Minimizing Cholesterol-Induced Aggregation of Membrane-Interacting DNA Origami Nanostructures
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Design and Folding of DNA Origami Nanostructures
2.2. PEG-Precipitation of DOB
2.3. Purification of DNA Origami Nanostructures for Membrane Binding Experiments
2.4. Formation of Small Unilamellar Vesicles (SUVs)
2.5. Evaluation of Membrane Binding Using Gel Shift Assay
2.6. Transmission Electron Microscopy
3. Results and Discussion
3.1. Effect of Number and Separation of Cholesterols, Spacer Length and Overhang on Aggregation of DOT
3.2. Effect of Spacer Length and Overhang on Aggregation of DOB
3.3. Comparison of Yields of DOT and DOB
3.4. Comparison of Membrane Binding of DOT and DOB
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The Beauty and Utility of DNA Origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.K.D.; Luu, M.T.; Abbas, A.; Wickham, S.F.J. Switchable DNA-origami nanostructures that respond to their environment and their applications. Biophys. Rev. 2018, 10, 1283–1293. [Google Scholar] [CrossRef]
- Franquelim, H.G.; Khmelinskaia, A.; Sobczak, J.-P.; Dietz, H.; Schwille, P. Membrane sculpting by curved DNA origami scaffolds. Nat. Commun. 2018, 9, 811. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Deng, Z.; Yan, H.; Cabrini, S.; Zuckermann, R.N.; Bokor, J. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J. Am. Chem. Soc. 2010, 132, 3248–3249. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.A.; Zhang, C.; Bao, G.; Ke, Y. Programmable Assembly of Iron Oxide Nanoparticles Using DNA Origami. Nano Lett. 2020, 20, 2799–2805. [Google Scholar] [CrossRef] [PubMed]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338, 932–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darley, E.; Singh, J.K.D.; Surace, N.A.; Wickham, S.F.J.; Baker, M.A.B. The Fusion of Lipid and DNA Nanotechnology. Genes 2019, 10, 1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.; Chen, F.; Zhao, Y.; Yu, X.; Wei, J.; Zhao, Y. Functional and Biomimetic DNA Nanostructures on Lipid Membranes. Langmuir 2018, 34, 14721–14730. