Liposomes Bearing Non-Bilayer Phospholipid Arrangements Induce Specific IgG Anti-Lipid Antibodies by Activating NK1.1+, CD4+ T Cells in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
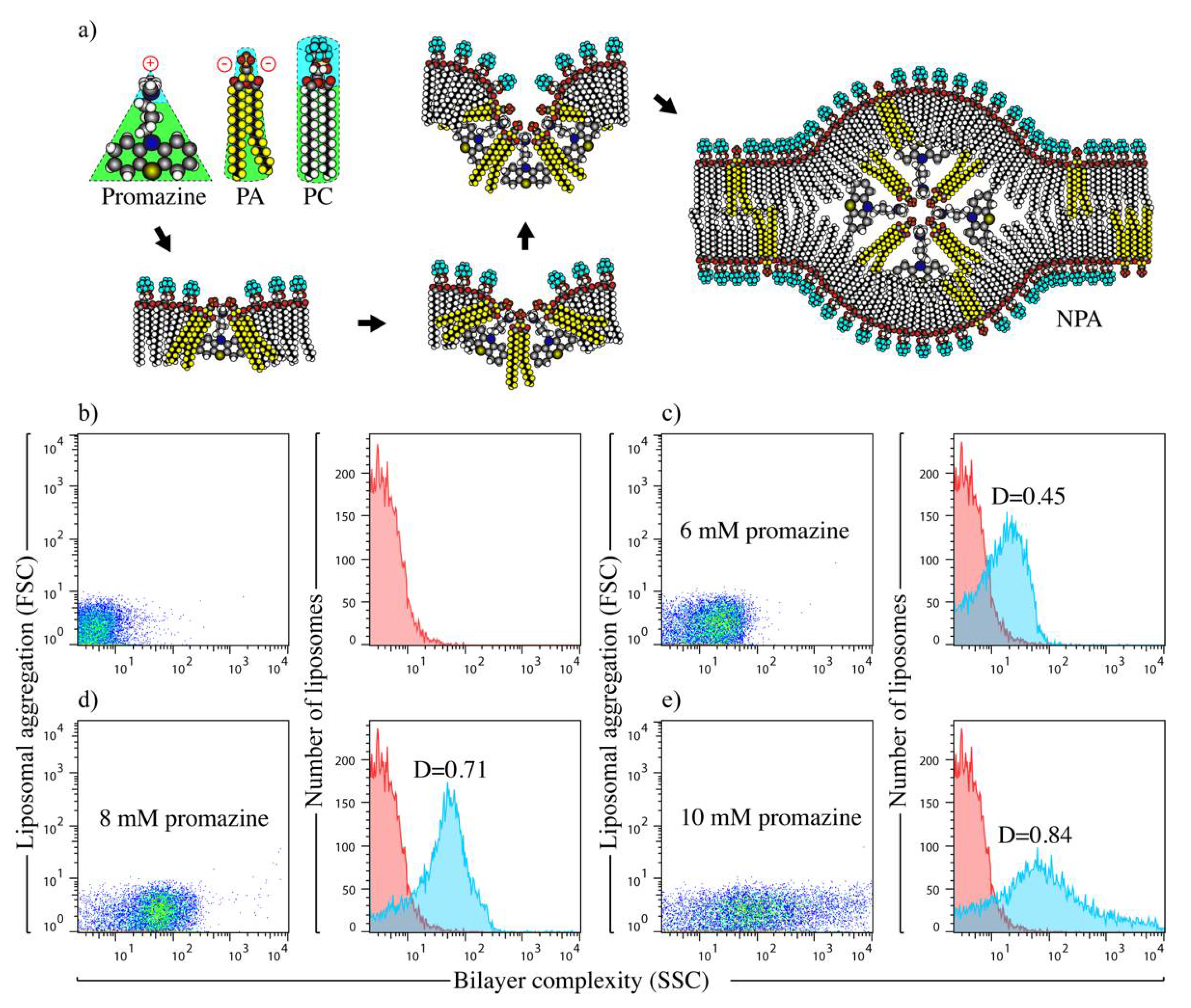
2.2. Preparation and Characterization of Liposomal Antigens
2.3. Induction of IgG Anti-NPA Antibodies in C57BL/6 Mice
2.4. Detection of IgG Anti-NPA, Anti-Cardiolipin and Anti-Histone Antibodies in Mice Sera
2.5. Analysis of Spleen and Mesenteric Lymph Node NK1.1+, CD4+ T Cells
2.6. In Vitro Re-Stimulation of NK1.1+, CD4+ T Cells
2.7. Gene Expression Analysis of NK1.1+, CD4+ T Cells
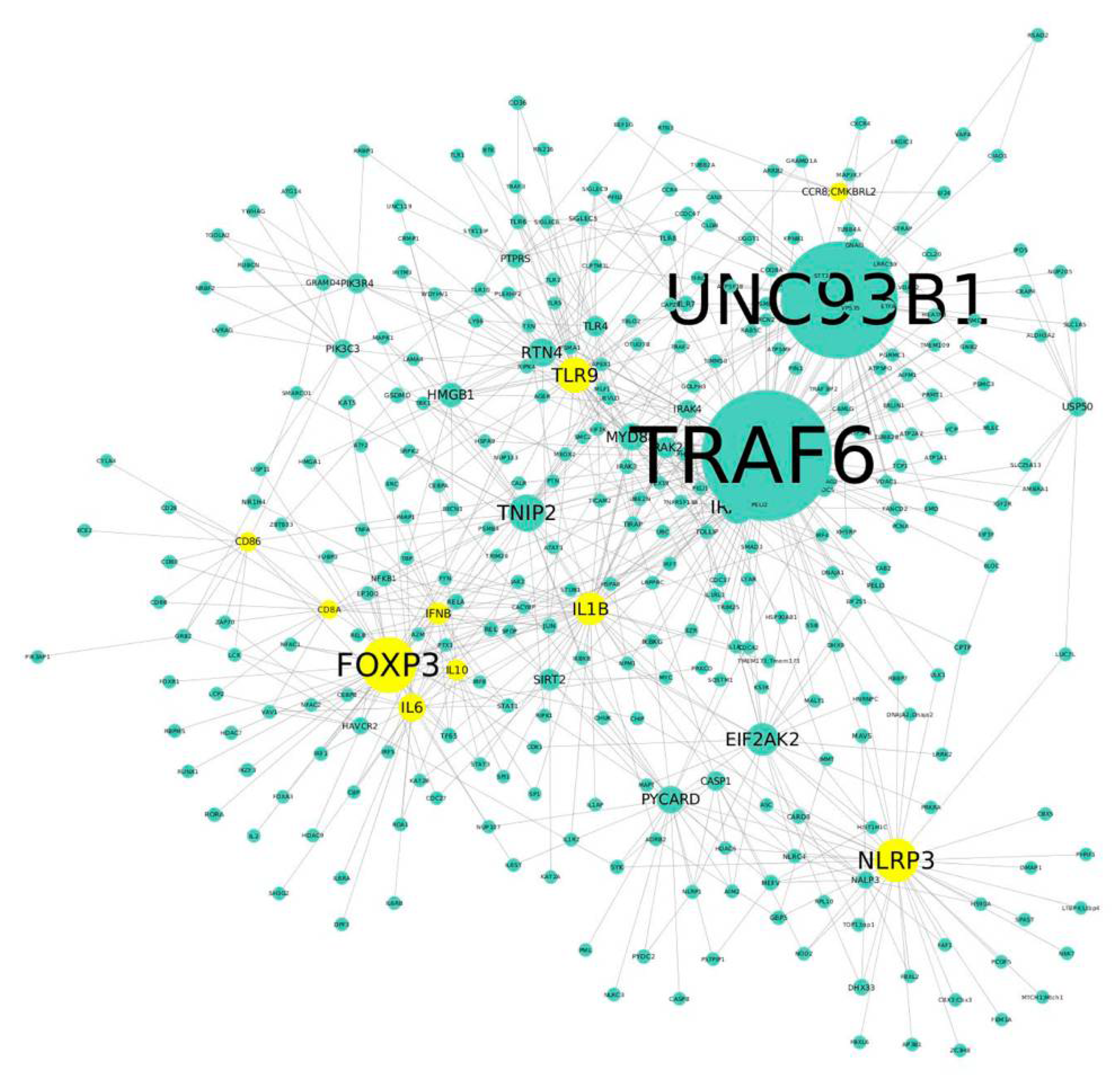
2.8. Protein–Protein Interaction (PPI) Network Analysis in NK1.1+, CD4+ T Cells
2.9. Statistical Analysis
3. Results
3.1. Liposomes Bearing Promazine-Induced NPAs Induce the Production of IgG Anti-NPA Antibodies in C57BL/6 Mice
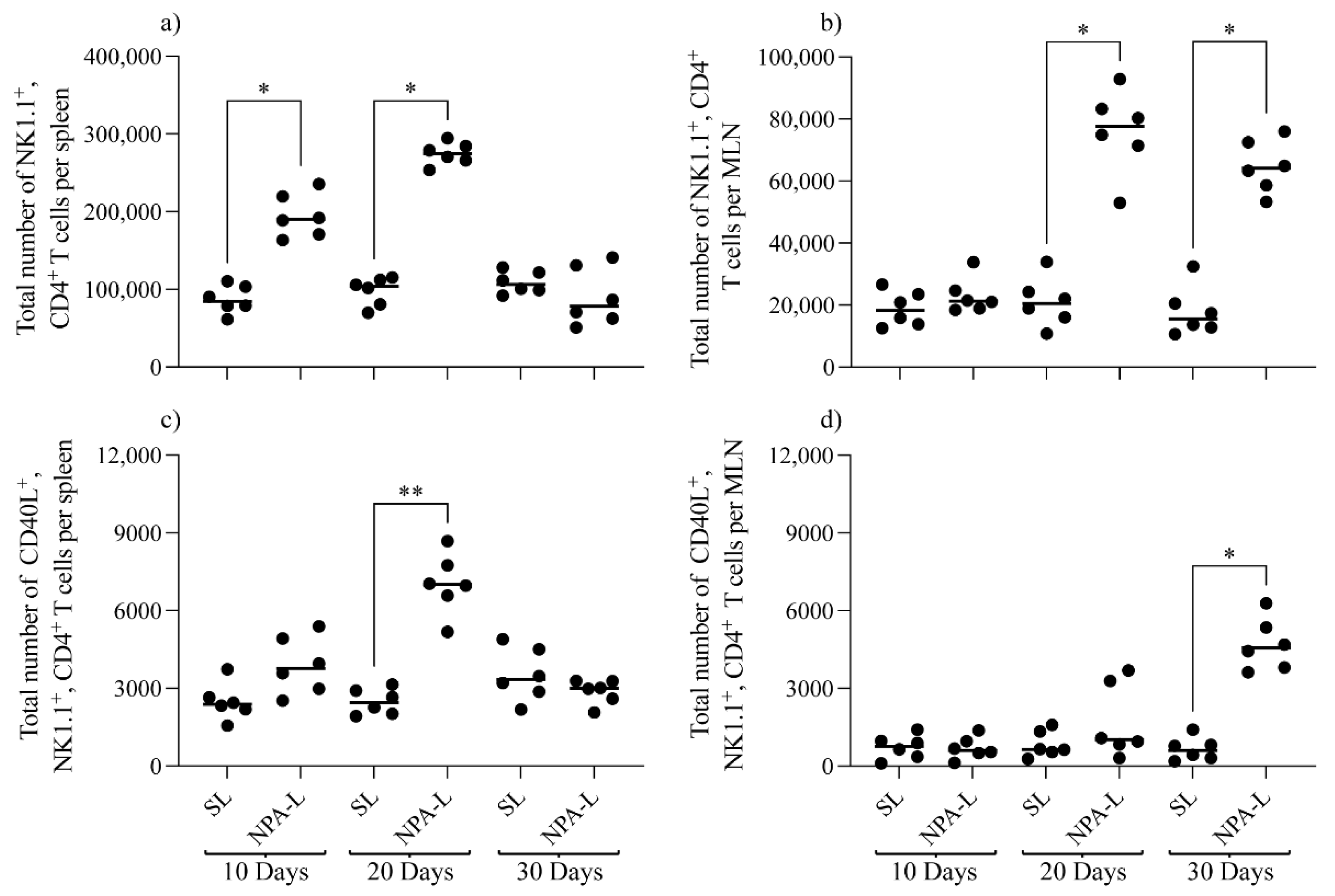
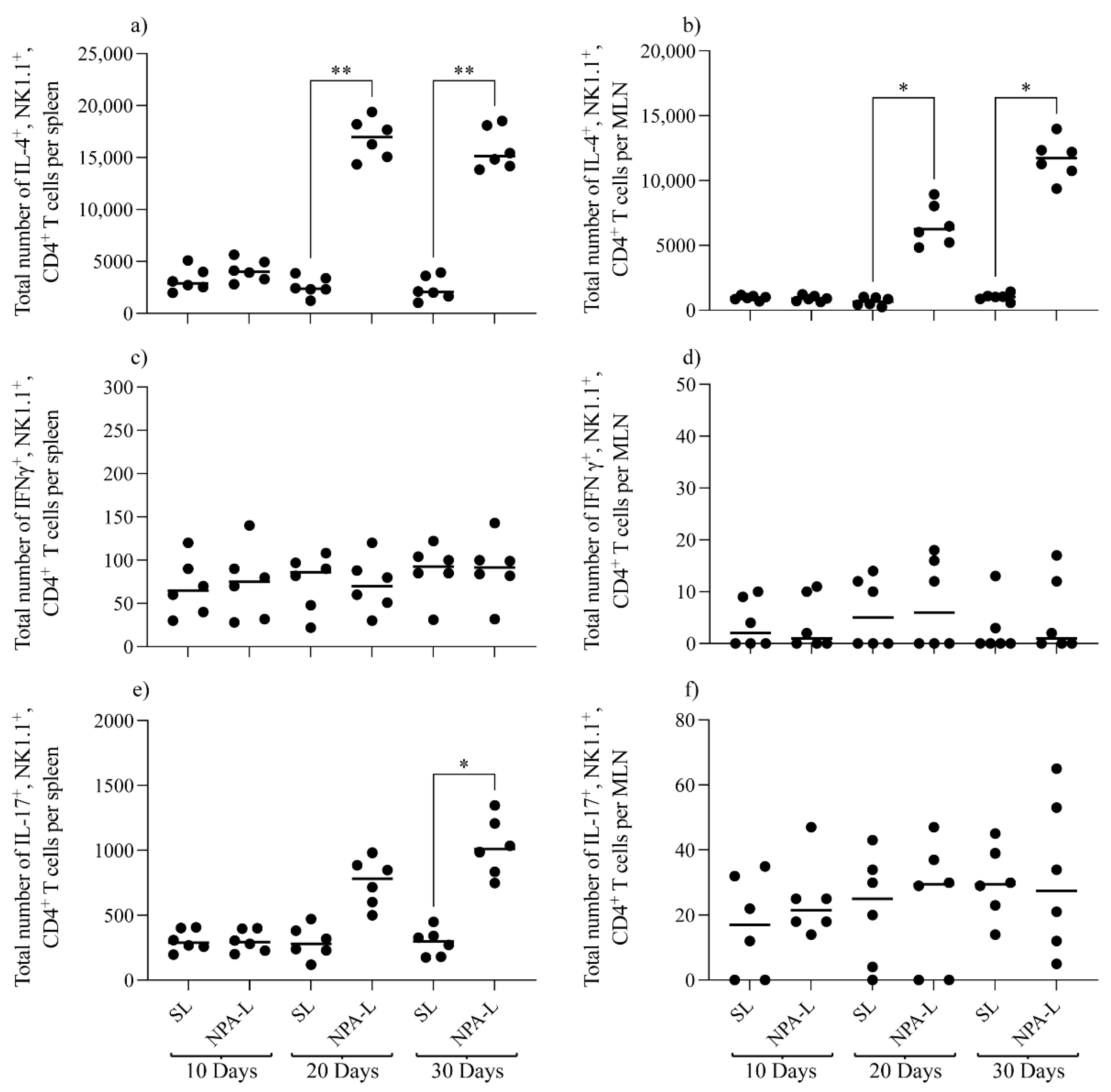
3.2. NK1.1+, CD4+ T Cells Are Activated in Mice That Produce Anti-NPA Antibodies, and These Cells Produce IL-4 in Response to NPAs
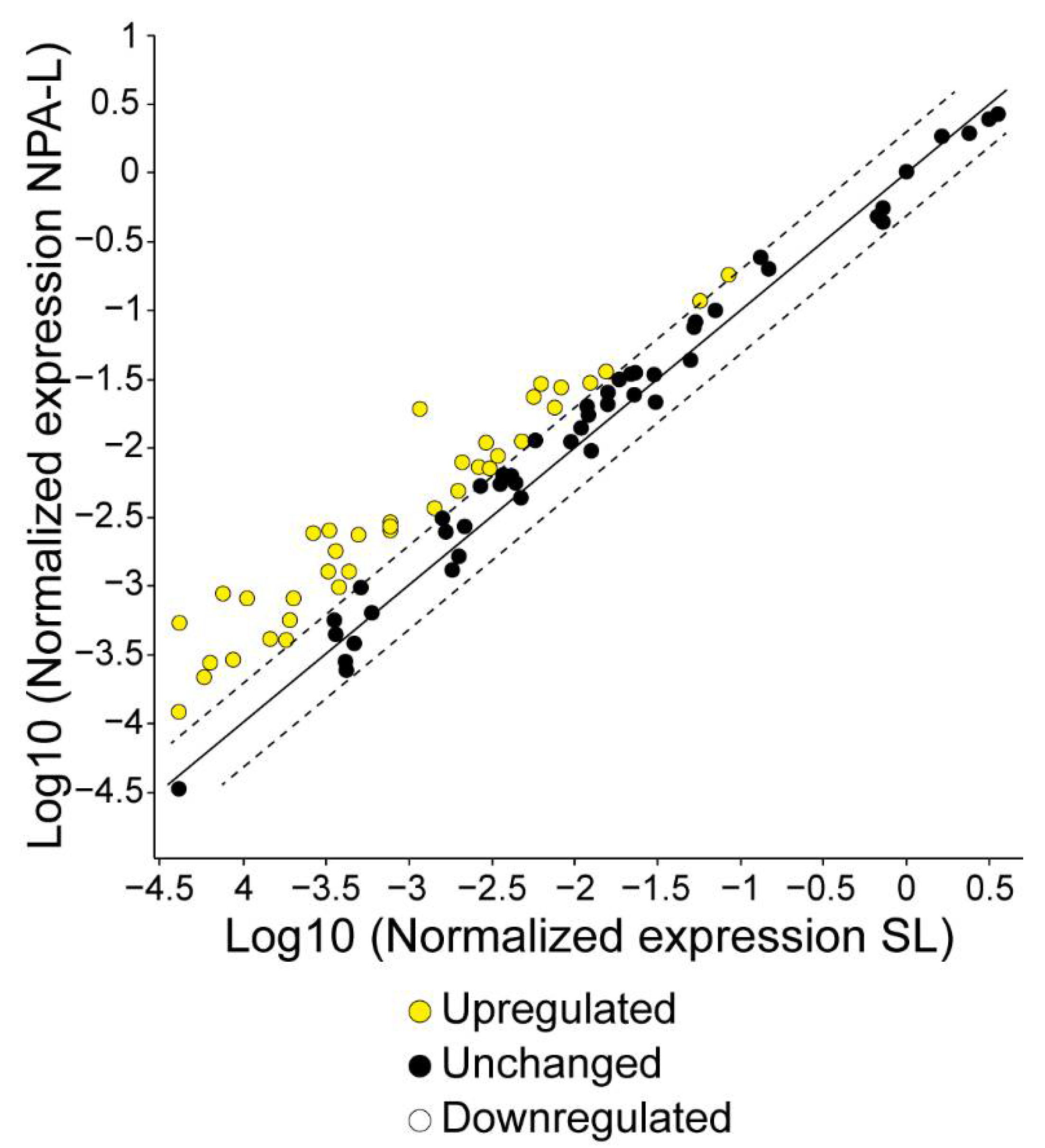
3.3. NK1.1+, CD4+ T Cells from Mice That Produce IgG Anti-NPA Antibodies Over-Express Genes Related to the Immune Response
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zylberberg, C.; Gaskill, K.; Pasley, S.; Matosevic, S. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 2017, 24, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Cheng, C.A.; Chen, W.; Li, Z.; Zink, J.I.; Lin, Y.Y. Magnetic Heating Stimulated Cargo Release with Dose Control using Multifunctional MR and Thermosensitive Liposome. Nanotheranostics 2019, 3, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Lee, C.; Singh, S.; Agrawal, N. Versatile Encapsulation and Synthesis of Potent Liposomes by Thermal Equilibration. Membranes 2022, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Nisini, R.; Poerio, N.; Mariotti, S.; De Santis, F.; Fraziano, M. The Multirole of Liposomes in Therapy and Prevention of Infectious Diseases. Front. Immunol. 2018, 9, 155. [Google Scholar] [CrossRef]
