Influence of Anoctamin-4 and -9 on ADAM10 and ADAM17 Sheddase Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Antibodies
2.2. Cell Culture
2.3. Expression Vectors and Transfection
2.4. AP-Substrate Shedding Assay
2.5. AREG ELISA
2.6. Proliferation Assay
2.7. Automated Western
2.8. Annexin V Staining
2.9. Image Analysis and Statistics
2.10. Statistical Analysis
3. Results
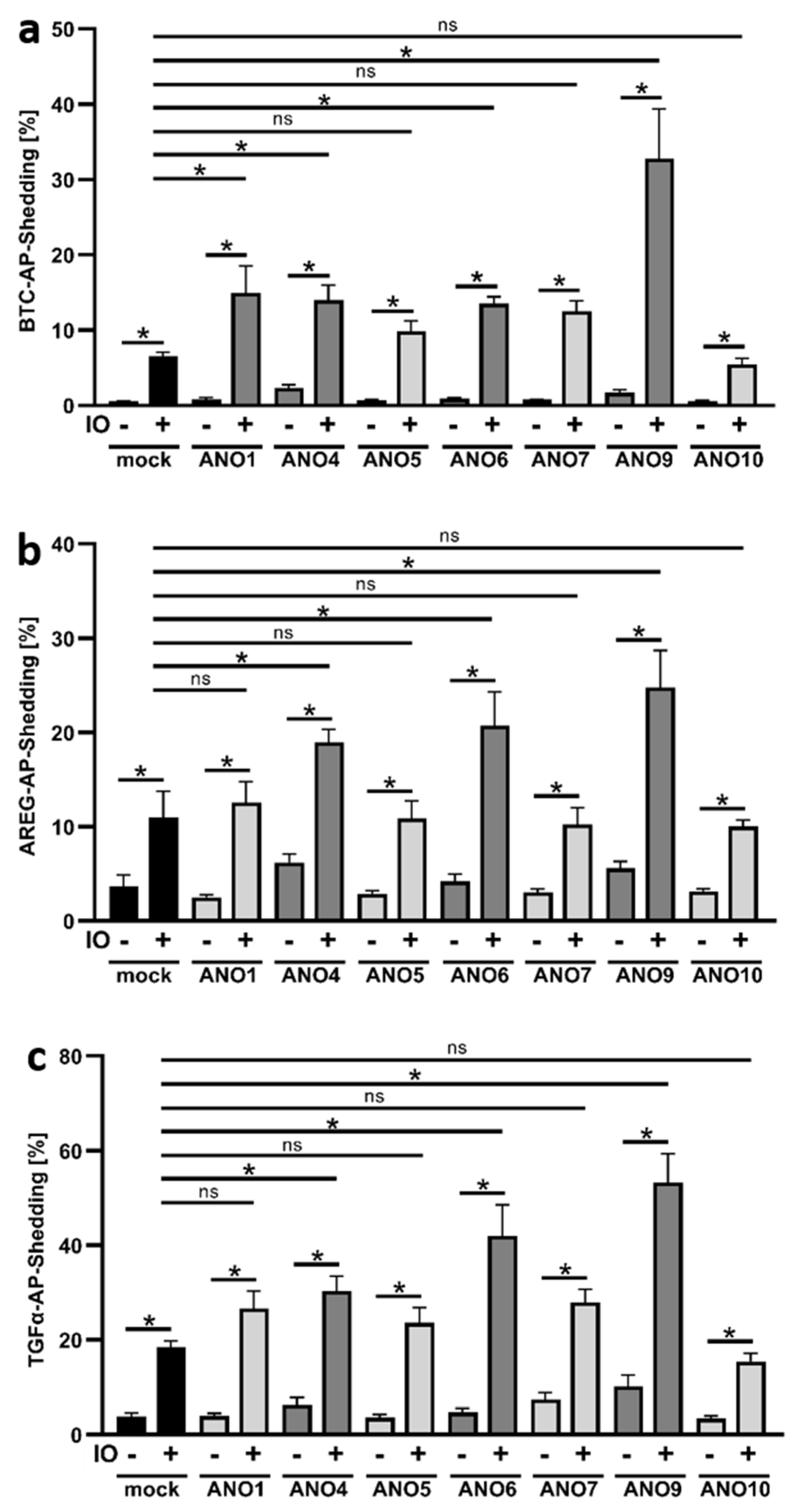
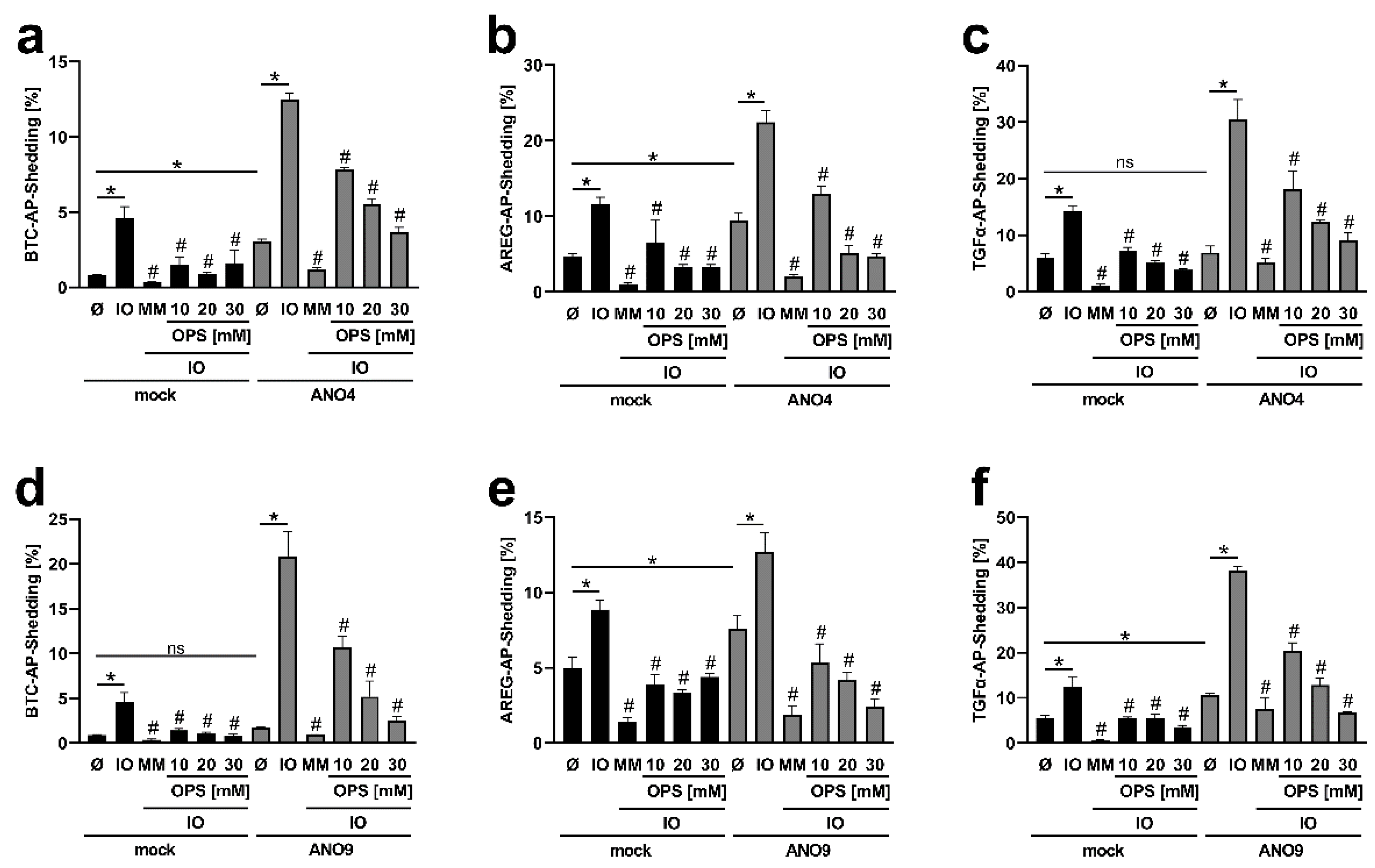
3.1. Ionomycin Stimulation Increases Shedding Activity in Cells Overexpressing ANO4, ANO6 and ANO9
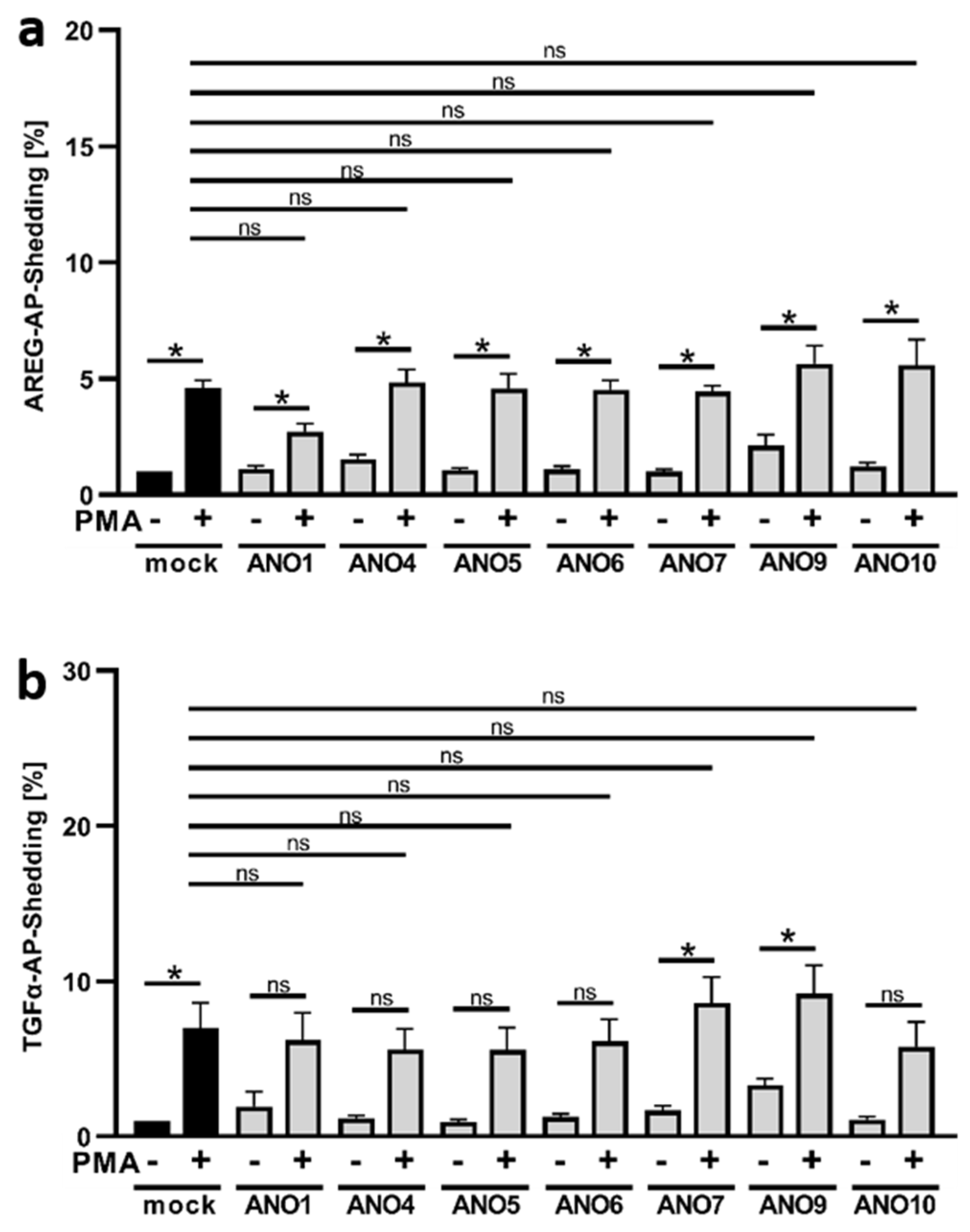
3.2. PMA Stimulation of ADAM17 Is Independent of Anoctamin Expression
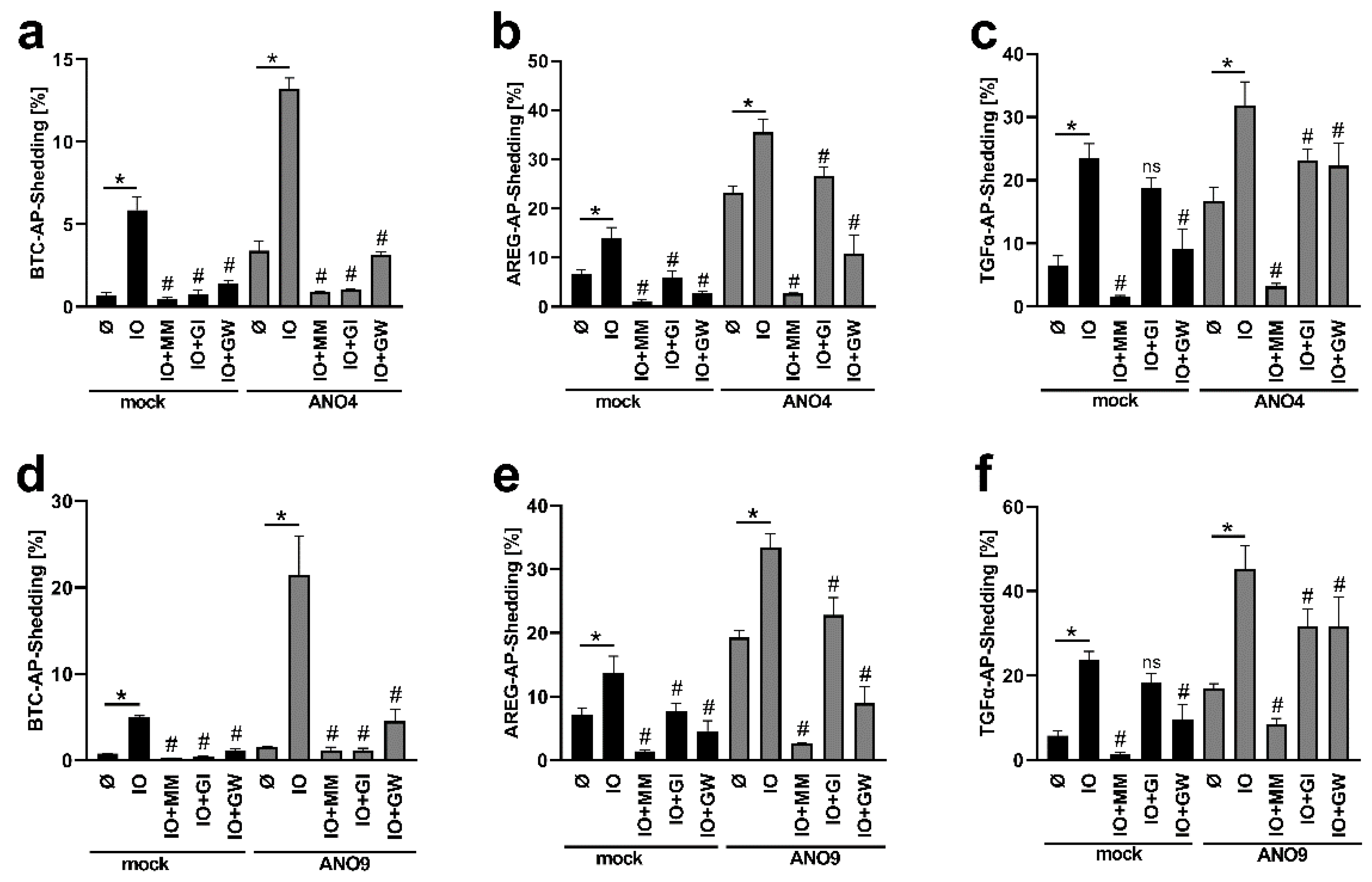
3.3. Enhanced TGFα Cleavage in ANO-Overexpressing Cells Involves Other Sheddases Than ADAM17
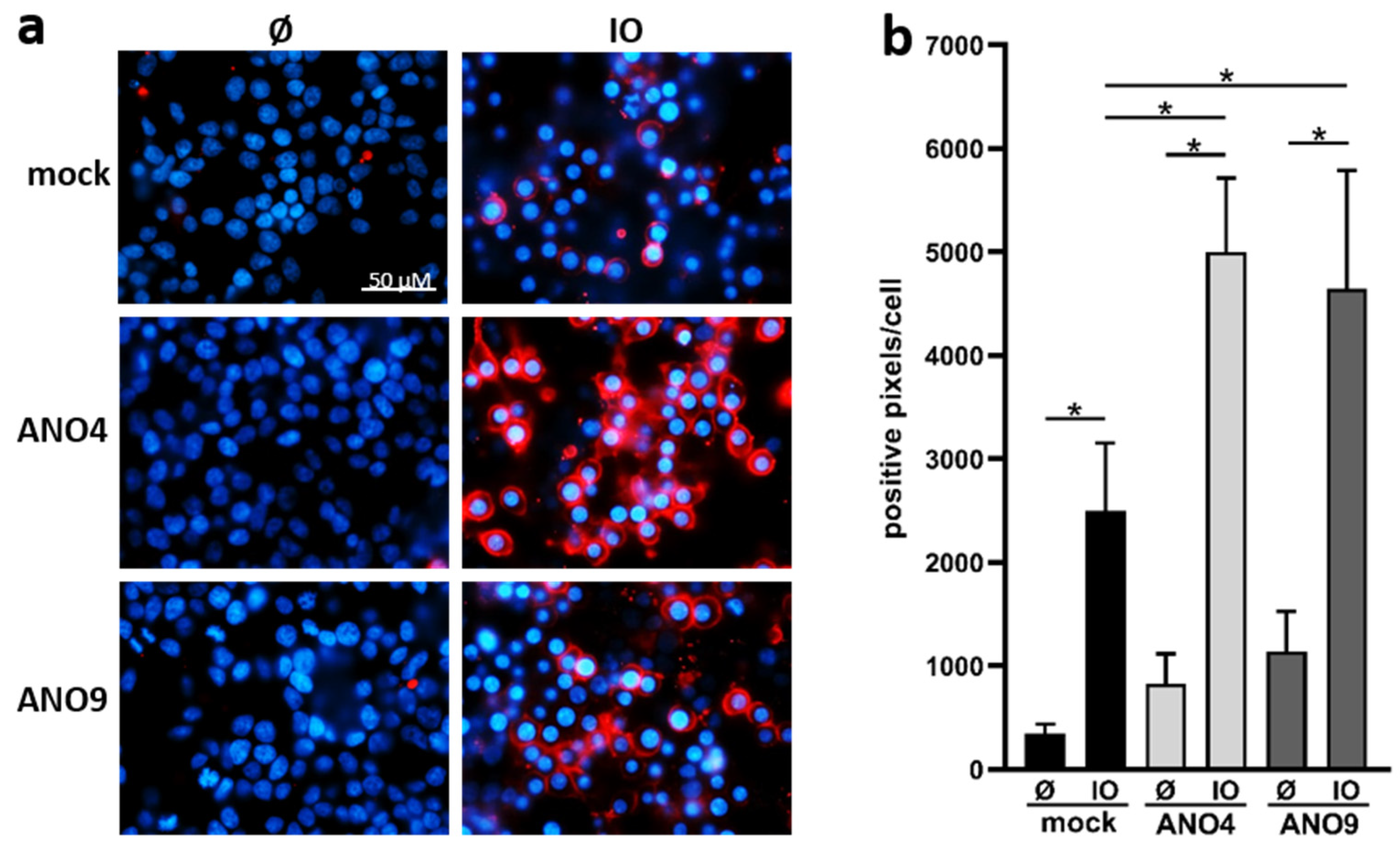
3.4. Overexpression of ANO4 or ANO9 Leads to Enhanced PS Exposure
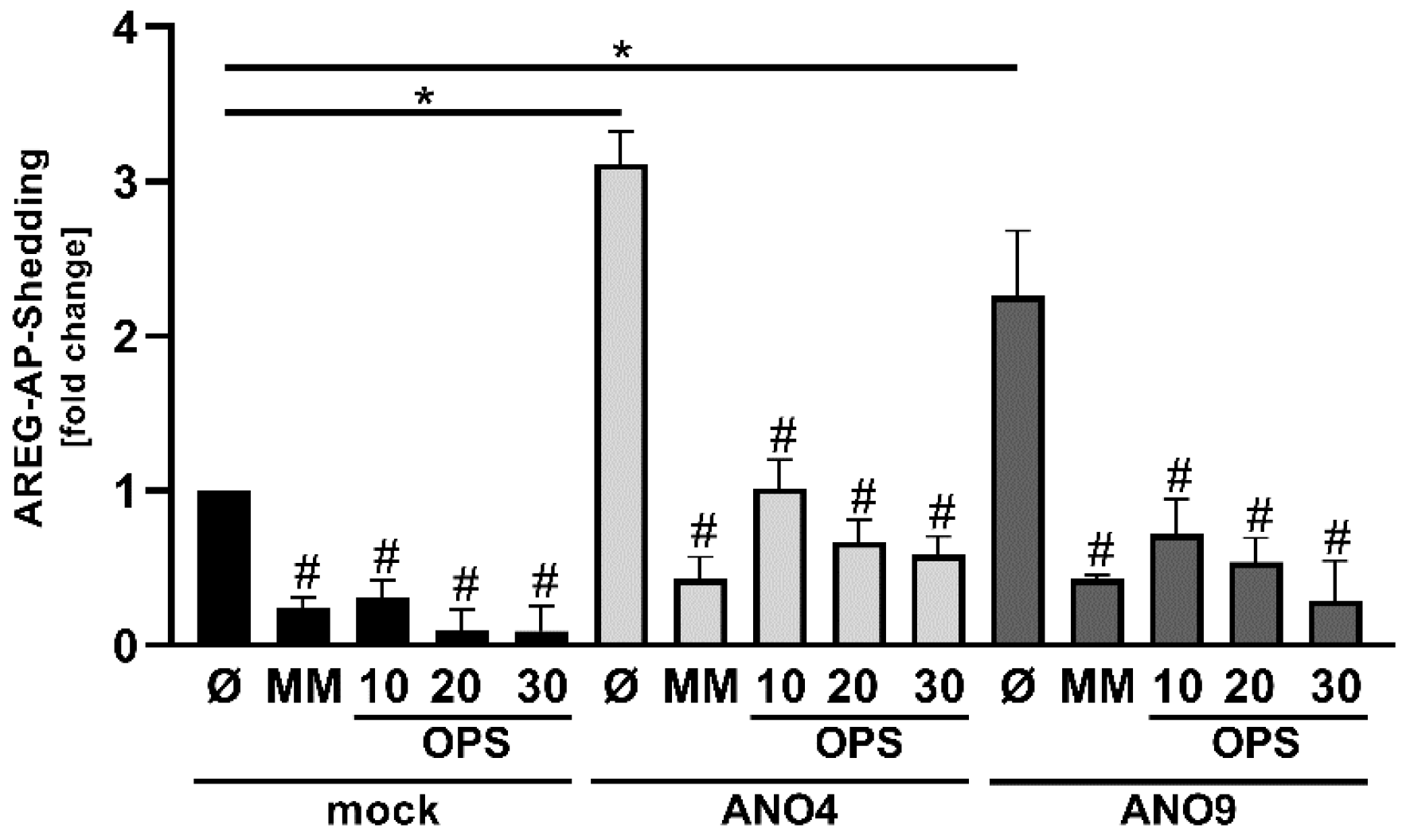
3.5. Increased Substrate Cleavage Depends on PS-Interaction
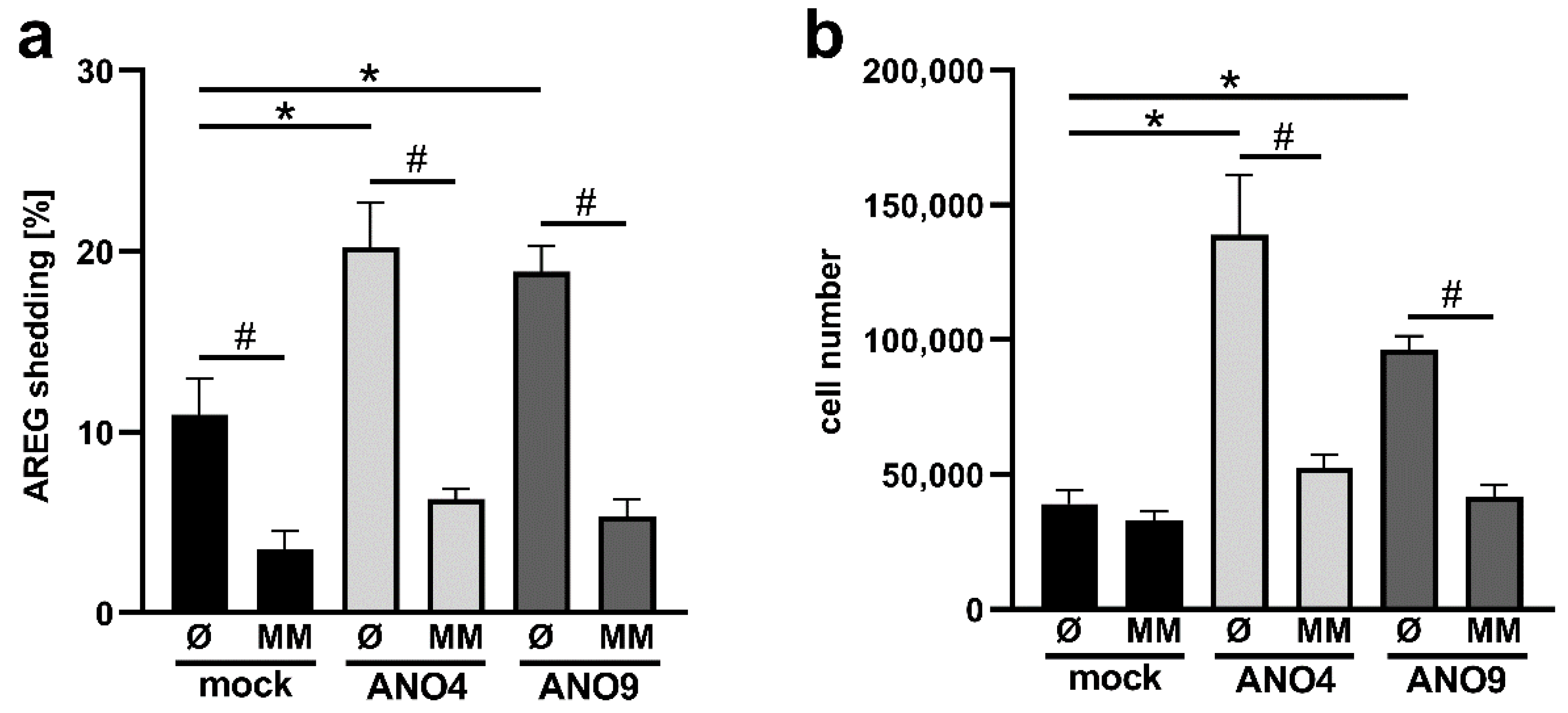
3.6. Increased Expression of ANO4 and ANO9 Stimulates Cell Proliferation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Moss, M.L.; Jin, S.L.; Milla, M.E.; Bickett, D.M.; Burkhart, W.; Carter, H.L.; Chen, W.J.; Clay, W.C.; Didsbury, J.R.; Hassler, D.; et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 1997, 385, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Peschon, J.J.; Slack, J.L.; Reddy, P.; Stocking, K.L.; Sunnarborg, S.W.; Lee, D.C.; Russell, W.E.; Castner, B.J.; Johnson, R.S.; Fitzner, J.N.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef]
- Sahin, U.; Blobel, C.P. Ectodomain shedding of the EGF-receptor ligand epigen is mediated by ADAM17. FEBS Lett. 2007, 581, 41–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, D.; de Strooper, B.; Serneels, L.; Craessaerts, K.; Herreman, A.; Annaert, W.; Umans, L.; Lübke, T.; Illert, A.L.; von Figura, K.; et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet. 2002, 11, 2615–2624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maretzky, T.; Reiss, K.; Ludwig, A.; Buchholz, J.; Scholz, F.; Proksch, E.; de Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc. Natl. Acad. Sci. USA 2005, 102, 9182–9187. [Google Scholar] [CrossRef] [Green Version]
