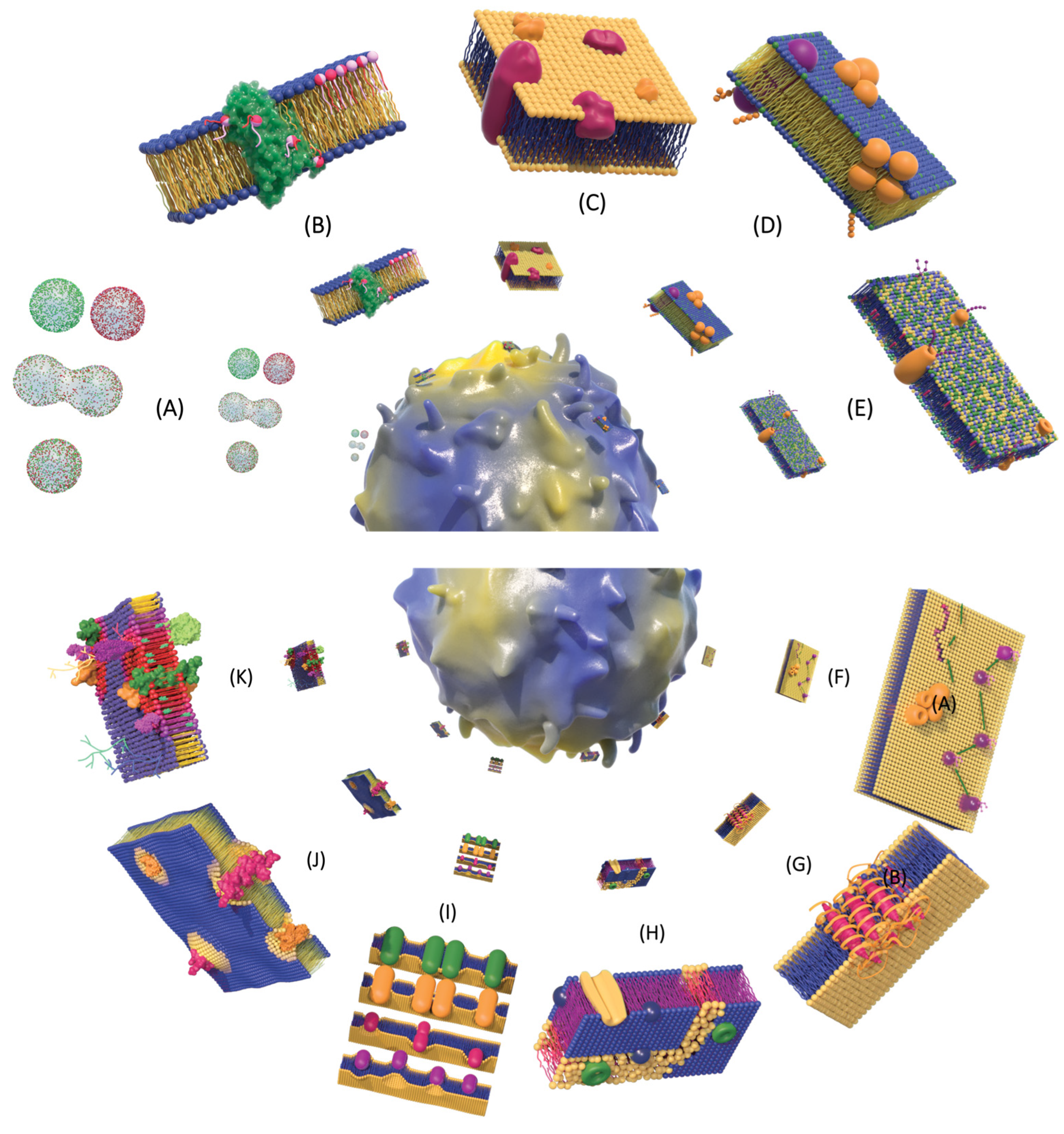
4.4. Membrane Platforms: Domains, Microdomains, Lipid Rafts and Membrane Rafts
4.5. Membrane Interaction: Glycocalyx–Membrane–Cytoskeleton
4.5.1. Membrane: A Three-Layer System Composed of Glycocalyx–Membrane–Cytoskeleton
4.5.2. Membrane–Cytoskeleton Interaction, Picket-Fence Model
4.5.3. Oligomerization-Induced Entrapment Model
4.5.4. The Vision of Escribá et al., (2008)
4.5.5. Protein Island Model
4.5.6. Active Membrane–Actin Composite Model
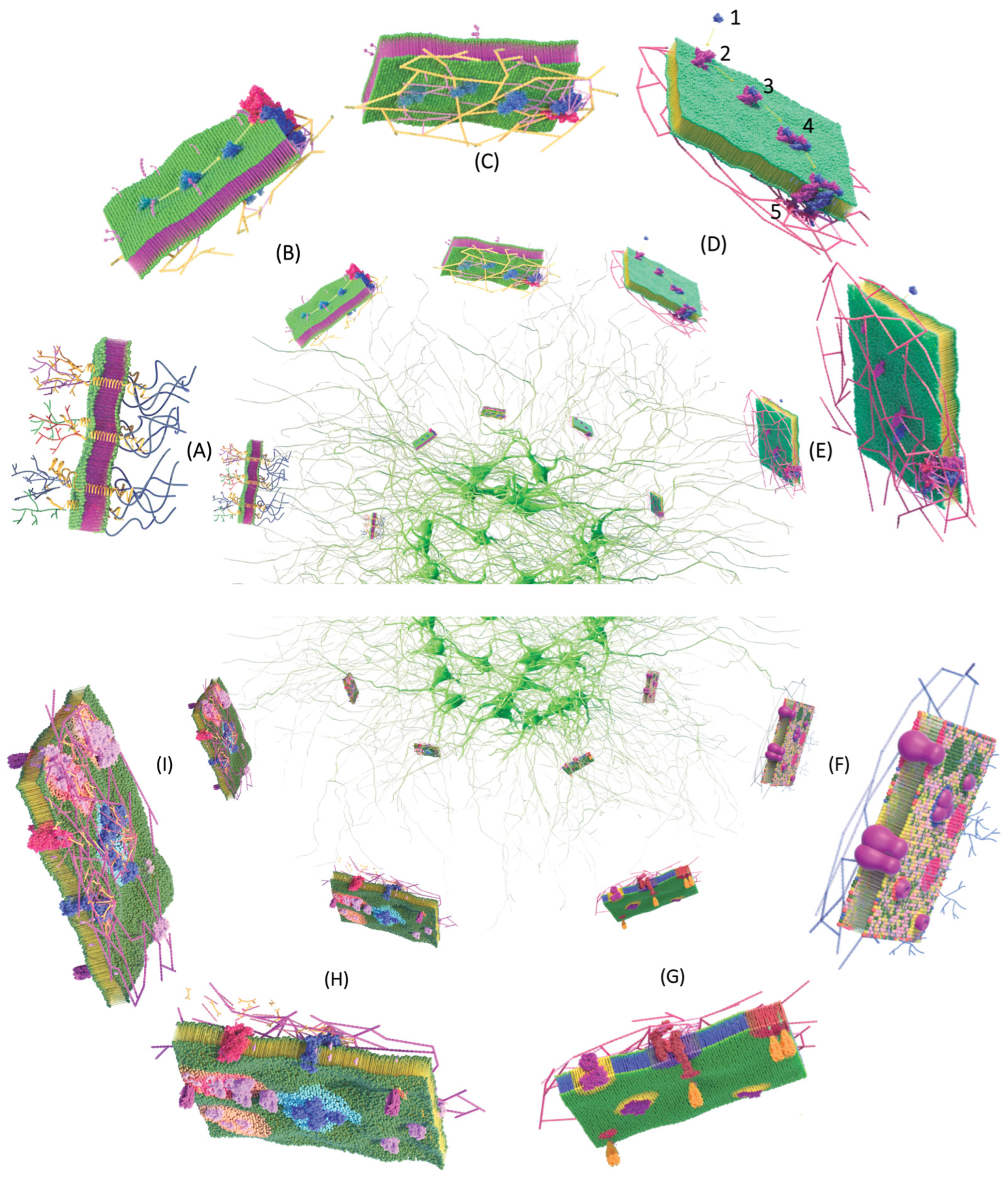
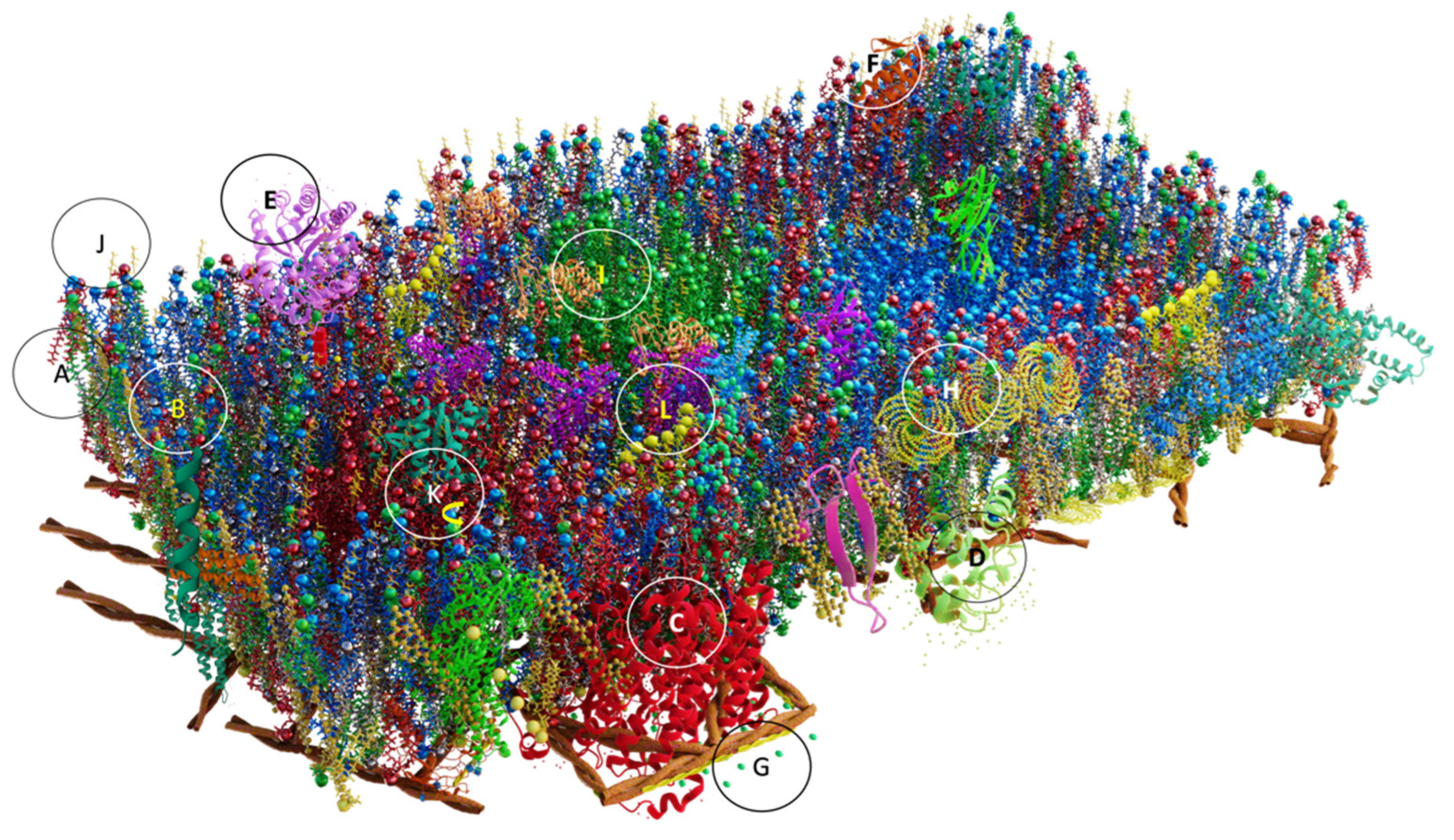
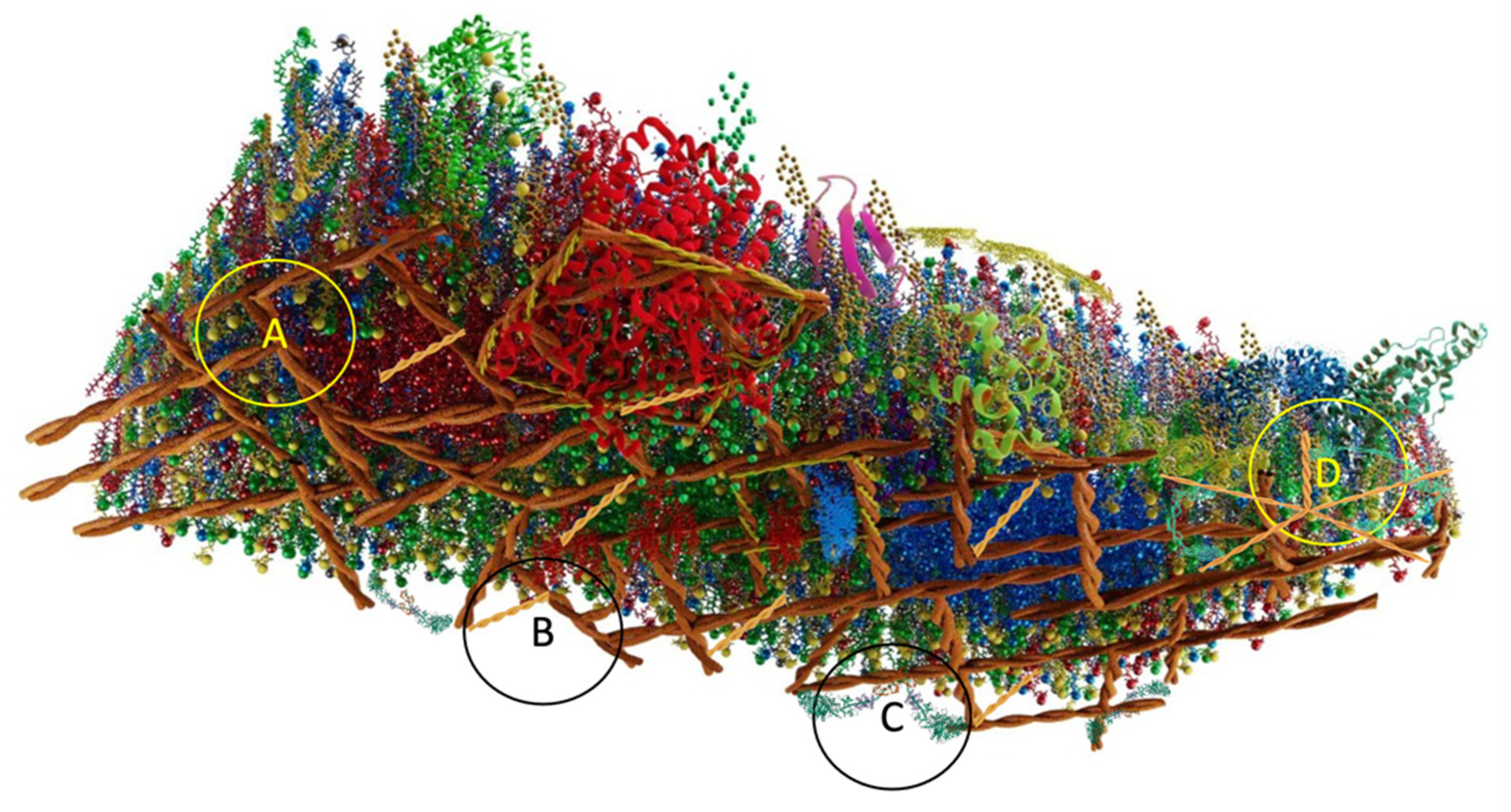
4.5.7. Griffié, Peters y Owen (2020)
5. Conclusions
6. Final Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honigmann, A.; Pralle, A. Compartmentalization of the Cell Membrane. J. Mol. Biol. 2016, 428, 4739–4748. [Google Scholar] [CrossRef] [PubMed]
- Stillwell, W. Introduction to Biological Membranes. In An Introduction to Biological Membranes; Elsevier: Amsterdam, The Netherlands, 2016; pp. 3–15. [Google Scholar]
- Nicolson, G.L.; Ferreira de Mattos, G. A Briefintroduction to Some Aspects of the Fluid-Mosaic Model of Cell Membrane Structure and Its Importance in Membrane Lipid Replacement. Membranes 2021, 11, 947. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L.; Ferreira de Mattos, G. Fifty Years of the Fluid-Mosaic Model of Biomembrane Structure and Organization and Its Importance in Biomedicine with Particular Emphasis on Membrane Lipid Replacement. Biomedicines 2022, 10, 1711. [Google Scholar] [CrossRef] [PubMed]
- Costa, J. La Esquemática; Paidós Ibérica, S A: Barcelona, Spain, 1998. [Google Scholar]
- Hewson, W. XXXIII. On the Figure and Composition of the Red Particles of the Blood, Commonly Called the Red Globules. Philos. Trans. R. Soc. Lond. 1773, 63, 303–323. [Google Scholar] [CrossRef]
- Franklin, B. Of the Stilling of Waves by Means of Oil. Philos. Trans. R. Soc. B Biol. Sci. 1774, 64, 445–460. [Google Scholar]
- Rayleigh L Surface Tension. Nature 1891, 43, 437–439. [CrossRef]
- Pockels, S.A. On the Spreading of Oil upon Water. Nature 1894, 50, 223–224. [Google Scholar] [CrossRef]
- Langmuir, I. The Constitution and Fundamental Properties of Solids and Liquids. II. Liquids. J. Am. Chem. Soc. 1917, 39, 1848–1906. [Google Scholar] [CrossRef]
- Nägeli, C.; Cramer, C. Pflanzeniphysiologische Untersuchungen; Fredrich Schultness: Zurich, Switzerland, 1855. [Google Scholar]
- Pfeffer, W. Osniotische Unterstuchunigen: Studieii Ztir Zellhiechanik; Wilhelm Engelmann: Leipzig, Germany, 1877. [Google Scholar]
- Jacobs, M.H. Early Osmotic Hstory of the Plasma Membrane. Circulation 1962, 26, 1013–1021. [Google Scholar] [CrossRef]
- de Vries, H. Eine Methode Zur Analyse Der Turgorkraft. In Jahrbücher für Wissenschaftliche Botanik; Pringsheim, N., Engelmann, W., Eds.; Bernstein: Berlin, Germany, 1884; Volume 14, pp. 427–440. [Google Scholar]
- Quincke, G. Ueber Periodische Ausbreitung an Flüssigkeitsoberflächen Und Dadurch Hervorgerufene Bewegungserscheinungen. Ann. Phys. Und Chem. 1888, 271, 580–642. [Google Scholar] [CrossRef]