- Poerio, N.; Bugli, F.; Taus, F.; Santucci, M.B.; Rodolfo, C.; Cecconi, F.; Torelli, R.; Varone, F.; Inchingolo, R.; Majo, F.; et al. Liposomes loaded with bioactive lipids enhance antibacterial innate immunity irrespective of drug resistance. Sci. Rep. 2017, 7, 45120. [Google Scholar] [CrossRef] [Green Version]
- Tandrup Schmidt, S.; Foged, C.; Korsholm, K.S.; Rades, T.; Christensen, D. Liposome-Based Adjuvants for Subunit Vaccines: Formulation Strategies for Subunit Antigens and Immunostimulators. Pharmaceutics 2016, 8, 7. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Baeza, I.; Leyva, E.; Campos, B.; Lara, M.; Ibanez, M.; Farfan, N.; Orozco, H.; Flores-Romo, L.; Hernandez-Pando, R.; Wong, C. Antibodies to non-bilayer phospholipid arrangements induce a murine autoimmune disease resembling human lupus. Eur. J. Immunol. 2004, 34, 576–586. [Google Scholar] [CrossRef]
- Wong-Baeza, C.; Resendiz-Mora, A.; Donis-Maturano, L.; Wong-Baeza, I.; Zarate-Neira, L.; Yam-Puc, J.C.; Calderon-Amador, J.; Medina, Y.; Wong, C.; Baeza, I.; et al. Anti-Lipid IgG Antibodies Are Produced via Germinal Centers in a Murine Model Resembling Human Lupus. Front Immunol. 2016, 7, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almlof, J.C.; Alexsson, A.; Imgenberg-Kreuz, J.; Sylwan, L.; Backlin, C.; Leonard, D.; Nordmark, G.; Tandre, K.; Eloranta, M.L.; Padyukov, L.; et al. Novel risk genes for systemic lupus erythematosus predicted by random forest classification. Sci. Rep. 2017, 7, 6236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawaf, M.; Dumortier, H.; Monneaux, F. Follicular Helper T Cells in Systemic Lupus Erythematosus: Why Should They Be Considered as Interesting Therapeutic Targets? J. Immunol. Res. 2016, 2016, 5767106. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, M.F.; Jordan, N.; D’Cruz, D.P. Systemic lupus erythematosus. Clin. Med. 2017, 17, 78–83. [Google Scholar] [CrossRef]
- Zharkova, O.; Celhar, T.; Cravens, P.D.; Satterthwaite, A.B.; Fairhurst, A.M.; Davis, L.S. Pathways leading to an immunological disease: Systemic lupus erythematosus. Rheumatology 2017, 56, i55–i66. [Google Scholar] [CrossRef] [Green Version]
- Perry, D.; Sang, A.; Yin, Y.; Zheng, Y.Y.; Morel, L. Murine models of systemic lupus erythematosus. J. Biomed. Biotechnol. 2011, 2011, 271694. [Google Scholar] [CrossRef] [Green Version]