- Schulz, B.; Pruessmeyer, J.; Maretzky, T.; Ludwig, A.; Blobel, C.P.; Saftig, P.; Reiss, K. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circ. Res. 2008, 102, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Reiss, K.; Maretzky, T.; Ludwig, A.; Tousseyn, T.; De Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 cleavage of N-cadherin and regulation of cell–cell adhesion and b-catenin nuclear signalling. EMBO J. 2005, 24, 742–752. [Google Scholar] [CrossRef]
- Sahin, U.; Weskamp, G.; Kelly, K.; Zhou, H.-M.; Higashiyama, S.; Peschon, J.; Hartmann, D.; Saftig, P.; Blobel, C.P. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 2004, 164, 769–779. [Google Scholar] [CrossRef] [Green Version]
- Hayashida, K.; Bartlett, A.H.; Chen, Y.; Park, P.W. Molecular and cellular mechanisms of ectodomain shedding. Anat. Rec. 2010, 293, 925–937. [Google Scholar] [CrossRef] [Green Version]
- Pupovac, A.; Sluyter, R. Roles of extracellular nucleotides and P2 receptors in ectodomain shedding. Cell. Mol. Life Sci. 2016, 73, 4159–4173. [Google Scholar] [CrossRef] [Green Version]
- Reiss, K.; Bhakdi, S. Pore-forming bacterial toxins and antimicrobial peptides as modulators of ADAM function. Med. Microbiol. Immunol. 2012, 201, 419–426. [Google Scholar] [CrossRef]
- Sommer, A.; Fries, A.; Cornelsen, I.; Speck, N.; Koch-Nolte, F.; Gimpl, G.; Andrä, J.; Bhakdi, S.; Reiss, K. Melittin modulates keratinocyte function through P2 receptor-dependent ADAM activation. J. Biol. Chem. 2012, 287, 23678–23689. [Google Scholar] [CrossRef] [Green Version]