- Overton, E. Ueber Die Osmotischen Eigenschaften Der Zelle in Ihrer Bedeutung Für Die Toxikologie Und Pharmakologie. Z. Für Phys. Chem. 1897, 22U, 189–209. [Google Scholar] [CrossRef]
- Overton, E. Über Die Allgemeinen Osmotischen Eigenschaften Der Zelle. Vierteljahrsschr. Nat. Ges. Zürich 1899, 44, 88–135. [Google Scholar]
- Gorter, E.; Grendel, F. On Bimolecular Layers of the Chromocytes of the Blood. J. Exp. Med. 1925, 41, 439–443. [Google Scholar] [CrossRef]
- Knoll, W. Oberflächenberechnungen Bei Menschlichen Erythrocyten. Pflug. Arch. Gesamte Physiol. Menschen Tiere 1923, 198, 367–372. [Google Scholar] [CrossRef]
- Fricke, H. The Electric Capacity of Suspensions of Red Corpuscles of a Dog. Phys. Rev. 1925, 26, 682–687. [Google Scholar] [CrossRef]
- Fricke, H. The Electric Capacity of Cell Suspensions. Phys. Rev. Ser. II 1923, 21, 708–709. [Google Scholar]
- Sperry, W.M. Lipid Excretion. J. Biol. Chem. 1926, 68, 357–383. [Google Scholar] [CrossRef]
- Gobley, T. Sur La Lécithine et La Cérébrine. J. Pharm. Chim. 1874, 19, 346. [Google Scholar]
- Wieland, H.O.; Windaus, A. Nobel Lectures Chemistry 1922–1941; Elsevier: Amsterdam, The Netherlands, 1966. [Google Scholar]
- Crowfoot, D.; Bernal, J.D. X-Ray Crystallography and the Chemistry of Sterols and Sex Hormones. Chem. Weekbl. 1937, 34, 19–22. [Google Scholar]
- Grendel, F. Über Die Lipoidschicht Der Chromocyten Beim Schaf. Biochem. Z. 1929, 214, 231–241. [Google Scholar]
- Danielli, J.F.; Davson, H. A Contribution to the Theory of Permeability of Thin Films. J. Cell. Comp. Physiol. 1935, 5, 495–508. [Google Scholar] [CrossRef]
- Cole, K.S.; Curtis, H.J. Electric Impedance of Nerve and Muscle. Cold Spring Harb. Symp. Quant. Biol. 1936, 4, 73–89. [Google Scholar] [CrossRef]
- Danielli, J.F.; Harvey, E.N. The Tension at the Surface of Mackerel Egg Oil, with Remarks on the Nature of the Cell Surface. J. Cell. Comp. Physiol. 1935, 5, 483–494. [Google Scholar] [CrossRef]
- Schmitt, F.O.; Palmer, K.J. X-Ray Diffraction Studies of Lipids and Lipide-Protein Systems. Cold Spring Harb. Symp. Quant. Biol. 1940, 8, 94–101. [Google Scholar] [CrossRef]
- Pangborn, M.C. A New Serologically Active Phospholipid from Beef Heart. Exp. Biol. Med. 1941, 48, 484–486. [Google Scholar] [CrossRef]
- Folch, J. Brain Cephalin, a Mixture of Phosphatides. Separation from It of Phosphatidyl Serine, Phosphatidyl Ethanolamine, and a Fraction Containing an Inositol Phosphatide. J. Biol. Chem. 1942, 146, 35–44. [Google Scholar] [CrossRef]
- Benson, A.A.; Maruo, B. Plant Phospholipids I. Identification of the Phosphatidyl Glycerols. Biochim Biophys Acta 1958, 27, 189–195. [Google Scholar] [CrossRef]
- Pizer, F.L.; Ballou, C.E. Studies on Myo-Inositol Phosphates of Natural Origin. J. Am. Chem. Soc. 1959, 81, 915–921. [Google Scholar] [CrossRef]
- Klenk, E.; Padberg, G. Über Die Ganglioside von Pferdeerythrocyten. Hoppe Seylers Z. Physiol. Chem. 1962, 327, 249–255. [Google Scholar] [CrossRef]
- Kuhn, R.; Egge, H. Über Ergebnisse Der Permethylierung Der Ganlioside G/Sub G/Sub. Chem. Ber. 1963, 96, 3338–3348. [Google Scholar] [CrossRef]
