Latest Update on Outer Membrane Vesicles and Their Role in Horizontal Gene Transfer: A Mini-Review
Abstract
:1. Introduction
2. Review Strategy
3. Biogenesis and Types of Extracellular Vesicles
3.1. Factors Associated with Vesicles Formation
3.2. Mechanisms of Vesicles Biogenesis
4. Components of OMVs and Their Biological Functions
4.1. OMV-Associated DNA
4.2. Potential Applications of OMVs
| Bacterial Species | Vesicle-Associated Virulence Factors | Activity | Cytotoxic Activity | Reference |
|---|---|---|---|---|
| Pseudomonas aeuriginosa (clinical strains DH1137) | Orp F, OrpH | Modulation of the host innate immune response: ↓ expression of several genes belonging to the Major Histocompatibility Complexes (MHC) class II ↓ proteins important for antigen presentation to T-helper lymphocytes, such as CD74 | ND | [30] |
| Helicobacter pylori (NCTC11637; Hp-400) | CagA, VacA, UreB | ↑ release of inflammatory factors related to bacterial infection including IL-6, IFN-γ, IL-8, and TNF-α | ND | [50] |
| Actinobacillus pleuropneumoniae (MIDG2331 and mutants) | ApxIII toxin, MomP2, OmlA, FkpA, OmpP1, LpoA | Immunomodulatory effects evaluated in vitro: ↓ non-specific immune response by inhibiting the expression of several genes normally overexpressed during the innate immune response (e.g., chemokines and IL-6) | ND | [51] |
| Bordetella pertussis (Tohama I strain CIP 81.32 and isogenic mutant BpΔCyaA) | Adenylate cyclase toxin (CyaA), pertussis toxin (Ptx), SodB, KatA, AhpC, AhpD | Direct interaction with macrophages: ↓ expression of genes important in the macrophage response to bacterial infection, which leads to the persistence of the producing bacterium within macrophages and thus increased survival of the pathogen itself | ND | [52] |
| Borrelia burgdorferi B31 (ATCC, 35210, and GCB726) | Outer membrane protein (OspA, OspB, OspC) | OMVs represent a vehicle to evade the immune system and could explain the persistence of the infection. | No cytotoxicity reported against non-immune cells (skin fibroblasts and chondrosarcoma cells) | [53] |
| Vibrio cholerae (WT and mutant strains) | CT toxin, OmpU, OmpT | OMVs act as a protective envelope for cholera toxin (CT), which, when internalized in the intestinal cells, undergoes degradation by intestinal proteases. | ND | [38] |
| Burkholderia cepacia (ATCC 25416) | OmpW, OmpA, Type 1 fimbrial protein, A chain, TPR repeat family protein, lipase, protease | Pro-inflammatory action at small doses in vitro: ↑ expression of genes coding for pro-inflammatory cytokines | Cytotoxic effects on human lung cells A549 | [54] |
| Escherichia coli O78:H11 (ATCC 35401) | Colonization factor I (CFA/I), heat-labile enterotoxin (LT), and non-classical factors: EtpA, EatA, and TibA | Stimulation of immune responses: ↑ release of neutralizing antibodies stimulated by the LT B immunogenic subunit expressed on OMVs. Stimulation of a Th1 immune response in macrophages: ↑ expression of CD40, MHCII, CD80, CD86; ↑ release of IL-6 and MCP-1 | No cytotoxicity detected in RAW 264.7 cells for 48 h | [55] |
| Treponema denticola | Msp | Inhibition of neutrophils chemotaxis; ↓ pPTEN levels; ↑ phosphatase activity of PTEN; ↓ PIP3 levels | ND | [56] |
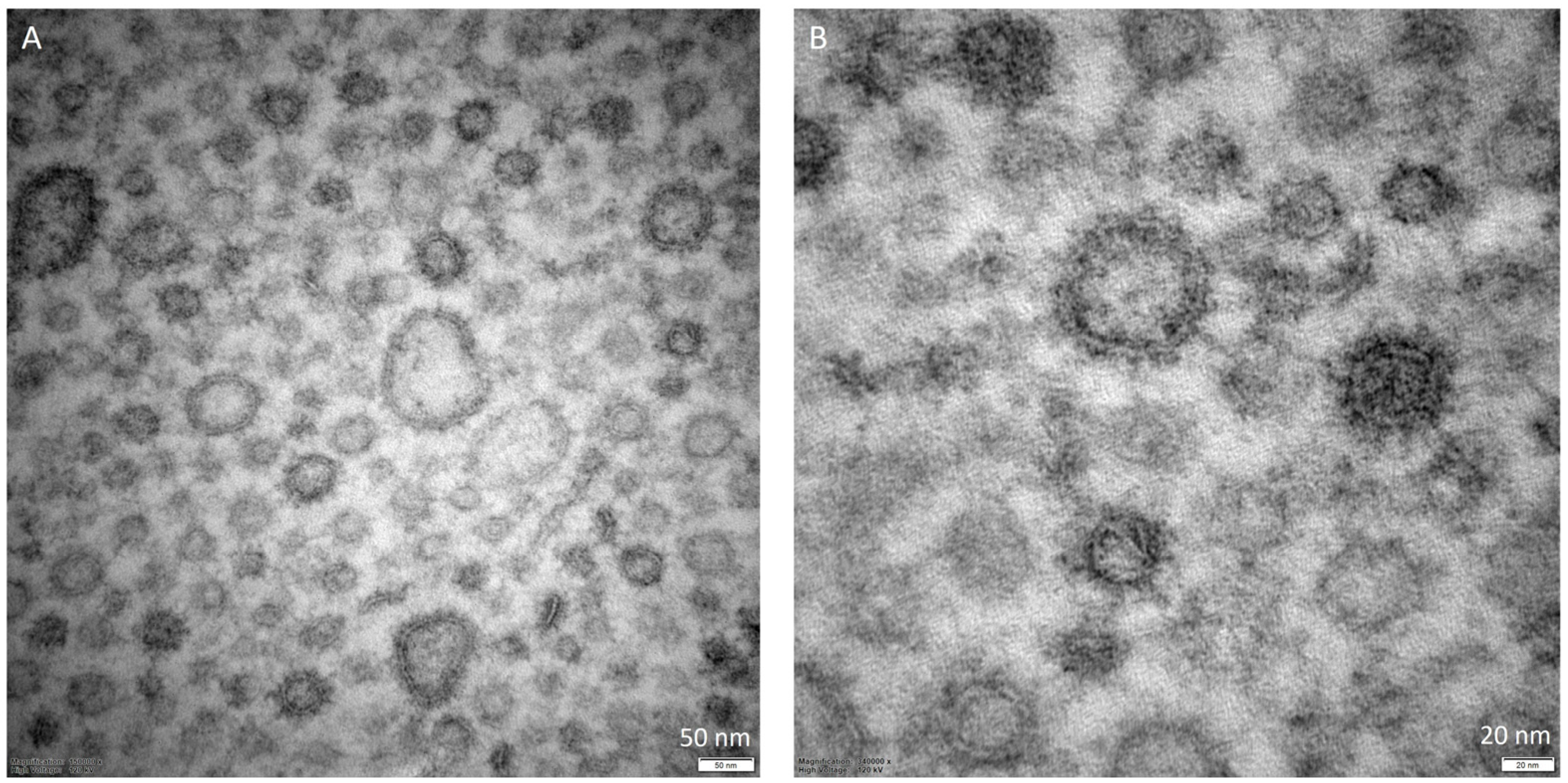
5. Characterization of OMVs
| Bacterial Producer | Technique | Diameter | Amount | Observations | Reference |
|---|---|---|---|---|---|
| Escherichia coli O104:H4 strain C227-11ϕcu | NTA | ~100–130 nm | 1 × 1011–1.5 × 1012 particles/mL (different growing conditions) | - | [62] |
| Klebsiella pneumoniae R1 | EM | 40–60 nm | - | Spherical morphology | [65] |
