Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Two-Electrode Voltage-Clamp (TEVC) in Xenopus laevis Oocytes
2.3. Transient Expression in N. benthamiana Leaves by Agro-Infiltration
2.4. Surface Potential Recordings with N. benthamiana Leaves
2.5. Transient Expression in N. tabacum Pollen Tubes
2.6. Light-Induced Voltage and Current Recording in Pollen Tubes
2.7. Confocal Images Processing
2.8. Data Analysis
3. Results
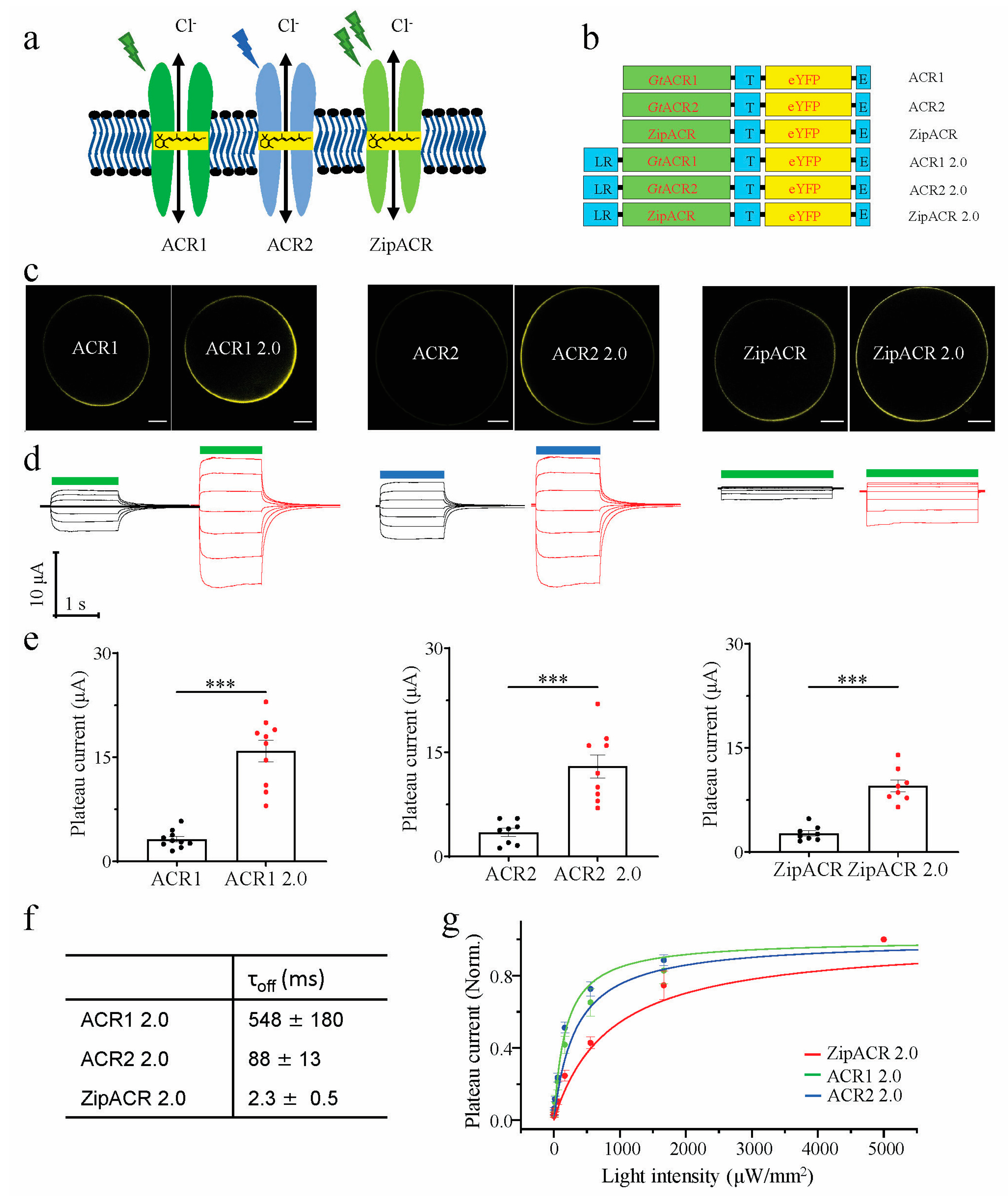
3.1. Characterization of Engineered ACRs in Xenopus laevis Oocytes
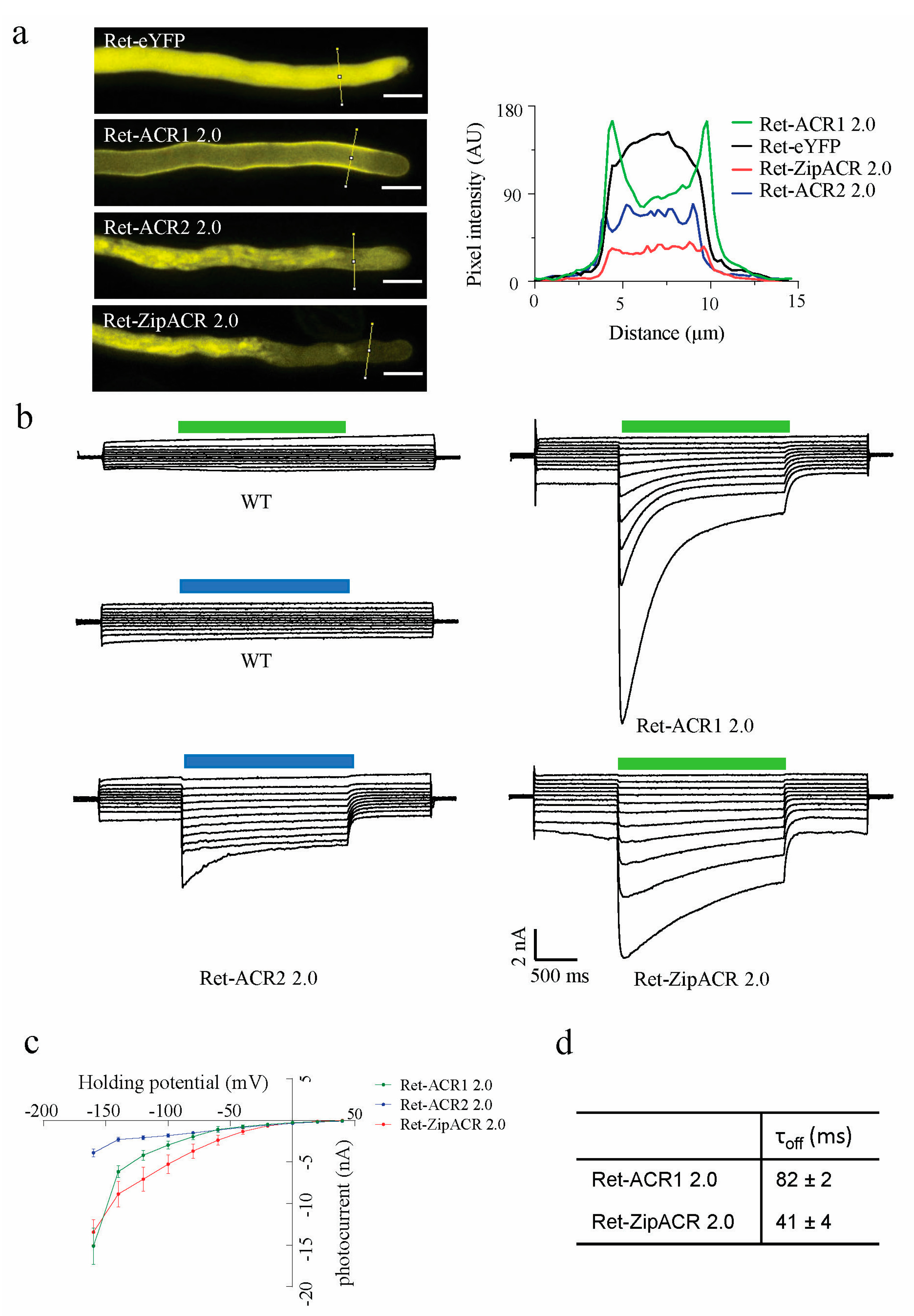
3.2. Functional Expression of Different ACRs in N. benthamiana Leaves
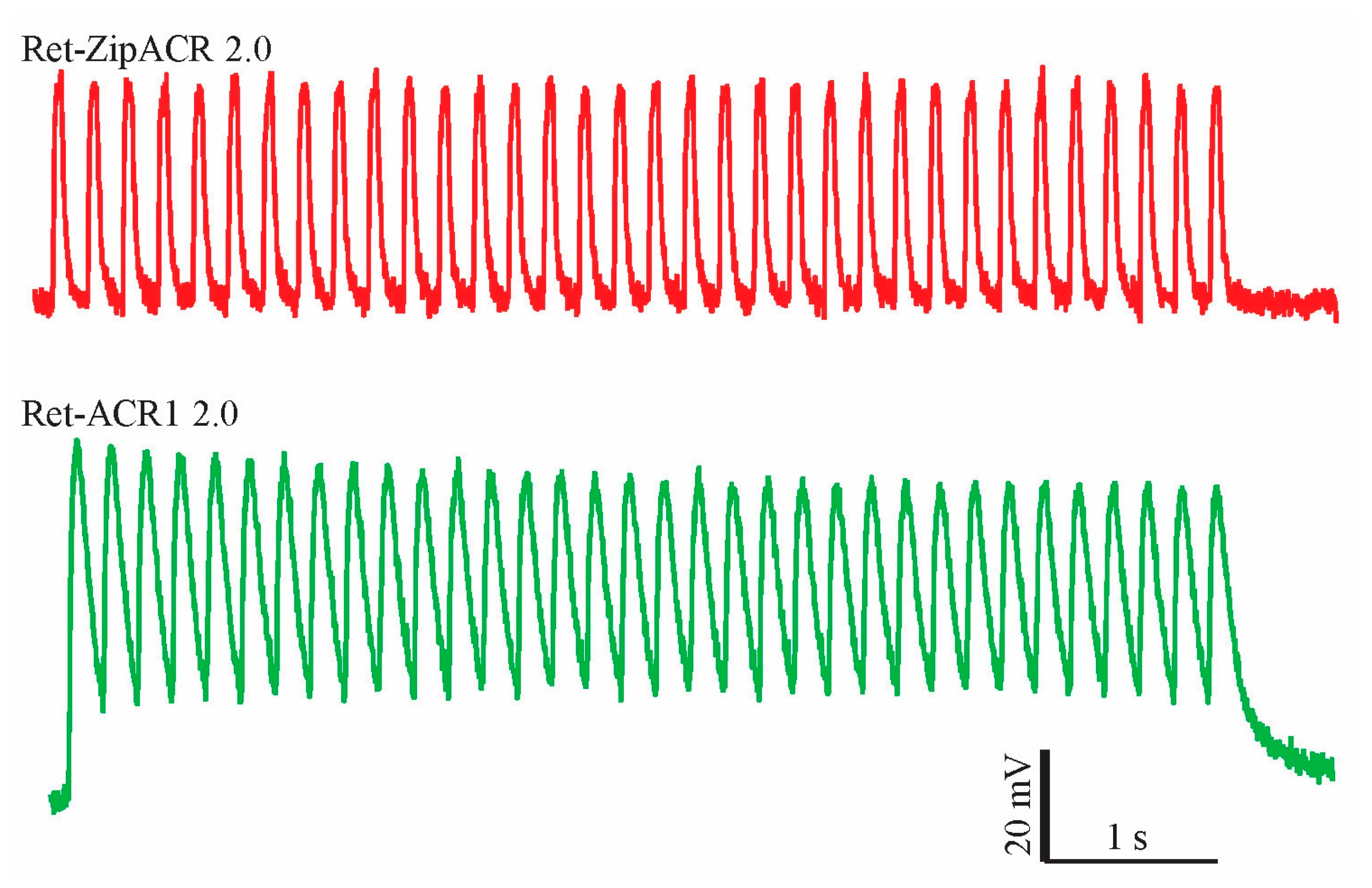
3.3. Functional Expression of ACR2 and ZipACR in N. tabacum Pollen Tubes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adamantidis, A.; Arber, S.; Bains, J.S.; Bamberg, E.; Bonci, A.; Buzsáki, G.; Cardin, J.A.; Costa, R.M.; Dan, Y.; Goda, Y.; et al. Optogenetics: 10 years after ChR2 in neurons—views from the community. Nat. Neurosci. 2015, 18, 1202–1212. [Google Scholar] [CrossRef]
- Guru, A.; Post, R.J.; Ho, Y.-Y.; Warden, M.R. Making Sense of Optogenetics. Int. J. Neuropsychopharmacol. 2015, 18, pyv079. [Google Scholar] [CrossRef] [PubMed]
- Terakita, A. The opsins. Genome Biol. 2005, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Bi, A.; Cui, J.; Ma, Y.-P.; Olshevskaya, E.; Pu, M.; Dizhoor, A.M.; Pan, Z.-H. Ectopic Expression of a Microbial-Type Rhodopsin Restores Visual Responses in Mice with Photoreceptor Degeneration. Neuron 2006, 50, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gutierrez, D.V.; Hanson, M.G.; Han, J.; Mark, M.D.; Chiel, H.; Hegemann, P.; Landmesser, L.T.; Herlitze, S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA 2005, 102, 17816–17821. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Brauner, M.; Liewald, J.F.; Adeishvili, N.; Bamberg, E.; Gottschalk, A. Light Activation of Channelrhodopsin-2 in Excitable Cells of Caenorhabditis elegans Triggers Rapid Behavioral Responses. Curr. Biol. 2005, 15, 2279–2284. [Google Scholar] [CrossRef]
- Nagel, G.; Szellas, T.; Kateriya, S.; Adeishvili, N.; Hegemann, P.; Bamberg, E. Channelrhodopsins: Directly light-gated cation channels. Biochem. Soc. Trans. 2005, 33, 863–866. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Janz, R.; Liu, X.; Spudich, J.L. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Stewart, J.C.; Ott, S.; Chlebikova, K.; Chua, J.Y.; Koh, T.-W.; Ho, J.; Claridge-Chang, A. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 2017, 14, 271–274. [Google Scholar] [CrossRef]
- Mauss, A.S.; Busch, C.; Borst, A. Optogenetic Neuronal Silencing in Drosophila during Visual Processing. Sci. Rep. 2017, 7, 13823. [Google Scholar] [CrossRef] [PubMed]
- Mahn, M.; Gibor, L.; Patil, P.; Malina, K.C.-K.; Oring, S.; Printz, Y.; Levy, R.; Lampl, I.; Yizhar, O. High-efficiency optogenetic silencing with soma-targeted anion-conducting channelrhodopsins. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.A.; Cheng, R.-K.; Ho, J.; Krishnan, S.; Farhan, M.; Claridge-Chang, A.; Jesuthasan, S. Optical inhibition of larval zebrafish behaviour with anion channelrhodopsins. BMC Biol. 2017, 15, 103. [Google Scholar] [CrossRef]