- Kuhn, R.; Wiegandt, H. Die Konstitution Der Ganglioside GII, GIII Und GIV. Z. Für Nat. B 1963, 18, 541–543. [Google Scholar] [CrossRef]
- de Gier, J.; van Deenen, L.L.M. Some Lipid Characteristics of Red Cell Membranes of Various Animal Species. Biochim. Biophys. Acta 1961, 49, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.D. New Observations on the Ultrastructure of the Membranes of Frog Peripheral Nerve Fibers. J. Biophys. Biochem. Cytol. 1957, 3, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Mühlethaler, K.; Moor, H.; Szarkowski, J.W. The Ultrastructure of the Chloroplast Lamellae. Planta 1965, 67, 305–323. [Google Scholar] [CrossRef]
- Bennett, H.S. Morphological Aspects of Extracellular Polysaccharides. J. Histochem. Cytochem. 1963, 11, 14–23. [Google Scholar] [CrossRef]
- Fawcett, D.W. Surface Specializations of Absorbing Cells. J. Histochem. Cytochem. 1965, 13, 75–91. [Google Scholar] [CrossRef]
- Benson, A.A. On the Orientation of Lipids in Chloroplast and Cell Membranes. J. Am. Oil Chem. Soc. 1966, 43, 265–270. [Google Scholar] [CrossRef]
- Green, D.E.; Tzagoloff, A. The Mitochondrial Electron Transfer Chain. Arch. Biochem. Biophys. 1966, 116, 293–304. [Google Scholar] [CrossRef]
- Green, D.E.; MacLennan, D.H. Structure and Function of the Mitochondrial Cristael Membrane. Bioscience 1969, 19, 213–222. [Google Scholar] [CrossRef]
- Green, D.E.; Perdue, J.F. Membranes as Expressions of Repeating Units. Proc. Natl. Acad. Sci. USA 1966, 55, 1295–1302. [Google Scholar] [CrossRef]
- Vanderkooi, G.; Green, D.E. Biological Membrane Structure, I. The Protein Crystal Model for Membranes. Proc. Natl. Acad. Sci. USA 1970, 66, 615–621. [Google Scholar] [CrossRef]
- Steere, R.L. Electron Microscopy of Structural Detail in Frozen Biological Specimens. J. Biophys. Biochem. Cytol. 1957, 3, 45–60. [Google Scholar] [CrossRef]
- Branton, D. Fracture Faces of Frozen Membranes. Proc. Natl. Acad. Sci. USA 1966, 55, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Branton, D. Freeze-Etching Studies of Membrane Structure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1971, 261, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Chapman, D.; Penkett, S.A. Nuclear Magnetic Resonance Spectroscopic Studies of the Interaction of Phospholipids with Cholesterol. Nature 1966, 211, 1304–1305. [Google Scholar] [CrossRef] [PubMed]
- Ladbrooke, B.D.; Williams, R.M.; Chapman, D. Studies on Lecithin-Cholesterol-Water Interactions by Differential Scanning Calorimetry and X-Ray Diffraction. Biochim. Biophys. Acta (BBA)-Biomembr. 1968, 150, 333–340. [Google Scholar] [CrossRef]
- Lenard, J.; Singer, S.J. Protein Conformation in Cell Membrane Preparations as Studied by Optical Rotatory Dispersion and Circular Dichroism. Proc. Natl. Acad. Sci. USA 1966, 56, 1828–1835. [Google Scholar] [CrossRef]
- Singer, S.J. A Fluid Lipid-Globular Protein Mosaic Model of Membrane Structure. Ann. N. Y. Acad. Sci. 1972, 195, 16–23. [Google Scholar] [CrossRef]
- Lucy, J.A. Ultrastructure of Membranes: Micellar Organization. Br. Med. Bull. 1968, 24, 127–129. [Google Scholar] [CrossRef]