| Klebsiella pneumoniae-pGR and Klebsiella pneumoniae-PRM | TEM; DLS | 113.8 ± 53.7 nm and 94.13 ± 41.10 nm | - | Uniform spherical morphology | [66] |
| Hypervirulent Klebsiella pneumoniae (hvKp) and ESBL-producing classical K. pneumoniae (cKp) | TEM; NTA | 54–634 nm (median size 112 nm) and 17–523 nm (median size 78 nm) | ~6.5 × 107 particles/mL, ~3.5 × 107 particles/ml | Oval and spherical morphologies | [67] |
| Helicobacter pylori NCTC11637 and Hp-400 | TEM; NTA | 50–250 nm | - | Spherical bilayerd morphology and cup-shaped structure | [50] |
| Carbapenem-resistant and hypervirulent Klebsiella pneumoniae NUHL30457 | DLS; TEM | 50–250 nm (median size of 132 nm) | - | Spherical bilayered structures | [68] |
| Bordetella pertussis BpAR106 | TEM | 50–25 nm | - | - | [28] |
| Avian pathogenic Escherichia coli SCAO22 | TEM; nFCM | 79.42 nm (control); 0.14 nm and 64.18 nm (under antibiotic treatment) | 2.26 ± 0.78 × 1010 particles/mL (control)–5.66 ± 1.2 × 1012 particles/mL and 8.89 ± 0.36 × 1011 particles/mL (under antibiotic treatment) | Classic saucer-like vesicles | [63] |
| Carbapenem-resistant Klebsiella pneumoniae | TEM; DLS | 68.1 to 396 nm (control); 78.8 to 396 nm (under antibiotic treatment) | - | Spherical morphology | [69] |
| Escherichia coli ATCC8739 | DLS; TEM | 48 ± 3 nm (at 37 °C); 37 ± 4 nm (at 27 °C); 24 ± 2 nm (at 20 °C) | - | Spherical morphology | [25] |
| Klebsiella pneumoniae hvK2115 and CRK3022 | NTA; TEM | 50–200 nm | 9.1 × 1011 particles/mL and 2.6 × 1011 particles/mL | Spherical morphology | [70] |
| Avibacterium paragallinarum P4chr1 | TEM | 30–100 nm | - | Spherical morphology | [71] |
| Pseudomonas aeruginosa PAO1; PAO1 Δlys and PAO1 Δlys pJN105 lys | TEM; NTA | 50–400 nm | Lower amount produced by bubbling compared with explosive cell lysis | Spherical morphology | [34] |
| Bordetella pertussis Tohama I strain CIP 81.32 (Bp) and BpΔCyaA (ΔCyaA) | TEM | 10–240 nm (median size of 92.8 nm) | - | Spherical morphology with a uniform size distribution | [52] |
| Pseudomonas aeruginosa PAO1 and PW2884 | NTA;TEM | 178 nm, median size 119 nm (WT); 144 nm, median size 160 nm (PW2884) | 1.29 × 109 particles/mL (WT); 0.58 × 109 particles/mL (PW2884) | - | [33] |
| Helicobacter pylori 26695 (ATCC 700392) | SEM | 10–300 nm | - | Spherical morphology | [32] |
| Burkholderia cepacia ATCC 25416 | NTA | 129.7 ± 0.8 nm (control); under subinhibitory concentrations of antibiotics: MEM = 127.6 ± 1.2 nm; CAZ = 123.4 ± 2.5 nm; SXT = 154.9 ± 7.2 nm | 2.79 × 109 particles/mL (control); 2.45 × 1010 particles/mL (MEM); 1.91 × 1010 particles/mL (CAZ); 3.58 × 109 particles/mL (SXT) | - | [64] |
| Bordetella pertussis B213 and Bordetella bronchiseptica BB-D09-SR | TEM | 10–80 nm (after heat shock) | - | - | [26] |
| Borrelia burgdorferi B31 (ATCC, 35210) and GCB726 | TEM | Four size categories: 0–20, 20.1–60, 60.1–100, and 100.1–140 nm | - | - | [53] |
| Escherichia coli O78:H11 (ATCC 35401) | PCS | 50–300 nm | - | - | [55] |
| Pseudomonas aeruginosa DH1137 | TEM; NTA | 30–600 nm | - | Concave aspect | [30] |
| Actinobacillus pleuropneumoniae WT and mutant strains | Cryo-TEM | 20–200 nm | - | Some WTs OMVs show a stick shape; OMVs of irregular shape in mutants | [51] |
| Pseudomonas aeruginosa PAO9503 and PAO9505 | NTA; TEM | 50–500 nm; in larger quantities 100–200 nm | - | - | [27] |
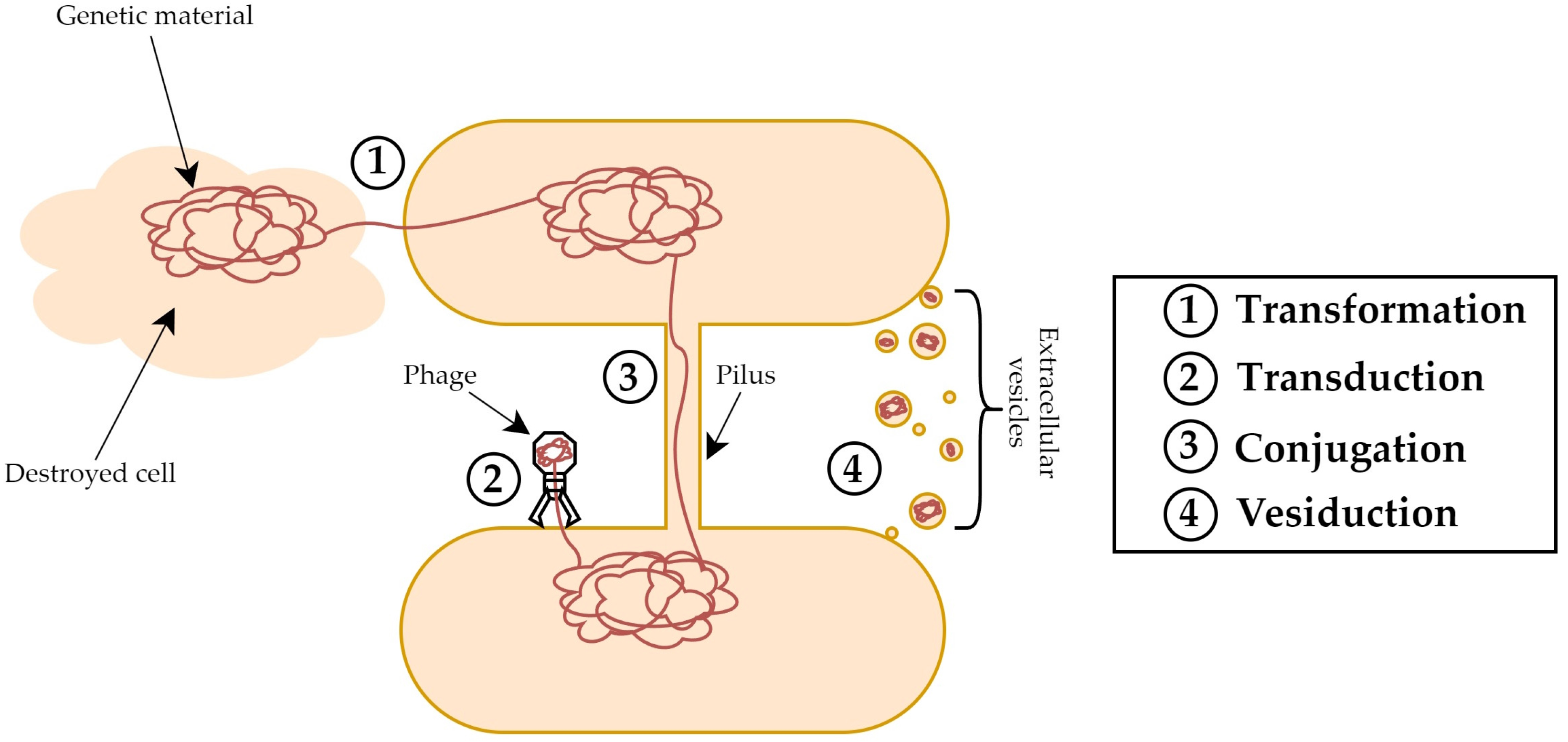
6. OMV-Mediated Horizontal Gene Transfer
| Bacterial Producer | Genetic Material | Recipient Bacteria | Observations | References |
|---|---|---|---|---|