- Horiuchi, K.; Le Gall, S.; Schulte, M.; Yamaguchi, T.; Reiss, K.; Murphy, G.; Toyama, Y.; Hartmann, D.; Saftig, P.; Blobel, C.P. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol. Biol. Cell 2007, 18, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Sommer, A.; Kordowski, F.; Büch, J.; Maretzky, T.; Evers, A.; Andrä, J.J.J.; Düsterhöft, S.; Michalek, M.; Lorenzen, I.; Somasundaram, P.; et al. Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat. Commun. 2016, 7, 11523. [Google Scholar] [CrossRef] [Green Version]
- Bleibaum, F.; Sommer, A.; Veit, M.; Rabe, B.; Andrä, J.; Kunzelmann, K.; Nehls, C.; Correa, W.; Gutsmann, T.; Grötzinger, J.; et al. ADAM10 sheddase activation is controlled by cell membrane asymmetry. J. Mol. Cell Biol. 2019, 11, 979–993. [Google Scholar] [CrossRef] [Green Version]
- Bevers, E.M.; Williamson, P.L. Getting to the outer leaflet: Physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 2016, 96, 605–645. [Google Scholar] [CrossRef]
- Kodigepalli, K.M.; Bowers, K.; Sharp, A.; Nanjundan, M. Roles and regulation of phospholipid scramblases. FEBS Lett. 2015, 589, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Falzone, M.E.; Malvezzi, M.; Lee, B.C.; Accardi, A. Known structures and unknown mechanisms of TMEM16 scramblases and channels. J. Gen. Physiol. 2018, 150, 933–947. [Google Scholar] [CrossRef] [Green Version]
- Kalienkova, V.; Clerico Mosina, V.; Paulino, C. The Groovy TMEM16 Family: Molecular Mechanisms of Lipid Scrambling and Ion Conduction. J. Mol. Biol. 2021, 433, 166941. [Google Scholar] [CrossRef]
- Suzuki, J.; Fujii, T.; Imao, T.; Ishihara, K.; Kuba, H.; Nagata, S. Calcium-dependent phospholipid scramblase activity of TMEM 16 protein family members. J. Biol. Chem. 2013, 288, 13305–13316. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, J.; Imanishi, E.; Nagata, S. Exposure of phosphatidylserine by Xkrelated protein family members during apoptosis. J. Biol. Chem. 2014, 289, 30257–30267. [Google Scholar] [CrossRef] [Green Version]
- Sakuragi, T.; Kosako, H.; Nagata, S. Phosphorylation-mediated activation of mouse Xkr8 scramblase for phosphatidylserine exposure. Proc. Natl. Acad. Sci. USA 2019, 116, 2907–2912. [Google Scholar] [CrossRef] [Green Version]