- Frye, L.D.; Edidin, M. The Rapid Intermixing of Cell Surface Antigens after Formation of Mouse-Human Heterokaryons. J. Cell Sci. 1970, 7, 319–335. [Google Scholar] [CrossRef]
- Papahadjopoulos, D.; Ohki, S. Stability of Asymmetric Phospholipid Membranes. Science 1969, 164, 1075–1077. [Google Scholar] [CrossRef] [PubMed]
- McConnell, H.M.; Kornberg, R.D. Inside-Outside Transitions of Phospholipids in Vesicle Membranes. Biochemistry 1971, 10, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.J.; Nicolson, G.L. The Fluid Mosaic Model of the Structure of Cell Membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.L. The Lipid Bilayer Membrane and Its Protein Constituents. J. Gen. Physiol. 2018, 150, 1472–1483. [Google Scholar] [CrossRef]
- Gitler, C. Microscopic Properties of Discrete Membrane Loci. In Biomembranes; Springer: Berlin, Germany, 1971; pp. 41–73. [Google Scholar]
- Gitler, C. Plasticity of Biological Membranes. Annu. Rev. Biophys. Bioeng. 1972, 1, 51–92. [Google Scholar] [CrossRef]
- Bretscher, M.S. Asymmetrical Lipid Bilayer Structure for Biological Membranes. Nat. New Biol 1972, 236, 11–12. [Google Scholar] [CrossRef]
- Bretscher, M.S. Phosphatidyl-Ethanolamine: Differential Labelling in Intact Cells and Cell Ghosts of Human Erythrocytes by a Membrane-Impermeable Reagent. J. Mol. Biol. 1972, 71, 523–528. [Google Scholar] [CrossRef]
- Saffman, P.G.; Delbrück, M. Brownian Motion in Biological Membranes. Proc. Natl. Acad. Sci. USA 1975, 72, 3111–3113. [Google Scholar] [CrossRef]
- Naji, A.; Levine, A.J.; Pincus, P.A. Corrections to the Saffman-Delbrück Mobility for Membrane Bound Proteins. Biophys. J. 2007, 93, L49–L51. [Google Scholar] [CrossRef]
- Gambin, Y.; Lopez-Esparza, R.; Reffay, M.; Sierecki, E.; Gov, N.S.; Genest, M.; Hodges, R.S.; Urbach, W. Lateral Mobility of Proteins in Liquid Membranes Revisited. Proc. Natl. Acad. Sci. USA 2006, 103, 2098–2102. [Google Scholar] [CrossRef]
- Venable, R.M.; Brown, F.L.H.; Pastor, R.W. Mechanical Properties of Lipid Bilayers from Molecular Dynamics Simulation. Chem. Phys. Lipids 2015, 192, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Unwin, N.; Henderson, R. Molecular Structure Determination by Electron Microscopy of Unstained Crystalline Specimens. J. Mol. Biol. 1975, 94, 425–440. [Google Scholar] [CrossRef]
- Unwin, N.; Henderson, R. The Structure of Proteins in Biological Membranes. Sci. Am. 1984, 250, 78–94. [Google Scholar] [CrossRef]
- Jain, M.K.; White, H.B. Long-Range Order in Biomembranes. In Advances in Lipid Research; Elsevier: Amsterdam, The Netherlands, 1977; pp. 1–60. [Google Scholar]
- Israelachvili, J.N. Refinement of the Fluid-Mosaic Model of Membrane Structure. Biochim. Biophys. Acta (BBA)-Biomembr. 1977, 469, 221–225. [Google Scholar] [CrossRef]
- Mouritsen, O.G.; Bloom, M. Mattress Model of Lipid-Protein Interactions in Membranes. Biophys. J. 1984, 46, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Karnovsky, M.J.; Kleinfeld, A.M.; Hoover, R.L.; Klausner, R.D. The Concept of Lipid Domains in Membranes. J. Cell Biol. 1982, 94, 1–6. [Google Scholar] [CrossRef]