| Avibacterium paragallinarum P4chr1 | ARGs: bl2d_oxa1; aph33ib; cml_e3; tetB | A. paragallinarum Modesto |
| [71] |
| Escherichia coli O104:H4 strain C227-11ϕcu | pESBL plasmid: blaCTX-M-15 and blaTEM-1 | Clinical Enterobacteriaceae isolates and E. coli K-12 C600 |
| [62] |
| Escherichia coli strains | pUC19; pCP20 | - |
| [78] |
| Klebsiella pneumoniae R1 | blaKPC-2 | K. pneumoniae S1; E. coli S1 |
| [65] |
| Hypervirulent Klebsiella pneumoniae (hvKp) | pLVPK-like plasmid: prmpA and iroB | ESBL-producing classical K. pneumoniae (cKp) |
| [67] |
| Klebsiella pneumoniae-pGR and Klebsiella pneumoniae-PRM | Plasmids containing genes for β-lactamase: pGR and PRM | K. pneumoniae ATCC 10031; E. coli ATCC 25922; S. enterica ATCC 14028; P. aeruginosa ATCC 13388; and B. cepacia ATCC 25416 |
| [66] |
| Carbapenem-resistant and hypervirulent Klebsiella pneumoniae NUHL30457 | Plasmids containing virulence and antimicrobial resistance genes | K. pneumoniae ATCC 700603 |
| [68] |
| Avian pathogenic Escherichia coli SCAO22 | IncI2 plasmid: blaCTX-M-55 | E. coli C600 |
| [63] |
| Escherichia coli DH5α | pET28a plasmid: nirS | E. coli BL21 |
| [75] |
| Carbapenem-resistant Klebsiella pneumoniae | IncFIBpKPHS1 plasmid: blaNDM-1 | K. pneumoniae ATCC 10031, ESBL-producing K. pneumoniae ATCC 700603, and hypervirulent K. pneumoniae NTUH-K2044 |
| [69] |
| Klebsiella pneumoniae hvK2115 and CRK3022 | phvK2115 plasmid: rmpAp, rmpA2p and iroB; pCRK3022 plasmid | E. coli EC600 and K. pneumoniae K20809 |
| [70] |
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jahromi, L.P.; Fuhrmann, G. Bacterial Extracellular Vesicles: Understanding Biology Promotes Applications as Nanopharmaceuticals. Adv. Drug Deliv. Rev. 2021, 173, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Celia, C.; Mincione, G.; Stringaro, A.; Di Marzio, L.; Colone, M.; Di Marcantonio, M.C.; Savino, L.; Puca, V.; Santoliquido, R.; et al. Detection and Physicochemical Characterization of Membrane Vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front. Microbiol. 2017, 8, 1040. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Carradori, S.; Puca, V.; Vitale, I.; Angeli, A.; Nocentini, A.; Bonardi, A.; Gratteri, P.; Lanuti, P.; Bologna, G.; et al. Selective Inhibition of Helicobacter pylori Carbonic Anhydrases by Carvacrol and Thymol Could Impair Biofilm Production and the Release of Outer Membrane Vesicles. Int. J. Mol. Sci. 2021, 22, 11583. [Google Scholar] [CrossRef]
- Gamazo, C.; Moriyon, I. Release of Outer Membrane Fragments by Exponentially Growing Brucella melitensis Cells. Infect. Immun. 1987, 55, 609. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Choi, D.Y.; Kim, D.K.; Kim, J.W.; Park, J.O.; Kim, S.; Kim, S.H.; Desiderio, D.M.; Kim, Y.K.; Kim, K.P.; et al. Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus aureus-Derived Membrane Vesicles. Proteomics 2009, 9, 5425–5436. [Google Scholar] [CrossRef]
- Krzyżek, P.; Marinacci, B.; Vitale, I.; Grande, R. Extracellular Vesicles of Probiotics: Shedding Light on the Biological Activity and Future Applications. Pharmaceutics 2023, 15, 522. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, D.; Liu, S.; Zhang, S.; Pan, Y. The Role of Porphyromonas gingivalis Outer Membrane Vesicles in Periodontal Disease and Related Systemic Diseases. Front. Cell. Infect. Microbiol. 2021, 10, 585917. [Google Scholar] [CrossRef]
- Toyofuku, M.; Schild, S.; Kaparakis-Liaskos, M.; Eberl, L. Composition and Functions of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2023, 21, 415–430. [Google Scholar] [CrossRef]