- Veit, M.; Koyro, K.I.; Ahrens, B.; Bleibaum, F.; Munz, M.; Rövekamp, H.; Andrä, J.; Schreiber, R.; Kunzelmann, K.; Sommer, A.; et al. Anoctamin-6 regulates ADAM sheddase function. Biochim. Biophys. Acta-Mol. Cell Res. 2018, 1865, 1598–1610. [Google Scholar] [CrossRef]
- Ousingsawat, J.; Wanitchakool, P.; Kmit, A.; Romao, A.M.; Jantarajit, W.; Schreiber, R.; Kunzelmann, K. Anoctamin 6 mediates effects essential for innate immunity downstream of P2X7 receptors in macrophages. Nat. Commun. 2015, 6, 6245. [Google Scholar] [CrossRef]
- Seidel, J.; Leitzke, S.; Ahrens, B.; Sperrhacke, M.; Bhakdi, S.; Reiss, K. Role of adam10 and adam17 in regulating cd137 function. Int. J. Mol. Sci. 2021, 22, 2730. [Google Scholar] [CrossRef]
- Kunzelmann, K.; Ousingsawat, J.; Benedetto, R.; Cabrita, I.; Schreiber, R. Contribution of anoctamins to cell survival and cell death. Cancers 2019, 11, 382. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.W.; Takatsu, H. Phosphatidylserine exposure in living cells. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G. The ADAMs: Signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 2008, 8, 929–941. [Google Scholar] [CrossRef]
- Hundhausen, C.; Misztela, D.; Berkhout, T.A.; Broadway, N.; Saftig, P.; Reiss, K.; Hartmann, D.; Fahrenholz, F.; Postina, R.; Matthews, V.; et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 2003, 102, 1186–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, A.; Hundhausen, C.; Lambert, M.H.; Broadway, N.; Andrews, R.C.; Bickett, D.M.; Leesnitzer, M.A.; David Becherer, J. Metalloproteinase Inhibitors for the Disintegrin-Like Metalloproteinases ADAM10 and ADAM17 that Differentially Block Constitutive and Phorbol Ester-Inducible Shedding of Cell Surface Molecules. Comb. Chem. High Throughput Screening 2005, 8, 161–171. [Google Scholar] [CrossRef]