- Simons, K.; van Meer, G. Lipid Sorting in Epithelial Cells. Biochemistry 1988, 27, 6197–6202. [Google Scholar] [CrossRef]
- Simons, K.; Ikonen, E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Pike, L.J. Rafts Defined: A Report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006, 47, 1597–1598. [Google Scholar] [CrossRef]
- Simons, K.; Sampaio, J.L. Membrane Organization and Lipid Rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef]
- van Meer, G.; Stelzer, E.; Wijnaendts-van-Resandt, R.; Simons, K. Sorting of Sphingolipids in Epithelial (Madin–Darby Canine Kidney) Cells. J. Cell Biol. 1987, 105, 1623–1635. [Google Scholar] [CrossRef]
- Ipsen, J.H.; Karlström, G.; Mourtisen, O.G.; Wennerström, H.; Zuckermann, M.J. Phase Equilibria in the Phosphatidylcholine-Cholesterol System. Biochim. Biophys. Acta (BBA)-Biomembr. 1987, 905, 162–172. [Google Scholar] [CrossRef]
- Sackmann, E. Biological Membranes: Architecture and Function. In Handbook of Biological Physics; Lipowsky, R., Sackmann, E., Eds.; Elsevier: Amsterdam, The Netherlands, 1995; Volume 1A. [Google Scholar]
- Isenberg, G.; Goldmann, W.H. Actin-Membrane Coupling: A Role for Talin. J. Muscle Res. Cell Motil. 1992, 13, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Koyama-Honda, I.; Suzuki, K. Molecular Dynamics and Interactions for Creation of Stimulation-Induced Stabilized Rafts from Small Unstable Steady-State Rafts. Traffic 2004, 5, 213–230. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Kusumi, A. Dynamics of Raft Molecules in the Cell and Artificial Membranes: Approaches by Pulse EPR Spin Labeling and Single Molecule Optical Microscopy. Biochim. Biophys. Acta (BBA)-Biomembr. 2003, 1610, 231–243. [Google Scholar] [CrossRef]
- Fujiwara, T.; Ritchie, K.; Murakoshi, H.; Jacobson, K.; Kusumi, A. Phospholipids Undergo Hop Diffusion in Compartmentalized Cell Membrane. J. Cell Biol. 2002, 157, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Nakada, C.; Ritchie, K.; Murase, K.; Suzuki, K.; Murakoshi, H.; Kasai, R.S.; Kondo, J.; Fujiwara, T. Paradigm Shift of the Plasma Membrane Concept from the Two-Dimensional Continuum Fluid to the Partitioned Fluid: High-Speed Single-Molecule Tracking of Membrane Molecules. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 351–378. [Google Scholar] [CrossRef]
- Sako, Y.; Kusumi, A. Compartmentalized Structure of the Plasma Membrane for Receptor Movements as Revealed by a Nanometer-Level Motion Analysis. J. Cell Biol. 1994, 125, 1251–1264. [Google Scholar] [CrossRef]
- Sako, Y.; Kusumi, A. Barriers for Lateral Diffusion of Transferrin Receptor in the Plasma Membrane as Characterized by Receptor Dragging by Laser Tweezers: Fence versus Tether. J. Cell Biol. 1995, 129, 1559–1574. [Google Scholar] [CrossRef]
- Briegel, A.; Ortega, D.R.; Tocheva, E.I.; Wuichet, K.; Li, Z.; Chen, S.; Müller, A.; Iancu, C.V.; Murphy, G.E.; Dobro, M.J.; et al. Universal Architecture of Bacterial Chemoreceptor Arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 17181–17186. [Google Scholar] [CrossRef]