- McMillan, H.M.; Kuehn, M.J. The Extracellular Vesicle Generation Paradox: A Bacterial Point of View. EMBO J. 2021, 40, e108174. [Google Scholar] [CrossRef]
- Zlatkov, N.; Nadeem, A.; Uhlin, B.E.; Wai, S.N. Eco-Evolutionary Feedbacks Mediated by Bacterial Membrane Vesicles. FEMS Microbiol. Rev. 2021, 45, fuaa047. [Google Scholar] [CrossRef] [PubMed]
- Nagakubo, T.; Nomura, N.; Toyofuku, M. Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 2020, 10, 3026. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An Emerging Focus on Lipids in Extracellular Vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Cassilly, C.D.; Reynolds, T.B. PS, It’s Complicated: The Roles of Phosphatidylserine and Phosphatidylethanolamine in the Pathogenesis of Candida albicans and Other Microbial Pathogens. J. Fungi 2018, 4, 28. [Google Scholar] [CrossRef]
- Lind, T.K.; Skoda, M.W.A.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Phosphatidylethanolamine and Phosphatidylglycerol by Vesicle Fusion, a Simple but Relevant Model for Bacterial Membranes. ACS Omega 2019, 4, 10687–10694. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Cell. Infect. Microbiol. 2018, 8, 366060. [Google Scholar] [CrossRef]
- Cooke, A.C.; Florez, C.; Dunshee, E.B.; Lieber, A.D.; Terry, M.L.; Light, C.J.; Schertzer, J.W. Pseudomonas Quinolone Signal-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa. mSphere 2020, 5, e01109-20. [Google Scholar] [CrossRef]
- Graham, C.L.B.; Newman, H.; Gillett, F.N.; Smart, K.; Briggs, N.; Banzhaf, M.; Roper, D.I. A Dynamic Network of Proteins Facilitate Cell Envelope Biogenesis in Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 12831. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical Properties of the Bacterial Outer Membrane. Nat. Rev. Microbiol. 2022, 20, 248. [Google Scholar] [CrossRef]
- Mozaheb, N.; Mingeot-Leclercq, M.-P. Membrane Vesicle Production as a Bacterial Defense Against Stress. Front. Microbiol. 2020, 11, 3120. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive Cell Lysis as a Mechanism for The Biogenesis of Bacterial Membrane Vesicles and Biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Ballerin, G.; Nolan, L.M.; Petty, N.K.; Whitchurch, C.B. Bacteriophage Infection of Escherichia coli Leads to the Formation of Membrane Vesicles via Both Explosive Cell Lysis and Membrane Blebbing. Microbiology 2021, 167, 001021. [Google Scholar] [CrossRef] [PubMed]
- Baeza, N.; Delgado, L.; Comas, J.; Mercade, E. Phage-Mediated Explosive Cell Lysis Induces the Formation of a Different Type of O-IMV in Shewanella vesiculosa M7T. Front. Microbiol. 2021, 12, 713669. [Google Scholar] [CrossRef] [PubMed]
- Sarra, A.; Celluzzi, A.; Bruno, S.P.; Ricci, C.; Sennato, S.; Ortore, M.G.; Casciardi, S.; Del Chierico, F.; Postorino, P.; Bordi, F.; et al. Biophysical Characterization of Membrane Phase Transition Profiles for the Discrimination of Outer Membrane Vesicles (OMVs) From Escherichia coli Grown at Different Temperatures. Front. Microbiol. 2020, 11, 514720. [Google Scholar] [CrossRef]
- de Jonge, E.F.; Balhuizen, M.D.; van Boxtel, R.; Wu, J.; Haagsman, H.P.; Tommassen, J. Heat Shock Enhances Outer-Membrane Vesicle Release in Bordetella Spp. Curr. Res. Microb. Sci. 2021, 2, 100009. [Google Scholar] [CrossRef]
- Johnston, E.L.; Zavan, L.; Bitto, N.J.; Petrovski, S.; Hill, A.F.; Kaparakis-Liaskos, M. Planktonic and Biofilm-Derived Pseudomonas aeruginosa Outer Membrane Vesicles Facilitate Horizontal Gene Transfer of Plasmid DNA. Microbiol. Spectr. 2023, 11, e05179-22. [Google Scholar] [CrossRef]