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Göpfrich, K.; Wood, J.W.; Thacker, V.V.; Stulz, E.; Keyser, U.F.; Howorka, S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew. Chem. Int. Ed. 2013, 52, 12069–12072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson-Buck, A.; Jiang, S.; Yan, H.; Walter, N.G. DNA–Cholesterol Barges as Programmable Membrane-Exploring Agents. ACS Nano 2014, 8, 5641–5649. [Google Scholar] [CrossRef] [PubMed]
- List, J.; Weber, M.; Simmel, F.C. Hydrophobic Actuation of a DNA Origami Bilayer Structure. Angew. Chem. Int. Ed. 2014, 53, 4236–4239. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Endo, M.; Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 2015, 6, 8052. [Google Scholar] [CrossRef] [Green Version]
- Göpfrich, K.; Li, C.-Y.; Ricci, M.; Bhamidimarri, S.P.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10, 8207–8214. [Google Scholar] [CrossRef] [PubMed]
- Ohmann, A.; Li, C.-Y.; Maffeo, C.; Nahas, K.A.; Baumann, K.N.; Göpfrich, K.; Yoo, J.; Keyser, U.F.; Aksimentiev, A. A synthetic enzyme built from DNA flips 10 7 lipids per second in biological membranes. Nat. Commun. 2018, 9, 2426. [Google Scholar] [CrossRef] [Green Version]
- Czogalla, A.; Kauert, D.J.; Franquelim, H.G.; Uzunova, V.; Zhang, Y.; Seidel, R.; Schwille, P. Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles. Angew. Chem. Int. Ed. 2015, 54, 6501–6505. [Google Scholar] [CrossRef]
- Kocabey, S.; Kempter, S.; List, J.; Xing, Y.; Bae, W.; Schiffels, D.; Shih, W.M.; Simmel, F.C.; Liedl, T. Membrane-Assisted Growth of DNA Origami Nanostructure Arrays. ACS Nano 2015, 9, 3530–3539. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Kauert, D.J.; Seidel, R.; Schwille, P.; Petrov, E.P. DNA origami nanoneedles on freestanding lipid membranes as a tool to observe isotropic-nematic transition in two dimensions. Nano Lett. 2015, 15, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Ziegler, D.; Arnaut, V.; Martin, T.G.; Kapsner, K.; Henneberg, K.; Bausch, A.R.; Dietz, H.; Simmel, F.C. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016, 7, 12787. [Google Scholar] [CrossRef] [PubMed]