- Hegemann, P.; Nagel, G. From channelrhodopsins to optogenetics. EMBO Mol. Med. 2013, 5, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Fernandez, R.; Samodelov, S.L.; Brandl, S.M.; Wehinger, E.; Müller, K.; Weber, W.; Zurbriggen, M.D. Optogenetics in Plants: Red/Far-Red Light Control of Gene Expression. In Methods in Molecular Biology; Metzler, J.B., Ed.; Humana Press: Totowa, NJ, USA, 2016; Volume 1408, pp. 125–139. [Google Scholar]
- Chatelle, C.; Ochoa-Fernandez, R.; Engesser, R.; Schneider, N.; Beyer, H.M.; Jones, A.R.; Timmer, J.; Zurbriggen, M.D.; Weber, W. A Green-Light-Responsive System for the Control of Transgene Expression in Mammalian and Plant Cells. ACS Synth. Biol. 2018, 7, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Fernandez, R.; Abel, N.B.; Wieland, F.-G.; Schlegel, J.; Koch, L.-A.; Miller, J.B.; Engesser, R.; Giuriani, G.; Brandl, S.M.; Timmer, J.; et al. Optogenetic control of gene expression in plants in the presence of ambient white light. Nat. Methods 2020, 17, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Papanatsiou, M.; Petersen, J.; Henderson, L.; Wang, Y.; Christie, J.M.; Blatt, M.R. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 2019, 363, 1456–1459. [Google Scholar] [CrossRef]
- Dawydow, A.; Gueta, R.; Ljaschenko, D.; Ullrich, S.; Hermann, M.; Ehmann, N.; Gao, S.Q.; Fiala, A.; Langenhan, T.; Nagel, G.; et al. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. USA 2014, 111, 13972–13977. [Google Scholar] [CrossRef]
- Reyer, A.; Häßler, M.; Scherzer, S.; Huang, S.; Pedersen, J.T.; Al-Rasheid, K.A.S.; Bamberg, E.; Palmgren, M.; Dreyer, I.; Nagel, G.; et al. Channelrhodopsin-mediated optogenetics highlights a central role of depolarization-dependent plant proton pumps. Proc. Natl. Acad. Sci. USA 2020, 117, 20920–20925. [Google Scholar] [CrossRef]