- Schlöndorff, J.; Becherer, J.D.; Blobel, C.P. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE). Biochem. J. 2000, 347, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.D.; Cho, H.; Koo, J.Y.; Tak, M.H.; Cho, Y.; Shim, W.S.; Park, S.P.; Lee, J.; Lee, B.; Kim, B.M.; et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 2008, 455, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, S.; Hoischen, A.; Meijer, R.P.P.; Gilissen, C.; Neveling, K.; Wieskamp, N.; De Brouwer, A.; Koenig, M.; Anheim, M.; Assoum, M.; et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 2010, 87, 813–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, T.; Cheng, J.; Tatematsu, T.; Ebata, A.; Kamikawa, H.; Fujita, A.; Gyobu, S.; Segawa, K.; Arai, H.; Taguchi, T.; et al. Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc. Natl. Acad. Sci. USA 2019, 116, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Marconi, C.; Binello, P.B.; Badiali, G.; Caci, E.; Cusano, R.; Garibaldi, J.; Pippucci, T.; Merlini, A.; Marchetti, C.; Rhoden, K.J.; et al. A novel missense mutation in ANO5/TMEM16E is causative for gnathodiaphyseal dyplasia in a large Italian pedigree. Eur. J. Hum. Genet. 2013, 21, 613–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreeva, T.V.; Tyazhelova, T.V.; Rykalina, V.N.; Gusev, F.E.; Goltsov, A.Y.; Zolotareva, O.I.; Aliseichik, M.P.; Borodina, T.A.; Grigorenko, A.P.; Reshetov, D.A.; et al. Whole exome sequencing links dental tumor to an autosomal-dominant mutation in ANO5 gene associated with gnathodiaphyseal dysplasia and muscle dystrophies. Sci. Rep. 2016, 6, 26440. [Google Scholar] [CrossRef] [Green Version]
- Gyobu, S.; Ishihara, K.; Suzuki, J.; Segawa, K.; Nagata, S. Characterization of the scrambling domain of the TMEM16 family. Proc. Natl. Acad. Sci. USA 2017, 114, 6274–6279. [Google Scholar] [CrossRef] [Green Version]
- Gyobu, S.; Miyata, H.; Ikawa, M.; Yamazaki, D.; Takeshima, H.; Suzuki, J.; Nagata, S. A Role of TMEM16E Carrying a Scrambling Domain in Sperm Motility. Mol. Cell. Biol. 2015, 36, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Di Zanni, E.; Gradogna, A.; Scholz-Starke, J.; Boccaccio, A. Gain of function of TMEM16E/ANO5 scrambling activity caused by a mutation associated with gnathodiaphyseal dysplasia. Cell. Mol. Life Sci. 2018, 75, 1657–1670. [Google Scholar] [CrossRef] [Green Version]