- Iric, K.; Subramanian, M.; Oertel, J.; Agarwal, N.P.; Matthies, M.; Periole, X.; Sakmar, T.P.; Huber, T.; Fahmy, K.; Schmidt, T.L. DNA-encircled lipid bilayers. Nanoscale 2018, 10, 18463–18467. [Google Scholar] [CrossRef] [PubMed]
- Banchelli, M.; Gambinossi, F.; Durand, A.; Caminati, G.; Brown, T.; Berti, D.; Baglioni, P. Modulation of Density and Orientation of Amphiphilic DNA on Phospholipid Membranes. II. Vesicles. J. Phys. Chem. B 2010, 114, 7348–7358. [Google Scholar] [CrossRef]
- Bunge, A.; Loew, M.; Pescador, P.; Arbuzova, A.; Brodersen, N.; Kang, J.; Dähne, L.; Liebscher, J.; Herrmann, A.; Stengel, G.; et al. Lipid Membranes Carrying Lipophilic Cholesterol-Based Oligonucleotides—Characterization and Application on Layer-by-Layer Coated Particles. J. Phys. Chem. B 2009, 113, 16425–16434. [Google Scholar] [CrossRef] [PubMed]
- Akbari, E.; Shahhosseini, M.; Baetz, J.C.; Song, J.W.; Castro, C.E. Functionalizing Cell Membranes with DNA Origami for Multiplexed Biomolecular Sensing. FASEB J. 2019, 33, 785.1. [Google Scholar] [CrossRef]
- Akbari, E.; Mollica, M.Y.; Lucas, C.R.; Bushman, S.M.; Patton, R.A.; Shahhosseini, M.; Song, J.W.; Castro, C.E. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29, 1703632. [Google Scholar] [CrossRef]
- Burns, J.R.; Howorka, S. Structural and Functional Stability of DNA Nanopores in Biological Media. Nanomaterials 2019, 9, 490. [Google Scholar] [CrossRef] [Green Version]
- Chidchob, P.; Offenbartl-Stiegert, D.; McCarthy, D.; Luo, X.; Li, J.; Howorka, S.; Sleiman, H.F. Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions. J. Am. Chem. Soc. 2019, 141, 1100–1108. [Google Scholar] [CrossRef]
- Ohmann, A.; Göpfrich, K.; Joshi, H.; Thompson, R.F.; Sobota, D.; Ranson, N.A.; Aksimentiev, A.; Keyser, U.F. Controlling aggregation of cholesterol-modified DNA nanostructures. Nucleic Acids Res. 2019, 47, 11441–11451. [Google Scholar] [CrossRef]
- Diederichs, T.; Pugh, G.; Dorey, A.; Xing, Y.; Burns, J.R.; Nguyen, Q.H.; Tornow, M.; Tampé, R.; Howorka, S. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 2019, 10, 5018. [Google Scholar] [CrossRef]
- Khmelinskaia, A.; Franquelim, H.G.; Petrov, E.P.; Schwille, P. Effect of anchor positioning on binding and diffusion of elongated 3D DNA nanostructures on lipid membranes. J. Phys. D Appl. Phys. 2016, 49, 194001. [Google Scholar] [CrossRef]