- Carriquiriborde, F.; Martin Aispuro, P.; Ambrosis, N.; Zurita, E.; Bottero, D.; Gaillard, M.E.; Castuma, C.; Rudi, E.; Lodeiro, A.; Hozbor, D.F. Pertussis Vaccine Candidate Based on Outer Membrane Vesicles Derived From Biofilm Culture. Front. Immunol. 2021, 12, 730434. [Google Scholar] [CrossRef]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. Biomed Res. Int. 2021, 2021, 1490732. [Google Scholar] [CrossRef]
- Armstrong, D.A.; Lee, M.K.; Hazlett, H.F.; Dessaint, J.A.; Mellinger, D.L.; Aridgides, D.S.; Hendricks, G.M.; Abdalla, M.A.K.; Christensen, B.C.; Ashare, A. Extracellular Vesicles from Pseudomonas aeruginosa Suppress MHC-Related Molecules in Human Lung Macrophages. ImmunoHorizons 2020, 4, 508. [Google Scholar] [CrossRef]
- Turner, L.; Bitto, N.J.; Steer, D.L.; Lo, C.; D’Costa, K.; Ramm, G.; Shambrook, M.; Hill, A.F.; Ferrero, R.L.; Kaparakis-Liaskos, M. Helicobacter pylori Outer Membrane Vesicle Size Determines Their Mechanisms of Host Cell Entry and Protein Content. Front. Immunol. 2018, 9, 1466. [Google Scholar] [CrossRef] [PubMed]
- Chew, Y.; Chung, H.Y.; Lin, P.Y.; Wu, D.C.; Huang, S.K.; Kao, M.C. Outer Membrane Vesicle Production by Helicobacter pylori Represents an Approach for the Delivery of Virulence Factors CagA, VacA and UreA into Human Gastric Adenocarcinoma (AGS) Cells. Int. J. Mol. Sci. 2021, 22, 3942. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Kragh, K.N.; Su, Y.C.; Sandblad, L.; Singh, B.; Bjarnsholt, T.; Riesbeck, K. Peptidoglycan-Binding Anchor Is a Pseudomonas aeruginosa OmpA Family Lipoprotein With Importance for Outer Membrane Vesicles, Biofilms, and the Periplasmic Shape. Front. Microbiol. 2021, 12, 639582. [Google Scholar] [CrossRef] [PubMed]
- Zavan, L.; Fang, H.; Johnston, E.L.; Whitchurch, C.; Greening, D.W.; Hill, A.F.; Kaparakis-Liaskos, M. The Mechanism of Pseudomonas aeruginosa Outer Membrane Vesicle Biogenesis Determines Their Protein Composition. Proteomics 2023, 23, e2200464. [Google Scholar] [CrossRef]
- Jefferies, D.; Khalid, S. To Infect or Not To Infect: Molecular Determinants of Bacterial Outer Membrane Vesicle Internalization by Host Membranes. J. Mol. Biol. 2020, 432, 1251–1264. [Google Scholar] [CrossRef]
- Caruana, J.C.; Walper, S.A. Bacterial Membrane Vesicles as Mediators of Microbe-Microbe and Microbe-Host Community Interactions. Front. Microbiol. 2020, 11, 432. [Google Scholar] [CrossRef]
- Kudryakova, I.V.; Afoshin, A.S.; Ivashina, T.V.; Suzina, N.E.; Leontyevskaya, E.A.; Leontyevskaya, N.V. Deletion of AlpB Gene Influences Outer Membrane Vesicles Biogenesis of Lysobacter Sp. XL1. Front. Microbiol. 2021, 12, 715802. [Google Scholar] [CrossRef]
- Zingl, F.G.; Thapa, H.B.; Scharf, M.; Kohl, P.; Müller, A.M.; Schild, S. Outer Membrane Vesicles of Vibrio cholerae Protect and Deliver Active Cholera Toxin to Host Cells via Porin-Dependent Uptake. MBio 2021, 12, e0053421. [Google Scholar] [CrossRef]
- Ronci, M.; Del Prete, S.; Puca, V.; Carradori, S.; Carginale, V.; Muraro, R.; Mincione, G.; Aceto, A.; Sisto, F.; Supuran, C.T.; et al. Identification and Characterization of the α-CA In The Outer Membrane Vesicles Produced by Helicobacter pylori. J. Enzyme Inhib. Med. Chem. 2019, 34, 189. [Google Scholar] [CrossRef]
- Salvachúa, D.; Werner, A.Z.; Pardo, I.; Michalska, M.; Black, B.A.; Donohoe, B.S.; Haugen, S.J.; Katahira, R.; Notonier, S.; Ramirez, K.J.; et al. Outer Membrane Vesicles Catabolize Lignin-Derived Aromatic Compounds in Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 2020, 117, 9302–9310. [Google Scholar] [CrossRef]