- Whitlock, J.M.; Yu, K.; Cui, Y.Y.; Hartzell, H.C. Anoctamin 5/TMEM16E facilitates muscle precursor cell fusion. J. Gen. Physiol. 2018, 150, 1498–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bera, T.K.; Das, S.; Maeda, H.; Beers, R.; Wolfgang, C.D.; Kumar, V.; Hahn, Y.; Lee, B.; Pastan, I. NGEP, a gene encoding a membrane protein detected only in prostate cancer and normal prostate. Proc. Natl. Acad. Sci. USA 2004, 101, 3059–3064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaikkonen, E.; Rantapero, T.; Zhang, Q.; Taimen, P.; Laitinen, V.; Kallajoki, M.; Jambulingam, D.; Ettala, O.; Knaapila, J.; Boström, P.J.; et al. ANO7 is associated with aggressive prostate cancer. Int. J. Cancer 2018, 143, 2479–2487. [Google Scholar] [CrossRef] [PubMed]
- Mohsenzadegan, M.; Shekarabi, M.; Madjd, Z.; Asgari, M.; Abolhasani, M.; Tajik, N.; Farajollahi, M.M. Study of NGEP expression pattern in cancerous tissues provides novel insights into prognostic marker in prostate cancer. Biomark. Med. 2015, 9, 391–401. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Ruiz-Paz, S.; Baselga, J.; Arribas, J. Metalloprotease-dependent protransforming growth factor-alpha ectodomain shedding in the absence of tumor necrosis factor-alpha-converting enzyme. J. Biol. Chem. 2001, 276, 48510–48517. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, B.; Gschwind, A.; Ullrich, A. Multiple G-protein-coupled receptor signals converge on the epidermal growth factor receptor to promote migration and invasion. Oncogene 2004, 23, 991–999. [Google Scholar] [CrossRef] [Green Version]
- Sommer, A.; Düppe, M.; Baumecker, L.; Kordowski, F.; Büch, J.; Chico, J.F.; Fritsch, J.; Schütze, S.; Adam, D.; Sperrhacke, M.; et al. Extracellular sphingomyelinase activity impairs TNF-α-induced endothelial cell death via ADAM17 activation and TNF receptor 1 shedding. Oncotarget 2017, 8, 72584–72596. [Google Scholar] [CrossRef] [Green Version]