- Khmelinskaia, A.; Mücksch, J.; Petrov, E.P.; Franquelim, H.G.; Schwille, P. Control of Membrane Binding and Diffusion of Cholesteryl-Modified DNA Origami Nanostructures by DNA Spacers. Langmuir 2018, 34, 14921–14931. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, W.L.; Noble, J.E.; Ryadnov, M.G.; Howorka, S. Cholesterol Anchors Enable Efficient Binding and Intracellular Uptake of DNA Nanostructures. Bioconjug. Chem. 2019, 30, 1836–1844. [Google Scholar] [CrossRef]
- Liu, Y.; Wijesekara, P.; Kumar, S.; Wang, W.; Ren, X.; Taylor, R.E. The effects of overhang placement and multivalency on cell labeling by DNA origami. Nanoscale 2021, 13, 6819–6828. [Google Scholar] [CrossRef] [PubMed]
- Perrault, S.D.; Shih, W.M. Virus-Inspired Membrane Encapsulation of DNA Nanostructures To Achieve In Vivo Stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef] [PubMed]
- Wickham, S.F.J.; Auer, A.; Min, J.; Ponnuswamy, N.; Woehrstein, J.B.; Schueder, F.; Strauss, M.T.; Schnitzbauer, J.; Nathwani, B.; Zhao, Z.; et al. Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard. Nat. Commun. 2020, 11, 5768. [Google Scholar] [CrossRef]
- Singh, J.K.D.; Darley, E.; Ridone, P.; Gaston, J.P.; Abbas, A.; Wickham, S.F.J.; Baker, M.A.B. Binding of DNA origami to lipids: Maximizing yield and switching via strand displacement. Nucleic Acids Res. 2021, 49, 10835–10850. [Google Scholar] [CrossRef]
- Schnitzbauer, J.; Strauss, M.T.; Schlichthaerle, T.; Schueder, F.; Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017, 12, 1198–1228. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef] [Green Version]
- Stahl, E.; Martin, T.G.; Praetorius, F.; Dietz, H. Facile and Scalable Preparation of Pure and Dense DNA Origami Solutions. Angew. Chem. Int. Ed. 2014, 53, 12735–12740. [Google Scholar] [CrossRef] [Green Version]
- Bellot, G.; McClintock, M.A.; Lin, C.; Shih, W.M. Recovery of intact DNA nanostructures following agarose-gel—Based separation. Nat. Methods 2011, 8, 192–194. [Google Scholar] [CrossRef]
- Arnott, P.M.; Joshi, H.; Aksimentiev, A.; Howorka, S. Dynamic Interactions between Lipid-Tethered DNA and Phospholipid Membranes. Langmuir 2018, 34, 15084–15092. [Google Scholar] [CrossRef] [PubMed]