- Minguet, S.; Swamy, M.; Alarcón, B.; Luescher, I.F.; Schamel, W.W.A. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Petrini, E.M.; Marchionni, I.; Zacchi, P.; Sieghart, W.; Cherubini, E. Clustering of Extrasynaptic GABAA Receptors Modulates Tonic Inhibition in Cultured Hippocampal Neurons. J. Biol. Chem. 2004, 279, 45833–45843. [Google Scholar] [CrossRef]
- Boggs, J.M.; Wang, H. Co-Clustering of Galactosylceramide and Membrane Proteins in Oligodendrocyte Membranes on Interaction with Polyvalent Carbohydrate and Prevention by an Intact Cytoskeleton. J. Neurosci. Res. 2004, 76, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Moutón, C.; Abad, J.L.; Mira, E.; Lacalle, R.A.; Gallardo, E.; Jiménez-Baranda, S.; Illa, I.; Bernad, A.; Mañes, S.; Martínez-A, C. Segregation of Leading-Edge and Uropod Components into Specific Lipid Rafts during T Cell Polarization. Proc. Natl. Acad. Sci. USA 2001, 98, 9642–9647. [Google Scholar] [CrossRef]
- Rodgers, W.; Zavzavadjian, J. Glycolipid-Enriched Membrane Domains Are Assembled into Membrane Patches by Associating with the Actin Cytoskeleton. Exp. Cell Res. 2001, 267, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, A.; Sako, Y. Cell Surface Organization by the Membrane Skeleton. Curr. Opin. Cell Biol. 1996, 8, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Tomishige, M.; Sako, Y.; Kusumi, A. Regulation Mechanism of the Lateral Diffusion of Band 3 in Erythrocyte Membranes by the Membrane Skeleton. J. Cell Biol. 1998, 142, 989–1000. [Google Scholar] [CrossRef]
- Kusumi, A.; Suzuki, K.; Koyasako, K. Mobility and Cytoskeletal Interactions of Cell Adhesion Receptors. Curr. Opin. Cell Biol. 1999, 11, 582–590. [Google Scholar] [CrossRef]
- Iino, R.; Koyama, I.; Kusumi, A. Single Molecule Imaging of Green Fluorescent Proteins in Living Cells: E-Cadherin Forms Oligomers on the Free Cell Surface. Biophys. J. 2001, 80, 2667–2677. [Google Scholar] [CrossRef]
- Andrews, N.L.; Lidke, K.A.; Pfeiffer, J.R.; Burns, A.R.; Wilson, B.S.; Oliver, J.M.; Lidke, D.S. Actin Restricts FcɛRI Diffusion and Facilitates Antigen-Induced Receptor Immobilization. Nat. Cell Biol. 2008, 10, 955–963. [Google Scholar] [CrossRef]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.J.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I.; Barceló-Coblijn, G. Membranes: A Meeting Point for Lipids, Proteins and Therapies. J. Cell. Mol. Med. 2008, 12, 829–875. [Google Scholar] [CrossRef] [PubMed]
- Lillemeier, B.F.; Pfeiffer, J.R.; Surviladze, Z.; Wilson, B.S.; Davis, M.M. Plasma Membrane-Associated Proteins Are Clustered into Islands Attached to the Cytoskeleton. Proc. Natl. Acad. Sci. USA 2006, 103, 18992–18997. [Google Scholar] [CrossRef] [PubMed]
- Lillemeier, B.F.; Mörtelmaier, M.A.; Forstner, M.B.; Huppa, J.B.; Groves, J.T.; Davis, M.M. TCR and Lat Are Expressed on Separate Protein Islands on T Cell Membranes and Concatenate during Activation. Nat. Immunol. 2010, 11, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Lillemeier, B.F.; Davis, M.M. Probing the Plasma Membrane Structure of Immune Cells through the Analysis of Membrane Sheets by Electron Microscopy. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 169–182. [Google Scholar]
