Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cohort
2.2. Extracellular Vesical Isolation
2.3. Transmission Electron Microscopy
2.4. Nanoparticle Tracking Analysis (NTA)
2.5. Western Blot
2.6. Extracellular Vesicle Protein Digestion and Labelling
2.7. Liquid Chromatography Tandem Mass Spectrometry Analysis
2.8. Data Analysis
3. Results
3.1. Patient and Clinical Characteristics
3.2. Validation of Successful Extracellular Vesicle Isolation
3.3. Quantitative Extracellular Vesical Proteomic Profiling
3.4. Analysis of Extracellular Vesical Associated Proteins of Interest
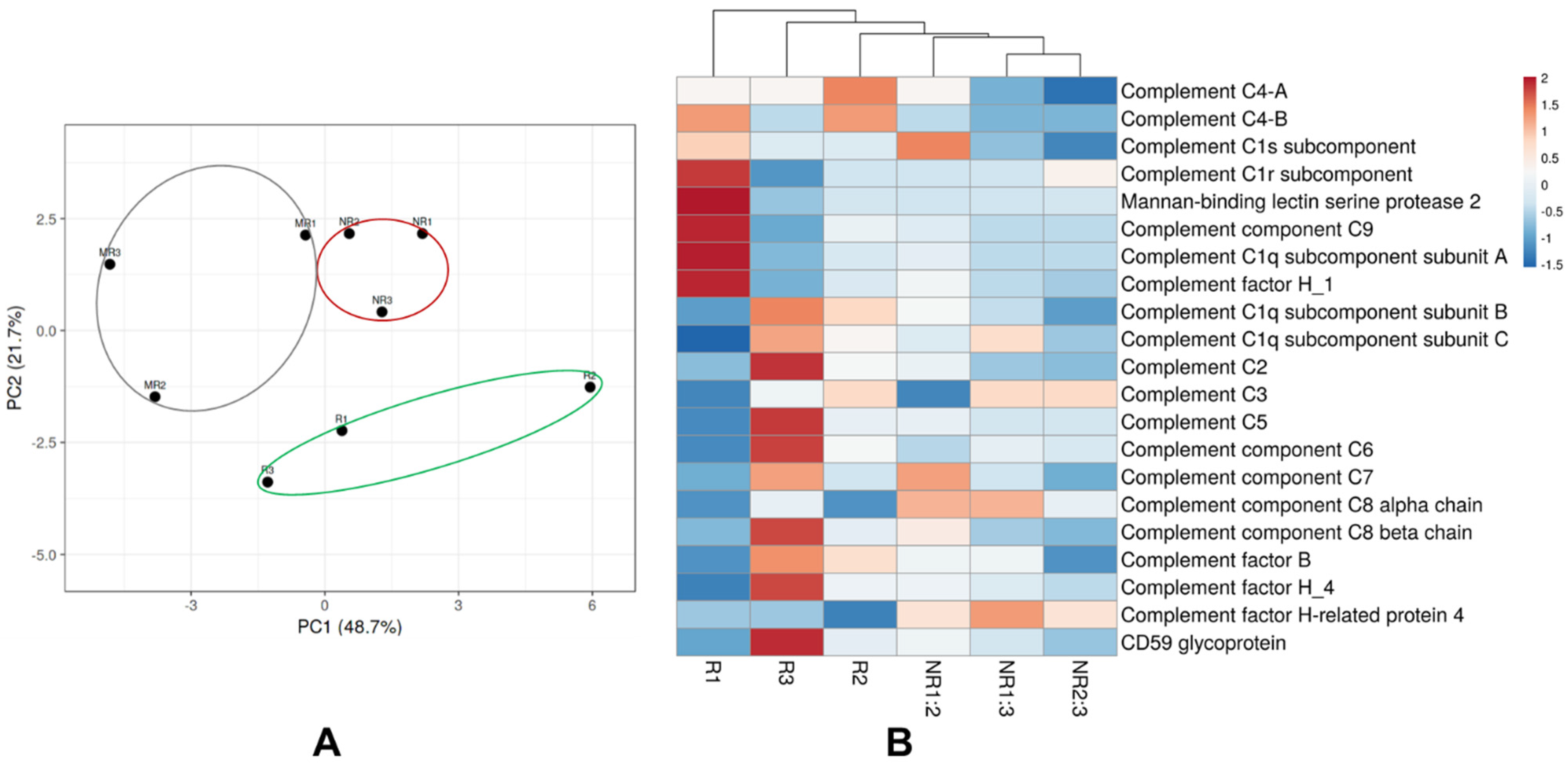
3.5. Extracellular Vesical Associated Proteins Involved in the Complement Cascade
3.6. Classical, High Abundant Extracellular Vesical Associated Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nguyen, B.; Meehan, K.; Pereira, M.R.; Mirzai, B.; Lim, S.H.; Leslie, C.; Clark, M.; Sader, C.; Friedland, P.; Lindsay, A.; et al. A comparative study of extracellular vesicle-associated and cell-free DNA and RNA for HPV detection in oropharyngeal squamous cell carcinoma. Sci. Rep. 2020, 10, 6083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhen, O.; Mirzai, B.; Clark, M.E.; Nguyen, B.; Salomon, C.; Erber, W.; Meehan, K. Comparison of Circulating Tumour DNA and Extracellular Vesicle DNA by Low-Pass Whole-Genome Sequencing Reveals Molecular Drivers of Disease in a Breast Cancer Patient. Biomedicines 2020, 9, 14. [Google Scholar] [CrossRef]
- Nixon, B.; De Iuliis, G.N.; Hart, H.M.; Zhou, W.; Mathe, A.; Bernstein, I.R.; Anderson, A.L.; Stanger, S.J.; Skerrett-Byrne, D.A.; Jamaluddin, M.F.B.; et al. Proteomic Profiling of Mouse Epididymosomes Reveals their Contributions to Post-testicular Sperm Maturation. Mol. Cell. Proteom. 2019, 18, S91–S108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almazi, J.G.; Pockney, P.; Gedye, C.; Smith, N.D.; Hondermarck, H.; Verrills, N.M.; Dun, M.D. Cell-Free DNA Blood Collection Tubes Are Appropriate for Clinical Proteomics: A Demonstration in Colorectal Cancer. Proteom. Clin. Appl. 2018, 12, e1700121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dun, M.D.; Chalkley, R.J.; Faulkner, S.; Keene, S.; Avery-Kiejda, K.A.; Scott, R.J.; Falkenby, L.G.; Cairns, M.J.; Larsen, M.R.; Bradshaw, R.A.; et al. Proteotranscriptomic Profiling of 231-BR Breast Cancer Cells: Identification of Potential Biomarkers and Therapeutic Targets for Brain Metastasis. Mol. Cell. Proteom. 2015, 14, 2316–2330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degryse, S.; de Bock, C.E.; Demeyer, S.; Govaerts, I.; Bornschein, S.; Verbeke, D.; Jacobs, K.; Binos, S.; Skerrett-Byrne, D.A.; Murray, H.C.; et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 2018, 32, 788–800. [Google Scholar] [CrossRef]