- Dorward, D.W.; Garon, C.F.; Judd, R.C. Export and Intercellular Transfer of DNA Via Membrane Blebs of Neisseria gonorrhoeae. J. Bacteriol. 1989, 171, 2499. [Google Scholar] [CrossRef] [PubMed]
- Bitto, N.J.; Chapman, R.; Pidot, S.; Costin, A.; Lo, C.; Choi, J.; D’Cruze, T.; Reynolds, E.C.; Dashper, S.G.; Turnbull, L.; et al. Bacterial Membrane Vesicles Transport Their DNA Cargo Into Host Cells. Sci. Rep. 2017, 7, 7072. [Google Scholar] [CrossRef] [PubMed]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-Mediated Transfer of Virulence Genes from Escherichia coli O157:H7 to Other Enteric Bacteria. Appl. Environ. Microbiol. 2000, 66, 4414. [Google Scholar] [CrossRef] [PubMed]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal Transfer of the OXA-24 Carbapenemase Gene Via Outer Membrane Vesicles: A New Mechanism of Dissemination of Carbapenem Resistance Genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Di Marcantonio, M.C.; Robuffo, I.; Pompilio, A.; Celia, C.; Marzio, L.D.; Paolino, D.; Codagnone, M.; Muraro, R.; Stoodley, P.; et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (EDNA). Front. Microbiol. 2015, 6, 1369. [Google Scholar] [CrossRef]
- Scaria, P.V.; Rowe, C.G.; Chen, B.B.; Muratova, O.V.; Fischer, E.R.; Barnafo, E.K.; Anderson, C.F.; Zaidi, I.U.; Lambert, L.E.; Lucas, B.J.; et al. Outer Membrane Protein Complex as a Carrier for Malaria Transmission Blocking Antigen Pfs230. NPJ Vaccines 2019, 4, 24. [Google Scholar] [CrossRef]
- Zare Banadkoki, E.; Rasooli, I.; Ghazanfari, T.; Siadat, S.D.; Shafiee Ardestani, M.; Owlia, P. Pseudomonas aeruginosa PAO1 Outer Membrane Vesicles-Diphtheria Toxoid Conjugate as a Vaccine Candidate In a Murine Burn Model. Sci. Rep. 2022, 12, 22324. [Google Scholar] [CrossRef]
- Weyant, K.B.; Oloyede, A.; Pal, S.; Liao, J.; Jesus, M.R.D.; Jaroentomeechai, T.; Moeller, T.D.; Hoang-Phou, S.; Gilmore, S.F.; Singh, R.; et al. A Modular Vaccine Platform Enabled by Decoration of Bacterial Outer Membrane Vesicles With Biotinylated Antigens. Nat. Commun. 2023, 14, 464. [Google Scholar] [CrossRef]
- Huang, W.; Meng, L.; Chen, Y.; Dong, Z.; Peng, Q. Bacterial Outer Membrane Vesicles as Potential Biological Nanomaterials for Antibacterial Therapy. Acta Biomater. 2022, 140, 102–115. [Google Scholar] [CrossRef]
- Wei, S.; Li, X.; Wang, J.; Wang, Y.; Zhang, C.; Dai, S.; Wang, X.; Deng, X.; Zhao, L.; Shan, B. Outer Membrane Vesicles Secreted by Helicobacter pylori Transmitting Gastric Pathogenic Virulence Factors. ACS Omega 2022, 7, 240–258. [Google Scholar] [CrossRef]
- Zhu, Z.; Antenucci, F.; Winther-Larsen, H.C.; Skovgaard, K.; Bojesen, A.M. Outer Membrane Vesicles of Actinobacillus pleuropneumoniae Exert Immunomodulatory Effects on Porcine Alveolar Macrophages. Microbiol. Spectr. 2022, 10, e01819-22. [Google Scholar] [CrossRef] [PubMed]
- Blancá, B.; Alvarez Hayes, J.; Surmann, K.; Hugo, V.; Hentschker, C.; Lamberti, Y.; Völker, U.; Rodriguez, M.E. Bordetella pertussis Outer Membrane Vesicles as Virulence Factor Vehicles That Influence Bacterial Interaction With Macrophages. Pathog. Dis. 2022, 80, ftac031. [Google Scholar] [CrossRef] [PubMed]
- Karvonen, K.; Tammisto, H.; Nykky, J.; Gilbert, L. Borrelia burgdorferi Outer Membrane Vesicles Contain Antigenic Proteins, but Do Not Induce Cell Death in Human Cells. Microorganisms 2022, 10, 212. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, S.I.; Yun, S.H.; Shin, M.; Lee, Y.C.; Lee, J.C. Proteins in Outer Membrane Vesicles Produced by Burkholderia cepacia Are Responsible for Pro-Inflammatory Responses in Epithelial Cells. J. Bacteriol. Virol. 2020, 50, 227–234. [Google Scholar] [CrossRef]