- Wong-Baeza, C.; Hernandez-Pando, R.; Resendiz, A.; Tescucano, A.; Bustos, I.; Ibanez, M.; Wong, C.; Baeza, I. Molecular organization of the non-bilayer phospholipid arrangements that induce an autoimmune disease resembling human lupus in mice. Mol. Membr. Biol. 2012, 29, 52–67. [Google Scholar] [CrossRef]
- Wong-Baeza, C.; Tescucano, A.; Astudillo, H.; Resendiz, A.; Landa, C.; Espana, L.; Serafin-Lopez, J.; Estrada-Garcia, I.; Estrada-Parra, S.; Flores-Romo, L.; et al. Nonbilayer Phospholipid Arrangements Are Toll-Like Receptor-2/6 and TLR-4 Agonists and Trigger Inflammation in a Mouse Model Resembling Human Lupus. J. Immunol. Res. 2015, 2015, 369462. [Google Scholar] [CrossRef]
- Tonti, E.; Fedeli, M.; Napolitano, A.; Iannacone, M.; von Andrian, U.H.; Guidotti, L.G.; Abrignani, S.; Casorati, G.; Dellabona, P. Follicular helper NKT cells induce limited B cell responses and germinal center formation in the absence of CD4(+) T cell help. J. Immunol. 2012, 188, 3217–3222. [Google Scholar] [CrossRef] [Green Version]
- Clerici, L.; Casorati, G.; Dellabona, P. B Cell Help by CD1d-Rectricted NKT Cells. Antibodies 2015, 4, 279–294. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.; Dhodapkar, M.V. Natural Killer T Cells in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, J.T.; Huang, J.R.; Yu, A.L. Tailored design of NKT-stimulatory glycolipids for polarization of immune responses. J. Biomed. Sci. 2017, 24, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Zhang, H.; Caimol, M.; Benike, C.J.; Chakravarty, E.F.; Strober, S.; Engleman, E.G. Invariant natural killer T cells in lupus patients promote IgG and IgG autoantibody production. Eur. J. Immunol. 2015, 45, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Wikenheiser, D.J.; Brown, S.L.; Lee, J.; Stumhofer, J.S. NK1.1 Expression Defines a Population of CD4(+) Effector T Cells Displaying Th1 and Tfh Cell Properties That Support Early Antibody Production During Plasmodium yoelii Infection. Front. Immunol. 2018, 9, 2277. [Google Scholar] [CrossRef]
- Brennan, P.J.; Brigl, M.; Brenner, M.B. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013, 13, 101–117. [Google Scholar] [CrossRef]