- Spudich, J.L.; Yang, C.-S.; Jung, K.-H.; Spudich, E.N. Retinylidene Proteins: Structures and Functions from Archaea to Humans. Annu. Rev. Cell Dev. Biol. 2000, 16, 365–392. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, M.; Gao, S.; Yu-Strzelczyk, J.; Krischke, M.; Duan, X.; Leide, J.; Riederer, M.; Mueller, M.J.; Hedrich, R.; et al. Optogenetic control of plant growth by a microbial rhodopsin. Nat. Plants 2021, 7, 144–151. [Google Scholar] [CrossRef]
- Urquiza-Garcia, U.; Zurbriggen, M.D. Biofortifying green optogenetics. Nat. Plants 2021, 7, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Govorunova, E.G.; Sineshchekov, O.A.; Rodarte, E.M.; Janz, R.; Morelle, O.; Melkonian, M.; Wong, G.K.-S.; Spudich, J.L. The Expanding Family of Natural Anion Channelrhodopsins Reveals Large Variations in Kinetics, Conductance, and Spectral Sensitivity. Sci. Rep. 2017, 7, srep43358. [Google Scholar] [CrossRef] [PubMed]
- Shepard, B.D.; Natarajan, N.; Protzko, R.J.; Acres, O.W.; Pluznick, J.L. A Cleavable N-Terminal Signal Peptide Promotes Widespread Olfactory Receptor Surface Expression in HEK293T Cells. PLoS ONE 2013, 8, e68758. [Google Scholar] [CrossRef] [PubMed]
- Gradinaru, V.; Zhang, F.; Ramakrishnan, C.; Mattis, J.; Prakash, R.; Diester, I.; Goshen, I.; Thompson, K.R.; Deisseroth, K. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell 2010, 141, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, A.L.; Vignali, D.A. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin. Biol. Ther. 2005, 5, 627–638. [Google Scholar] [CrossRef]
- Main, G.D.; Reynolds, S.; Gartland, J.S.; Gartland, K.M.A.; Davey, M.R. Electroporation Protocols for Agrobacterium. Agrobacterium Protoc. 2003, 44, 405–412. [Google Scholar] [CrossRef]