- Berzosa, M.; Delgado-López, A.; Irache, J.M.; Gamazo, C. Optimization of Enterotoxigenic Escherichia coli (ETEC) Outer Membrane Vesicles Production and Isolation Method for Vaccination Purposes. Microorganisms 2023, 11, 2088. [Google Scholar] [CrossRef]
- Jones, M.M.; Vanyo, S.T.; Visser, M.B. The Msp Protein of Treponema denticola Interrupts Activity of Phosphoinositide Processing in Neutrophils. Infect. Immun. 2019, 87, e00553-19. [Google Scholar] [CrossRef]
- Comfort, N.; Cai, K.; Bloomquist, T.R.; Strait, M.D.; Ferrante, A.W.; Baccarelli, A.A. Nanoparticle Tracking Analysis for the Quantification and Size Determination of Extracellular Vesicles. J. Vis. Exp. 2021, 2021, e62447. [Google Scholar] [CrossRef]
- Hilton, S.H.; White, I.M. Advances In the Analysis of Single Extracellular Vesicles: A Critical Review. Sens. Actuators Rep. 2021, 3, 100052. [Google Scholar] [CrossRef]
- Chan, M.Y.; Dowling, Q.M.; Sivananthan, S.J.; Kramer, R.M. Particle Sizing of Nanoparticle Adjuvant Formulations by Dynamic Light Scattering (DLS) and Nanoparticle Tracking Analysis (NTA). Methods Mol. Biol. 2017, 1494, 239–252. [Google Scholar]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Cizmar, P.; Yuana, Y. Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy. Methods Mol. Biol. 2017, 1660, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Bielaszewska, M.; Daniel, O.; Karch, H.; Mellmann, A. Dissemination of the BlaCTX-M-15 Gene Among Enterobacteriaceae Via Outer Membrane Vesicles. J. Antimicrob. Chemother. 2020, 75, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wen, R.; Mu, R.; Chen, X.; Ma, P.; Gu, K.; Huang, Z.; Ju, Z.; Lei, C.; Tang, Y.; et al. Outer Membrane Vesicles of Avian Pathogenic Escherichia coli Mediate the Horizontal Transmission of BlaCTX-M-55. Pathogens 2022, 11, 481. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, M.H.; Son, J.H.; Kim, S.I.; Yun, S.H.; Kim, K.; Kim, S.; Shin, M.; Lee, J.C. Outer Membrane Vesicles Produced by Burkholderia cepacia Cultured With Subinhibitory Concentrations of Ceftazidime Enhance Pro-Inflammatory Responses. Virulence 2020, 11, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Jing, X.P.; Meng, D.L.; Wu, T.T.; Zhou, H.; Sun, R.L.; Min, X.C.; Liu, R.; Zeng, J. Newly Detected Transmission of BlaKPC-2 by Outer Membrane Vesicles in Klebsiella pneumoniae. Curr. Med. Sci. 2023, 43, 80–85. [Google Scholar] [CrossRef]
- Dell’annunziata, F.; Dell’aversana, C.; Doti, N.; Donadio, G.; Dal Piaz, F.; Izzo, V.; De Filippis, A.; Galdiero, M.; Altucci, L.; Boccia, G.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Are a Driving Force For Horizontal Gene Transfer. Int. J. Mol. Sci. 2021, 22, 8732. [Google Scholar] [CrossRef]
- Hua, Y.; Wang, J.; Huang, M.; Huang, Y.; Zhang, R.; Bu, F.; Yang, B.; Chen, J.; Lin, X.; Hu, X.; et al. Outer Membrane Vesicles-Transmitted Virulence Genes Mediate the Emergence of New Antimicrobial-Resistant Hypervirulent Klebsiella pneumoniae. Emerg. Microbes Infect. 2022, 11, 1281–1292. [Google Scholar] [CrossRef]
- Li, P.; Luo, W.; Xiang, T.X.; Jiang, Y.; Liu, P.; Wei, D.D.; Fan, L.; Huang, S.; Liao, W.; Liu, Y.; et al. Horizontal Gene Transfer Via OMVs Co-Carrying Virulence and Antimicrobial-Resistant Genes Is a Novel Way for the Dissemination of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2022, 13, 945972. [Google Scholar] [CrossRef]
- Tang, B.; Yang, A.; Liu, P.; Wang, Z.; Jian, Z.; Chen, X.; Yan, Q.; Liang, X.; Liu, W. Outer Membrane Vesicles Transmitting BlaNDM-1 Mediate the Emergence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2023, 67, e01444-22. [Google Scholar] [CrossRef]