- Vomhof-DeKrey, E.E.; Yates, J.; Leadbetter, E.A. Invariant NKT cells provide innate and adaptive help for B cells. Curr. Opin. Immunol. 2014, 28, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Bezbradica, J.S.; Stanic, A.K.; Joyce, S. Characterization and Functional Analysis of Mouse Semi-invariant Natural T Cells. Curr. Protoc. Immunol. 2017, 117, 14-13. [Google Scholar] [CrossRef]
- National Research Council. Guide for the Care and Use of Laboratory Animals; National Academies Press: Washington, WA, USA, 2011; Volume XXV, p. 220.
- Loizou, S.; McCrea, J.D.; Rudge, A.C.; Reynolds, R.; Boyle, C.C.; Harris, E.N. Measurement of anti-cardiolipin antibodies by an enzyme-linked immunosorbent assay (ELISA): Standardization and quantitation of results. Clin. Exp. Immunol. 1985, 62, 738–745. [Google Scholar]
- Szklarczyk, R.; Heringa, J. AuberGene—A sensitive genome alignment tool. Bioinformatics 2006, 22, 1431–1436. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zhang, J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006, 2, e88. [Google Scholar] [CrossRef] [Green Version]
- Vedove, C.D.; Del Giglio, M.; Schena, D.; Girolomoni, G. Drug-induced lupus erythematosus. Arch. Dermatol. Res. 2009, 301, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.L. The importance of B cell CD1d expression for humoral immunity. Expert Rev. Vaccines 2014, 13, 1275–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellers, R.S.; Clifford, C.B.; Treuting, P.M.; Brayton, C. Immunological variation between inbred laboratory mouse strains: Points to consider in phenotyping genetically immunomodified mice. Vet. Pathol. 2012, 49, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Hannestad, K.; Scott, H. The MHC haplotype H2b converts two pure nonlupus mouse strains to producers of antinuclear antibodies. J. Immunol. 2009, 183, 3542–3550. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, S.C.; Bosma, A.; Bruckner-Tuderman, L.; Vukmanovic-Stejic, M.; Jury, E.C.; Isenberg, D.A.; Mauri, C. Invariant natural killer T cells are enriched at the site of cutaneous inflammation in lupus erythematosus. J. Dermatol. Sci. 2013, 71, 22–28. [Google Scholar] [CrossRef]