- Lanphere, C.; Offenbartl-Stiegert, D.; Dorey, A.; Pugh, G.; Georgiou, E.; Xing, Y.; Burns, J.R.; Howorka, S. Design, assembly, and characterization of membrane-spanning DNA nanopores. Nat. Protoc. 2021, 16, 86–130. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.A.B.; Tuckwell, A.J.; Berengut, J.F.; Bath, J.; Benn, F.; Duff, A.P.; Whitten, A.E.; Dunn, K.E.; Hynson, R.M.; Turberfield, A.J.; et al. Dimensions and Global Twist of Single-Layer DNA Origami Measured by Small-Angle X-ray Scattering. ACS Nano 2018, 9, 5791–5799. [Google Scholar] [CrossRef] [Green Version]
- Saenger, W. Principles of Nucleic Acid Structure; Springer Science & Business Media: New York, NY, USA, 2013; ISBN 978-1-4612-5190-3. [Google Scholar]
- Murphy, M.C.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef] [Green Version]
- Schröder, S.; Zhang, H.; Yeung, E.S.; Jänsch, L.; Zabel, C.; Wätzig, H. Quantitative gel electrophoresis: Sources of variation. J. Proteome Res. 2008, 7, 1226–1234. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, Y.; Pincet, F.; Llaguno, M.C.; Lin, C. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 2017, 9, 653–659. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, J.; Shigematsu, H.; Xu, W.; Shih, W.M.; Rothman, J.E.; Lin, C. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 2016, 8, 476. [Google Scholar] [CrossRef] [Green Version]
- Lichtenberg, D.; Ahyayauch, H.; Goñi, F.M. The Mechanism of Detergent Solubilization of Lipid Bilayers. Biophys. J. 2013, 105, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Morzy, D.; Rubio-Sánchez, R.; Joshi, H.; Aksimentiev, A.; Di Michele, L.; Keyser, U.F. Cations Regulate Membrane Attachment and Functionality of DNA Nanostructures. J. Am. Chem. Soc. 2021, 143, 7358–7367. [Google Scholar] [CrossRef] [PubMed]
- Bastings, M.M.C.; Anastassacos, F.M.; Ponnuswamy, N.; Leifer, F.G.; Cuneo, G.; Lin, C.; Ingber, D.E.; Ryu, J.H.; Shih, W.M. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18, 3557–3564. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Sánchez, R.; Barker, S.E.; Walczak, M.; Cicuta, P.; Michele, L.D. A Modular, Dynamic, DNA-Based Platform for Regulating Cargo Distribution and Transport between Lipid Domains. Nano Lett. 2021, 21, 2800–2808. [Google Scholar] [CrossRef] [PubMed]