- Gowrishankar, K.; Ghosh, S.; Saha, S.; Rumamol, C.; Mayor, S.; Rao, M. Active Remodeling of Cortical Actin Regulates Spatiotemporal Organization of Cell Surface Molecules. Cell 2012, 149, 1353–1367. [Google Scholar] [CrossRef]
- Raghupathy, R.; Anilkumar, A.; Polley, A.; Singh, P.; Yadav, M.; Johnson, C.; Suryawanshi, S.; Saikam, V.; Sawant, S.; Panda, A.; et al. Transbilayer Lipid Interactions Mediate Nanoclustering of Lipid-Anchored Proteins. Cell 2015, 161, 581–594. [Google Scholar] [CrossRef]
- Griffié, J.; Peters, R.; Owen, D.M. An Agent-Based Model of Molecular Aggregation at the Cell Membrane. PLoS ONE 2020, 15, e0226825. [Google Scholar] [CrossRef] [PubMed]
- Stoeckenius, W.; Engelman, D.M. Current Models for the Structure of Biological Membranes. J. Cell Biol. 1969, 42, 613–646. [Google Scholar] [CrossRef]
- de la Serna, J.B.; Schütz, G.J.; Eggeling, C.; Cebecauer, M. There Is No Simple Model of the Plasma Membrane Organization. Front. Cell. Dev. Biol. 2016, 4, 106. [Google Scholar] [CrossRef]
- Israelachvili, J.N. Intermolecular and Surface Forces; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 9780123751829. [Google Scholar]
- Levental, I.; Veatch, S.L. The Continuing Mystery of Lipid Rafts. J. Mol. Biol. 2016, 428, 4749–4764. [Google Scholar] [CrossRef]
- Lorent, J.H.; Levental, K.R.; Ganesan, L.; Rivera-Longsworth, G.; Sezgin, E.; Doktorova, M.; Lyman, E.; Levental, I. Plasma Membranes Are Asymmetric in Lipid Unsaturation, Packing and Protein Shape. Nat. Chem. Biol. 2020, 16, 644–652. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The Mystery of Membrane Organization: Composition, Regulation and Roles of Lipid Rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Curry, F.E.; Clark, J.F.; Adamson, R.H. Erythrocyte-Derived Sphingosine-1-Phosphate Stabilizes Basal Hydraulic Conductivity and Solute Permeability in Rat Microvessels. Am. J. Physiol.-Heart Circ. Physiol. 2012, 303, H825–H834. [Google Scholar] [CrossRef] [PubMed]
- Alphonsus, C.S.; Rodseth, R.N. The Endothelial Glycocalyx: A Review of the Vascular Barrier. Anaesthesia 2014, 69, 777–784. [Google Scholar] [CrossRef]
- Tarbell, J.M.; Pahakis, M.Y. Mechanotransduction and the Glycocalyx. J. Intern. Med. 2006, 259, 339–350. [Google Scholar] [CrossRef]
- Ozu, M.; Galizia, L.; Acuña, C.; Amodeo, G. Aquaporins: More than Functional Monomers in a Tetrameric Arrangement. Cells 2018, 7, 209. [Google Scholar] [CrossRef]
- Aimon, S.; Callan-Jones, A.; Berthaud, A.; Pinot, M.; Toombes, G.E.S.; Bassereau, P. Membrane Shape Modulates Transmembrane Protein Distribution. Dev Cell 2014, 28, 212–218. [Google Scholar] [CrossRef]
- Fribourg, P.F.; Chami, M.; Sorzano, C.O.S.; Gubellini, F.; Marabini, R.; Marco, S.; Jault, J.-M.; Lévy, D. 3D Cryo-Electron Reconstruction of BmrA, a Bacterial Multidrug ABC Transporter in an Inward-Facing Conformation and in a Lipidic Environment. J Mol Biol 2014, 426, 2059–2069. [Google Scholar] [CrossRef]
- MacKinnon, R. Potassium Channels. FEBS Lett 2003, 555, 62–65. [Google Scholar] [CrossRef]