- Yang, W.C.; Hwang, Y.S.; Chen, Y.Y.; Liu, C.L.; Shen, C.N.; Hong, W.H.; Lo, S.M.; Shen, C.R. Interleukin-4 Supports the Suppressive Immune Responses Elicited by Regulatory T Cells. Front. Immunol. 2017, 8, 1508. [Google Scholar] [CrossRef] [Green Version]
- Paul, W.E. History of interleukin-4. Cytokine 2015, 75, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Perry, D.; Peck, A.B.; Carcamo, W.C.; Morel, L.; Nguyen, C.Q. The current concept of T (h) 17 cells and their expanding role in systemic lupus erythematosus. Arthritis 2011, 2011, 810649. [Google Scholar] [CrossRef] [Green Version]
- Thedrez, A.; de Lalla, C.; Allain, S.; Zaccagnino, L.; Sidobre, S.; Garavaglia, C.; Borsellino, G.; Dellabona, P.; Bonneville, M.; Scotet, E.; et al. CD4 engagement by CD1d potentiates activation of CD4+ invariant NKT cells. Blood 2007, 110, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Resendiz-Mora, A.; Wong-Baeza, C.; Nevarez-Lechuga, I.; Landa-Saldivar, C.; Molina-Gomez, E.; Hernandez-Pando, R.; Wong-Baeza, I.; Escobar-Gutierrez, A.; Baeza, I. Interleukin 4 deficiency limits the development of a lupus-like disease in mice triggered by phospholipids in a non-bilayer arrangement. Scand. J. Immunol. 2021, 93, e13002. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, C.; Zhang, Z.; Liu, C.C.; Johnson, M.E.; Espinoza, C.A.; Edsall, L.E.; Ren, B.; Zhou, X.J.; Grant, S.F.; et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015, 43, 1268–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonsson, H.; Peng, S.L. Forkhead transcription factors in immunology. Cell Mol. Life Sci. 2005, 62, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, P.; Farkas, K.; Kis, J.; Richman, G.; Zhang, Z.; Liew, C.W.; Borowiec, M.; Niewczas, M.A.; Jalahej, H.; Orban, T. Characterization of human invariant natural killer T cells expressing FoxP3. Int. Immunol. 2011, 23, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Chow, M.T.; Duret, H.; Andrews, D.M.; Faveeuw, C.; Moller, A.; Smyth, M.J.; Paget, C. Type I NKT-cell-mediated TNF-alpha is a positive regulator of NLRP3 inflammasome priming. Eur. J. Immunol. 