- Reiss, K.; Bhakdi, S. The plasma membrane: Penultimate regulator of ADAM sheddase function. Biochim. Biophys. Acta-Mol. Cell Res. 2017, 1864, 2082–2087. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Maniero, C.; Zhou, J.; Shaikh, L.H.; Azizan, E.A.B.; McFarlane, I.; Neogi, S.; Scudieri, P.; Galietta, L.J.; Brown, M.J. Role of ANO4 in regulation of aldosterone secretion in the zona glomerulosa of the human adrenal gland. Lancet 2015, 385, S62. [Google Scholar] [CrossRef]
- Maniero, C.; Scudieri, P.; Haris Shaikh, L.; Zhao, W.; Gurnell, M.; Galietta, L.J.V.; Brown, M.J. ANO4 (Anoctamin 4) is a novel marker of zona glomerulosa that regulates stimulated aldosterone secretion. Hypertension 2019, 74, 1152–1159. [Google Scholar] [CrossRef]
- Sherva, R.; Tripodis, Y.; Bennett, D.A.; Chibnik, L.B.; Crane, P.K.; De Jager, P.L.; Farrer, L.A.; Saykin, A.J.; Shulman, J.M.; Naj, A.; et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.T.; Guo, A.Y.; Maher, B.S.; Zhao, Z.; Van Den Oord, E.J.; Kendler, K.S.; Riley, B.P.; Gillespie, N.A.; Prescott, C.A.; Middeldorp, C.M.; et al. Meta-analyses of genome-wide linkage scans of anxiety-related phenotypes. Eur. J. Hum. Genet. 2012, 20, 1078–1084. [Google Scholar] [CrossRef]
- Terracciano, A.; Sanna, S.; Uda, M.; Deiana, B.; Usala, G.; Busonero, F.; Maschio, A.; Scally, M.; Patriciu, N.; Chen, W.M.; et al. Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 2010, 15, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Athanasiu, L.; Mattingsdal, M.; Kähler, A.K.; Brown, A.; Europe PMC Funders Group. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J. Psychiatr. Res. 2011, 44, 748–753. [Google Scholar] [CrossRef] [Green Version]
- Wittkowski, K.M.; Dadurian, C.; Seybold, M.P.; Kim, H.S.; Hoshino, A.; Lyden, D. Complex polymorphisms in endocytosis genes suggest alpha-cyclodextrin as a treatment for breast cancer. PLoS ONE 2018, 13, e0199012. [Google Scholar] [CrossRef] [Green Version]
- Reichhart, N.; Milenkovic, V.M.; Wetzel, C.H.; Strauß, O. Prediction of functional consequences of missense mutations in ano4 gene. Int. J. Mol. Sci. 2021, 22, 2732. [Google Scholar] [CrossRef]