- Li, X. Infiltration of Nicotiana benthamiana Protocol for Transient Expression via Agrobacterium. BIO-PROTOCOL 2011, 1, 95. [Google Scholar] [CrossRef]
- Mousavi, S.A.R.; Nguyen, C.T.; Farmer, E.E.; Kellenberger, S. Measuring surface potential changes on leaves. Nat. Protoc. 2014, 9, 1997–2004. [Google Scholar] [CrossRef]
- Gutermuth, T.; Lassig, R.; Portes, M.-T.; Maierhofer, T.; Romeis, T.; Borst, J.-W.; Hedrich, R.; Feijó, J.A.; Konrad, K.R. Pollen Tube Growth Regulation by Free Anions Depends on the Interaction between the Anion Channel SLAH3 and Calcium-Dependent Protein Kinases CPK2 and CPK20. Plant Cell 2013, 25, 4525–4543. [Google Scholar] [CrossRef] [PubMed]
- Gutermuth, T.; Herbell, S.; Lassig, R.; Brosché, M.; Romeis, T.; Feijó, J.A.; Hedrich, R.; Konrad, K.R. Tip-localized Ca2+-permeable channels control pollen tube growth via kinase-dependent R- and S-type anion channel regulation. New Phytol. 2018, 218, 1089–1105. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Ding, M.; Duan, X.; Konrad, K.R.; Nagel, G.; Gao, S. Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics. Membranes 2021, 11, 287. https://doi.org/10.3390/membranes11040287
Zhou Y, Ding M, Duan X, Konrad KR, Nagel G, Gao S. Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics. Membranes. 2021; 11(4):287. https://doi.org/10.3390/membranes11040287
Chicago/Turabian StyleZhou, Yang, Meiqi Ding, Xiaodong Duan, Kai R. Konrad, Georg Nagel, and Shiqiang Gao. 2021. "Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics" Membranes 11, no. 4: 287. https://doi.org/10.3390/membranes11040287
APA StyleZhou, Y., Ding, M., Duan, X., Konrad, K. R., Nagel, G., & Gao, S. (2021). Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics. Membranes, 11(4), 287. https://doi.org/10.3390/membranes11040287