2014, 44, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.A. Developing Costimulatory Molecules for Immunotherapy of Diseases; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Reimer, M.K.; Brange, C.; Rosendahl, A. CCR8 signaling influences Toll-like receptor 4 responses in human macrophages in inflammatory diseases. Clin. Vaccine Immunol. 2011, 18, 2050–2059. [Google Scholar] [CrossRef] [Green Version]
- Oshio, T.; Kawashima, R.; Kawamura, Y.I.; Hagiwara, T.; Mizutani, N.; Okada, T.; Otsubo, T.; Inagaki-Ohara, K.; Matsukawa, A.; Haga, T.; et al. Chemokine receptor CCR8 is required for lipopolysaccharide-triggered cytokine production in mouse peritoneal macrophages. PLoS ONE 2014, 9, e94445. [Google Scholar] [CrossRef]
- Zingoni, A.; Soto, H.; Hedrick, J.A.; Stoppacciaro, A.; Storlazzi, C.T.; Sinigaglia, F.; D’Ambrosio, D.; O’Garra, A.; Robinson, D.; Rocchi, M.; et al. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J. Immunol. 1998, 161, 547–551. [Google Scholar]
- Kulkarni, R.R.; Villanueva, A.I.; Elawadli, I.; Jayanth, P.; Read, L.R.; Haeryfar, S.M.; Sharif, S. Costimulatory activation of murine invariant natural killer T cells by toll-like receptor agonists. Cell Immunol. 2012, 277, 33–43. [Google Scholar] [CrossRef]
- Villanueva, A.I.; Haeryfar, S.M.; Mallard, B.A.; Kulkarni, R.R.; Sharif, S. Functions of invariant NK T cells are modulated by TLR ligands and IFN-alpha. Innate. Immun. 2015, 21, 275–288. [Google Scholar] [CrossRef]
- Catalina, M.D.; Bachali, P.; Geraci, N.S.; Grammer, A.C.; Lipsky, P.E. Gene expression analysis delineates the potential roles of multiple interferons in systemic lupus erythematosus. Commun. Biol. 2019, 2, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.J.; Liang, J.Q.; Diao, L.H.; Altman, A.; Li, Y. TNFR-associated factor 6 regulates TCR signaling via interaction with and modification of LAT adapter. J. Immunol. 2013, 190, 4027–4036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, T.; Nagashima, H.; Ishii, N. TNF Receptor-Associated Factor (TRAF) Signaling Network in CD4(+) T-Lymphocytes. Tohoku J. Exp. Med. 2015, 236, 139–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dainichi, T.; Matsumoto, R.; Mostafa, A.; Kabashima, K. Immune Control by TRAF6-Mediated Pathways of Epithelial Cells in the EIME (Epithelial Immune Microenvironment). Front. Immunol. 