- Wang, Z.; Wen, Z.; Jiang, M.; Xia, F.; Wang, M.; Zhuge, X.; Dai, J. Dissemination of Virulence and Resistance Genes Among Klebsiella Pneumoniae Via Outer Membrane Vesicle: An Important Plasmid Transfer Mechanism to Promote The Emergence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae. Transbound. Emerg. Dis. 2022, 69, e2661–e2676. [Google Scholar] [CrossRef]
- Xu, J.; Mei, C.; Zhi, Y.; Liang, Z.; Zhang, X.; Wang, H. Comparative Genomics Analysis and Outer Membrane Vesicle-Mediated Horizontal Antibiotic-Resistance Gene Transfer in Avibacterium paragallinarum. Microbiol. Spectr. 2022, 10, e01379-22. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Soler, N.; Forterre, P. Vesiduction: The Fourth Way of HGT. Environ. Microbiol. 2020, 22, 2457–2460. [Google Scholar] [CrossRef]
- Kahn, M.E.; Barany, F.; Smith, H.O. Transformasomes: Specialized Membranous Structures That Protect DNA During Haemophilus Transformation. Proc. Natl. Acad. Sci. USA 1983, 80, 6927–6931. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Wang, L.; Luo, Y.; Miao, J. Outer Membrane Vesicles Mediated Horizontal Transfer of an Aerobic Denitrification Gene Between Escherichia coli. Biodegradation 2021, 32, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Hammes, W.; Schleifer, K.H.; Kandler, O. Mode of Action of Glycine on the Biosynthesis of Peptidoglycan. J. Bacteriol. 1973, 116, 1029. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, S.; Nakao, R. Glycine Significantly Enhances Bacterial Membrane Vesicle Production: A Powerful Approach for Isolation of LPS-Reduced Membrane Vesicles of Probiotic Escherichia coli. Microb. Biotechnol. 2020, 13, 1162–1178. [Google Scholar] [CrossRef]
- Aktar, S.; Okamoto, Y.; Ueno, S.; Tahara, Y.O.; Imaizumi, M.; Shintani, M.; Miyata, M.; Futamata, H.; Nojiri, H.; Tashiro, Y. Incorporation of Plasmid DNA Into Bacterial Membrane Vesicles by Peptidoglycan Defects in Escherichia coli. Front. Microbiol. 2021, 12, 747606. [Google Scholar] [CrossRef]
- Tran, F.; Boedicker, J.Q. Genetic Cargo and Bacterial Species Set Rhe Rate of Vesicle-Mediated Horizontal Gene Transfer. Sci. Rep. 2017, 7, 8813. [Google Scholar] [CrossRef]
- Li, C.; Zhu, L.; Wang, D.; Wei, Z.; Hao, X.; Wang, Z.; Li, T.; Zhang, L.; Lu, Z.; Long, M.; et al. T6SS Secretes An LPS-Binding Effector to Recruit OMVs for Exploitative Competition and Horizontal Gene Transfer. ISME J. 2021, 16, 500–510. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinacci, B.; Krzyżek, P.; Pellegrini, B.; Turacchio, G.; Grande, R. Latest Update on Outer Membrane Vesicles and Their Role in Horizontal Gene Transfer: A Mini-Review. Membranes 2023, 13, 860. https://doi.org/10.3390/membranes13110860
Marinacci B, Krzyżek P, Pellegrini B, Turacchio G, Grande R. Latest Update on Outer Membrane Vesicles and Their Role in Horizontal Gene Transfer: A Mini-Review. Membranes. 2023; 13(11):860. https://doi.org/10.3390/membranes13110860
Chicago/Turabian StyleMarinacci, Beatrice, Paweł Krzyżek, Benedetta Pellegrini, Gabriele Turacchio, and Rossella Grande. 2023. "Latest Update on Outer Membrane Vesicles and Their Role in Horizontal Gene Transfer: A Mini-Review" Membranes 13, no. 11: 860. https://doi.org/10.3390/membranes13110860
APA StyleMarinacci, B., Krzyżek, P., Pellegrini, B., Turacchio, G., & Grande, R. (2023). Latest Update on Outer Membrane Vesicles and Their Role in Horizontal Gene Transfer: A Mini-Review. Membranes, 13(11), 860. https://doi.org/10.3390/membranes13110860