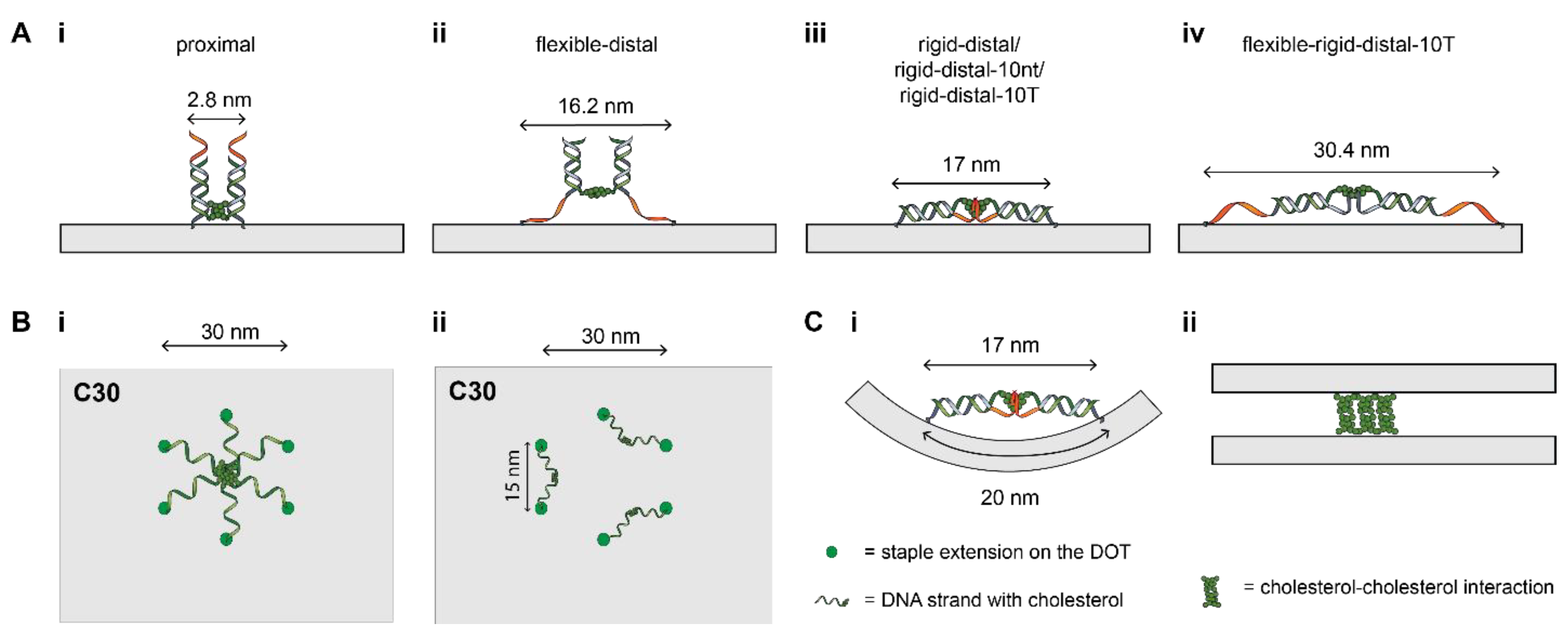
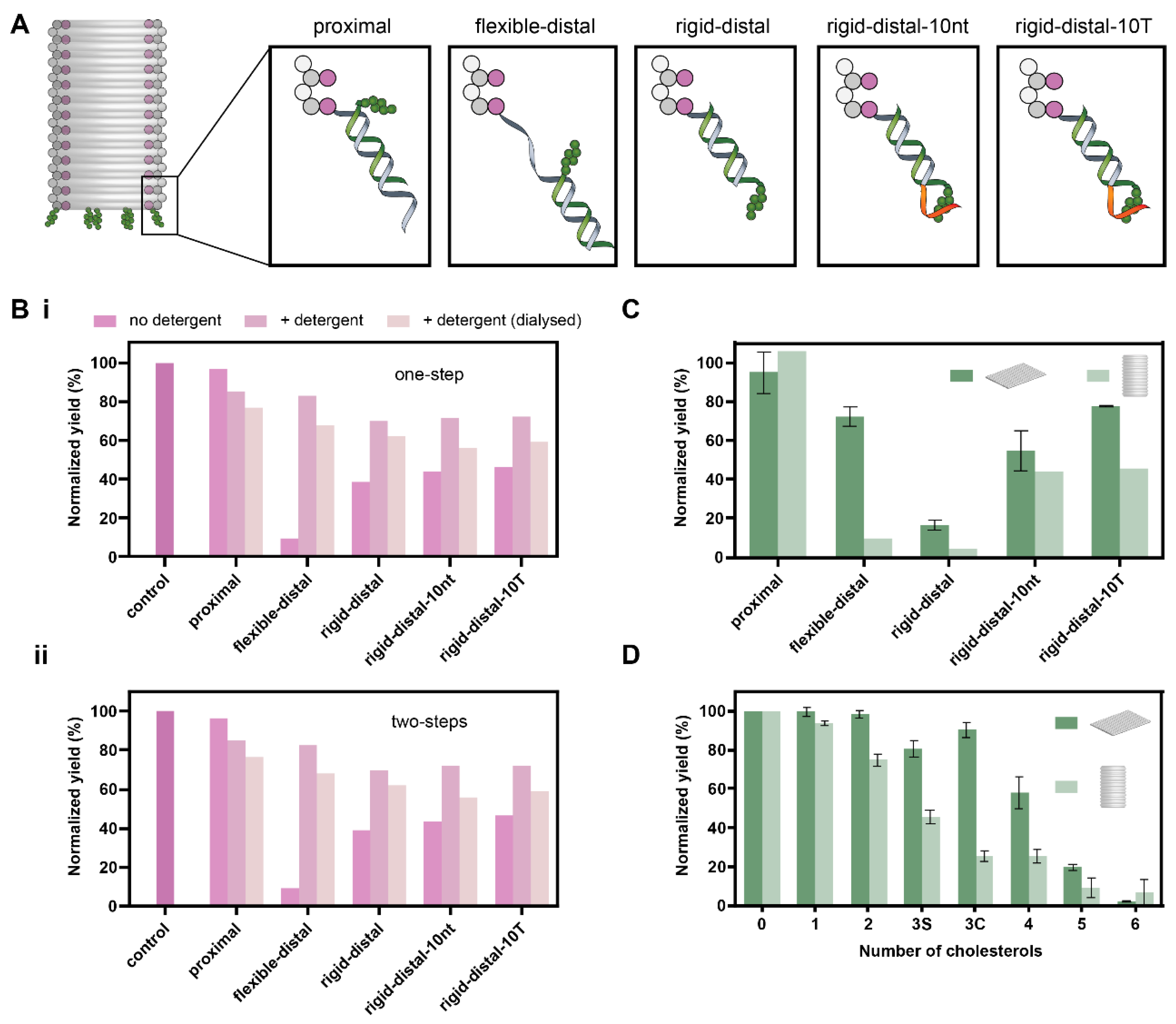
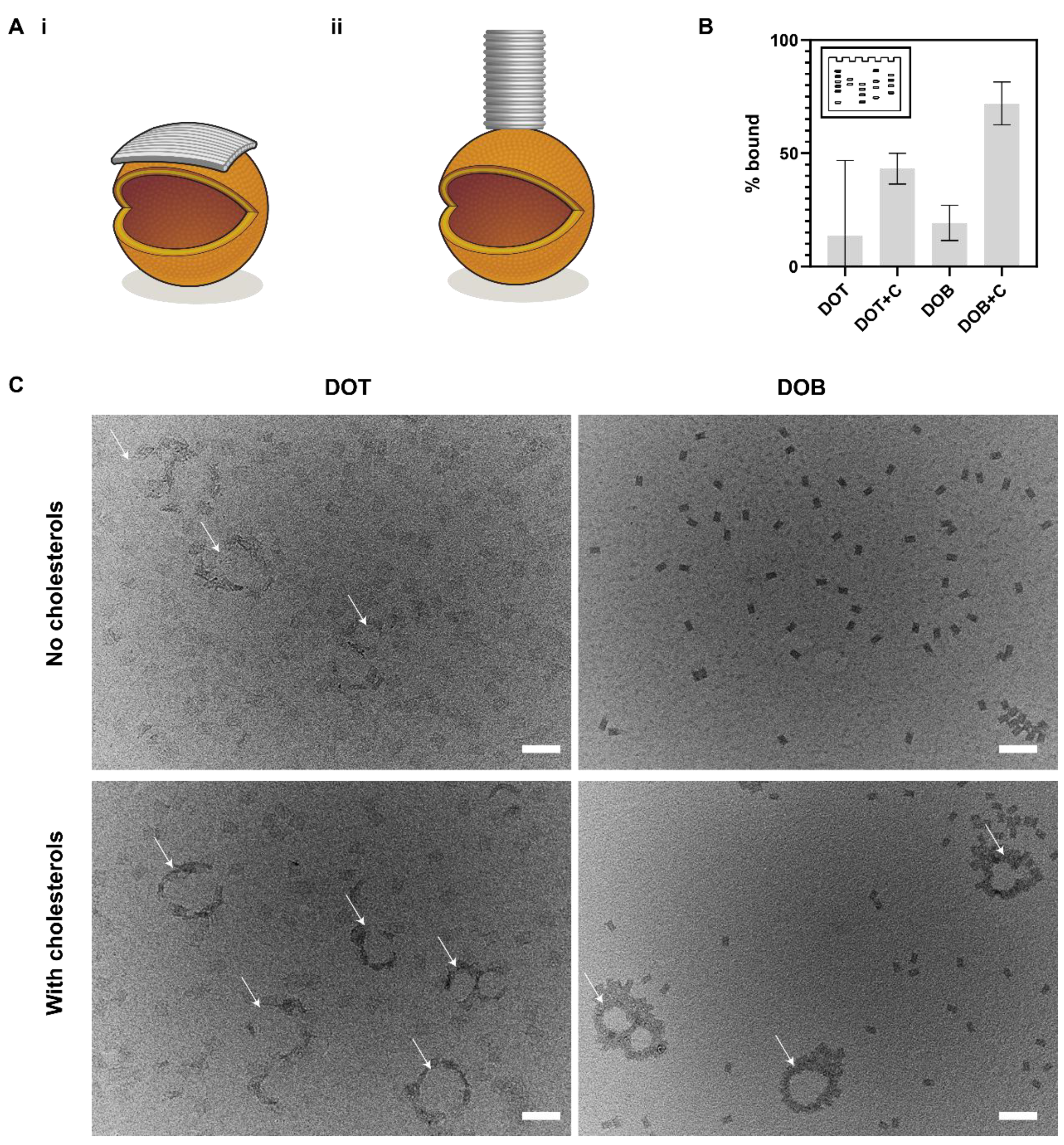
| Configuration | Maximum Separation (Diameter), nm | Minimum Separation, nm |
|---|---|---|
| C20 | 20 | 10 |
| C30 | 30 | 15 |
| C40 | 40 | 20 |
| C50 | 50 | 25 |
| C60 | 60 | 30 |
| Configuration | Maximum Separation, nm | Minimum Separation, nm |
|---|---|---|
| R20 | 20 | 10 |
| R30 | 30 | 15 |
| R40 | 40 | 20 |
| R50 | 50 | 25 |
| R60 | 60 | 30 |
| Spacer | Spacer Type | Maximum Spacing, nm | Overhang |
|---|---|---|---|
| proximal | - | 1.4 | - |
| flexible-distal | 10 nt ssDNA | 8.1 | - |
| rigid-distal | 21 bp dsDNA | 8.5 | - |
| rigid-distal-10nt | 21 bp dsDNA | 8.5 | 10-nt |
| rigid-distal-10T | 21 bp dsDNA | 8.5 | 10-T |
| flexible-rigid-distal-10T | 10 nt ssDNA + 21 bp dsDNA | 15.2 | 10-T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daljit Singh, J.K.; Luu, M.T.; Berengut, J.F.; Abbas, A.; Baker, M.A.B.; Wickham, S.F.J. Minimizing Cholesterol-Induced Aggregation of Membrane-Interacting DNA Origami Nanostructures. Membranes 2021, 11, 950. https://doi.org/10.3390/membranes11120950
Daljit Singh JK, Luu MT, Berengut JF, Abbas A, Baker MAB, Wickham SFJ. Minimizing Cholesterol-Induced Aggregation of Membrane-Interacting DNA Origami Nanostructures. Membranes. 2021; 11(12):950. https://doi.org/10.3390/membranes11120950
Chicago/Turabian StyleDaljit Singh, Jasleen Kaur, Minh Tri Luu, Jonathan F. Berengut, Ali Abbas, Matthew A. B. Baker, and Shelley F. J. Wickham. 2021. "Minimizing Cholesterol-Induced Aggregation of Membrane-Interacting DNA Origami Nanostructures" Membranes 11, no. 12: 950. https://doi.org/10.3390/membranes11120950
APA StyleDaljit Singh, J. K., Luu, M. T., Berengut, J. F., Abbas, A., Baker, M. A. B., & Wickham, S. F. J. (2021). Minimizing Cholesterol-Induced Aggregation of Membrane-Interacting DNA Origami Nanostructures. Membranes, 11(12), 950. https://doi.org/10.3390/membranes11120950