- Montigny, C.; Lyons, J.; Champeil, P.; Nissen, P.; Lenoir, G. On the Molecular Mechanism of Flippase- and Scramblase-Mediated Phospholipid Transport. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 767–783. [Google Scholar] [CrossRef]
- Hankins, H.M.; Baldridge, R.D.; Xu, P.; Graham, T.R. Role of Flippases, Scramblases and Transfer Proteins in Phosphatidylserine Subcellular Distribution. Traffic 2014, 16, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-W.; Takatsu, H. Substrates of P4-ATPases: Beyond Aminophospholipids (Phosphatidylserine and Phosphatidylethanolamine). FASEB J. 2018, 33, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular Mechanisms Controlling Actin Filament Dynamics in Nonmuscle Cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef] [PubMed]
- Bezanilla, M.; Gladfelter, A.S.; Kovar, D.R.; Lee, W.-L. Cytoskeletal Dynamics: A View from the Membrane. J. Cell Biol. 2015, 209, 329–337. [Google Scholar] [CrossRef]
- Yumura, S.; Itoh, G.; Kikuta, Y.; Kikuchi, T.; Kitanishi-Yumura, T.; Tsujioka, M. Cell-Scale Dynamic Recycling and Cortical Flow of the Actin–Myosin Cytoskeleton for Rapid Cell Migration. Biol. Open 2012, 2, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Yu, C.; Lieu, Z.Z.; Allard, J.; Mogilner, A.; Sheetz, M.P.; Bershadsky, A.D. Analysis of the Local Organization and Dynamics of Cellular Actin Networks. J. Cell Biol. 2013, 202, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shao, L.; Chen, B.-C.; Zhang, X.; Zhang, M.; Moses, B.; Milkie, D.E.; Beach, J.R.; Hammer, J.A.; Pasham, M.; et al. Extended-Resolution Structured Illumination Imaging of Endocytic and Cytoskeletal Dynamics. Science 2015, 349, aab3500. [Google Scholar] [CrossRef] [PubMed]
- Bovellan, M.; Romeo, Y.; Biro, M.; Boden, A.; Chugh, P.; Yonis, A.; Vaghela, M.; Fritzsche, M.; Moulding, D.; Thorogate, R.; et al. Cellular Control of Cortical Actin Nucleation. Curr. Biol. 2014, 24, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wu, X.S.; Crites, T.; Hammer, J.A. Actin Retrograde Flow and Actomyosin II Arc Contraction Drive Receptor Cluster Dynamics at the Immunological Synapse in Jurkat T Cells. Mol. Biol. Cell 2012, 23, 834–852. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational Modeling of Realistic Cell Membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos Muñiz, C.; Fernández Perrino, F.J. Evolution of the Concepts of Architecture and Supramolecular Dynamics of the Plasma Membrane. Membranes 2023, 13, 547. https://doi.org/10.3390/membranes13060547
Campos Muñiz C, Fernández Perrino FJ. Evolution of the Concepts of Architecture and Supramolecular Dynamics of the Plasma Membrane. Membranes. 2023; 13(6):547. https://doi.org/10.3390/membranes13060547
Chicago/Turabian StyleCampos Muñiz, Carolina, and Francisco José Fernández Perrino. 2023. "Evolution of the Concepts of Architecture and Supramolecular Dynamics of the Plasma Membrane" Membranes 13, no. 6: 547. https://doi.org/10.3390/membranes13060547
APA StyleCampos Muñiz, C., & Fernández Perrino, F. J. (2023). Evolution of the Concepts of Architecture and Supramolecular Dynamics of the Plasma Membrane. Membranes, 13(6), 547. https://doi.org/10.3390/membranes13060547