- Kunisaki, C. Role of the Anoctamin Family in Various Carcinomas. Ann. Surg. Oncol. 2020, 27, 3112–3114. [Google Scholar] [CrossRef] [PubMed]
- Jun, I.; Park, H.S.; Piao, H.; Han, J.W.; An, M.J.; Yun, B.G.; Zhang, X.; Cha, Y.H.; Shin, Y.K.; Yook, J.I.; et al. ANO9/TMEM16j promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br. J. Cancer 2017, 117, 1798–1809. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cai, S.; Wang, X.; Jiang, Z. Identification and characterization of ANO9 in stage II and III colorectal carcinoma. Oncotarget 2015, 6, 29324–29334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsurahara, K.; Shiozaki, A.; Kosuga, T.; Kudou, M.; Shoda, K.; Arita, T.; Konishi, H.; Komatsu, S.; Kubota, T.; Fujiwara, H.; et al. ANO9 Regulated Cell Cycle in Human Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2020, 27, 3218–3230. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitzke, S.; Seidel, J.; Ahrens, B.; Schreiber, R.; Kunzelmann, K.; Sperrhacke, M.; Bhakdi, S.; Reiss, K. Influence of Anoctamin-4 and -9 on ADAM10 and ADAM17 Sheddase Function. Membranes 2022, 12, 123. https://doi.org/10.3390/membranes12020123
Leitzke S, Seidel J, Ahrens B, Schreiber R, Kunzelmann K, Sperrhacke M, Bhakdi S, Reiss K. Influence of Anoctamin-4 and -9 on ADAM10 and ADAM17 Sheddase Function. Membranes. 2022; 12(2):123. https://doi.org/10.3390/membranes12020123
Chicago/Turabian StyleLeitzke, Sinje, Jana Seidel, Björn Ahrens, Rainer Schreiber, Karl Kunzelmann, Maria Sperrhacke, Sucharit Bhakdi, and Karina Reiss. 2022. "Influence of Anoctamin-4 and -9 on ADAM10 and ADAM17 Sheddase Function" Membranes 12, no. 2: 123. https://doi.org/10.3390/membranes12020123
APA StyleLeitzke, S., Seidel, J., Ahrens, B., Schreiber, R., Kunzelmann, K., Sperrhacke, M., Bhakdi, S., & Reiss, K. (2022). Influence of Anoctamin-4 and -9 on ADAM10 and ADAM17 Sheddase Function. Membranes, 12(2), 123. https://doi.org/10.3390/membranes12020123