2019, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.W.; Shibata, T.; Hwang, M.; Kwon, E.H.; Jang, M.S.; Fukui, R.; Kanno, A.; Jung, D.J.; Jang, M.H.; Miyake, K.; et al. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc. Natl. Acad. Sci. USA 2014, 111, 7072–7077. [Google Scholar] [CrossRef] [Green Version]
- Amadori, M.; Amadori, M. The Innate Immune Response to Non-Infectious Stressors: Human and Animal Models; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Panchanathan, R.; Liu, H.; Choubey, D. Expression of murine Unc93b1 is up-regulated by interferon and estrogen signaling: Implications for sex bias in the development of autoimmunity. Int. Immunol. 2013, 25, 521–529. [Google Scholar] [CrossRef] [Green Version]



| Genes | Fold Regulation | Genes | Fold Regulation | Genes | Fold Regulation | Genes | Fold Regulation |
|---|---|---|---|---|---|---|---|
| Il1b | 16.62 | Cxcl10 | 4.16 | Il18 | 3.34 | Tnf | 2.58 |
| Ifnb1 | 13.04 | Mpo | 4.08 | Tlr2 | 3.29 | Ifnar1 | 2.56 |
| Foxp3 | 11.82 | Cd40 | 3.88 | Il2 | 2.97 | Ly96 | 2.47 |
| Il10 | 9.08 | Il1a | 3.86 | Il23a | 2.97 | Cd4 | 2.38 |
| Tlr9 | 7.76 | Tlr4 | 3.82 | Myd88 | 2.94 | Icam1 | 2.33 |
| Nlrp3 | 7.71 | H2-Q10 | 3.74 | Stat6 | 2.81 | Casp1 | 2.32 |
| Cd8a | 4.92 | Slc11a1 | 3.72 | Tlr8 | 2.80 | Mapk8 | 2.30 |
| Ccr8 | 4.72 | Tlr7 | 3.72 | Irf7 | 2.61 | Csf2 | 2.24 |
| Cd86 | 4.68 | Cd80 | 3.51 | Nod2 | 2.58 | Nfkbia | 2.15 |
| Il6 | 4.38 | Tlr5 | 3.40 | Mx1 | 2.58 | Ifng | 2.04 |
| Protein Name | Betweenness Value | Connectivity Degree | Protein Name | Betweenness Value | Connectivity Degree | Protein Name | Betweenness Value | Connectivity Degree |
|---|---|---|---|---|---|---|---|---|
| TRAF6 | 0.45664152 | 393 | PYCARD | 0.06525022 | 49 | CASP1 | 0.01519920 | 22 |
| UNC93B1 | 0.37356461 | 348 | MYD88 | 0.03781771 | 48 | USP50 | 0.01586547 | 21 |
| FOXP3 | 0.13193952 | 142 | IL6 | 0.03542776 | 47 | PIK3C3 | 0.02003203 | 20 |
| NLRP3 | 0.0972960 | 101 | HMGB1 | 0.03793161 | 39 | CD86 | 0.01148624 | 20 |
| TNIP2 | 0.08172416 | 81 | SIRT2 | 0.03014661 | 29 | IRAK4 | 0.00940748 | 20 |
| TLR9 | 0.14978064 | 74 | IFNB | 0.01508672 | 29 | PTPRS | 0.01635204 | 19 |
| IRAK1 | 0.09756521 | 69 | IRAK2 | 0.02226735 | 28 | CCR8 | 0.01400334 | 19 |
| EIF2AK2 | 0.06857739 | 66 | TLR4 | 0.01940404 | 26 | CD8A | 0.01217994 | 18 |
| IL1B | 0.07191164 | 65 | IL10 | 0.01699116 | 25 | NALP3 | 0.00752642 | 15 |
| RTN4 | 0.06375998 | 52 | PIK3R4 | 0.02006215 | 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landa-Saldívar, C.; Reséndiz-Mora, A.; Sánchez-Barbosa, S.; Sotelo-Rodríguez, A.; Barrera-Aveleida, G.; Nevárez-Lechuga, I.; Galarce-Sosa, I.; Taniguchi-Ponciano, K.; Cruz-Guzmán, O.d.R.; Wong-Baeza, I.; et al. Liposomes Bearing Non-Bilayer Phospholipid Arrangements Induce Specific IgG Anti-Lipid Antibodies by Activating NK1.1+, CD4+ T Cells in Mice. Membranes 2022, 12, 643. https://doi.org/10.3390/membranes12070643
Landa-Saldívar C, Reséndiz-Mora A, Sánchez-Barbosa S, Sotelo-Rodríguez A, Barrera-Aveleida G, Nevárez-Lechuga I, Galarce-Sosa I, Taniguchi-Ponciano K, Cruz-Guzmán OdR, Wong-Baeza I, et al. Liposomes Bearing Non-Bilayer Phospholipid Arrangements Induce Specific IgG Anti-Lipid Antibodies by Activating NK1.1+, CD4+ T Cells in Mice. Membranes. 2022; 12(7):643. https://doi.org/10.3390/membranes12070643
Chicago/Turabian StyleLanda-Saldívar, Carla, Albany Reséndiz-Mora, Sandra Sánchez-Barbosa, Anahi Sotelo-Rodríguez, Giovanna Barrera-Aveleida, Irene Nevárez-Lechuga, Iván Galarce-Sosa, Keiko Taniguchi-Ponciano, Oriana del Rocío Cruz-Guzmán, Isabel Wong-Baeza, and et al. 2022. "Liposomes Bearing Non-Bilayer Phospholipid Arrangements Induce Specific IgG Anti-Lipid Antibodies by Activating NK1.1+, CD4+ T Cells in Mice" Membranes 12, no. 7: 643. https://doi.org/10.3390/membranes12070643
APA StyleLanda-Saldívar, C., Reséndiz-Mora, A., Sánchez-Barbosa, S., Sotelo-Rodríguez, A., Barrera-Aveleida, G., Nevárez-Lechuga, I., Galarce-Sosa, I., Taniguchi-Ponciano, K., Cruz-Guzmán, O. d. R., Wong-Baeza, I., Escobar-Gutiérrez, A., Baeza, I., & Wong-Baeza, C. (2022). Liposomes Bearing Non-Bilayer Phospholipid Arrangements Induce Specific IgG Anti-Lipid Antibodies by Activating NK1.1+, CD4+ T Cells in Mice. Membranes, 12(7), 643. https://doi.org/10.3390/membranes12070643