- Nixon, B.; Johnston, S.D.; Skerrett-Byrne, D.A.; Anderson, A.L.; Stanger, S.J.; Bromfield, E.G.; Martin, J.H.; Hansbro, P.M.; Dun, M.D. Modification of Crocodile Spermatozoa Refutes the Tenet That Post-testicular Sperm Maturation Is Restricted To Mammals. Mol. Cell. Proteom. 2019, 18, S58–S76. [Google Scholar] [CrossRef] [Green Version]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef] [PubMed]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhong, W.; Bu, J.; Li, Y.; Li, R.; Nie, R.; Xiao, C.; Ma, K.; Huang, X.; Li, Y. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol. Carcinog. 2019, 58, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Vinik, Y.; Ortega, F.G.; Mills, G.B.; Lu, Y.; Jurkowicz, M.; Halperin, S.; Aharoni, M.; Gutman, M.; Lev, S. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci. Adv. 2020, 6, eaba5714. [Google Scholar] [CrossRef] [PubMed]
- Tutanov, O.; Orlova, E.; Proskura, K.; Grigor’eva, A.; Yunusova, N.; Tsentalovich, Y.; Alexandrova, A.; Tamkovich, S. Proteomic Analysis of Blood Exosomes from Healthy Females and Breast Cancer Patients Reveals an Association between Different Exosomal Bioactivity on Non-tumorigenic Epithelial Cell and Breast Cancer Cell Migration in Vitro. Biomolecules 2020, 10, 495. [Google Scholar] [CrossRef] [Green Version]
- Koi, Y.; Tsutani, Y.; Nishiyama, Y.; Ueda, D.; Ibuki, Y.; Sasada, S.; Akita, T.; Masumoto, N.; Kadoya, T.; Yamamoto, Y.; et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, J.; Altman, M.C.; Fearnley, I.M.; Walker, J.E. Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc. Natl. Acad. Sci. USA 2007, 104, 14330–14335. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Rosenow, M.; Xiao, N.; Spetzler, D. Profiling plasma extracellular vesicle by pluronic block-copolymer based enrichment method unveils features associated with breast cancer aggression, metastasis and invasion. J. Extracell. Vesicles 2018, 7, 1458574. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.C.; Enjeti, A.K.; Kahl, R.G.S.; Flanagan, H.M.; Sillar, J.; Skerrett-Byrne, D.A.; Al Mazi, J.G.; Au, G.G.; de Bock, C.E.; Evans, K.; et al. Quantitative phosphoproteomics uncovers synergy between DNA-PK and FLT3 inhibitors in acute myeloid leukaemia. Leukemia 2021, 35, 1782–1787. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Suades, R.; Fuentes, E.; Palomo, I.; Padro, T. Role of Platelet-Derived Microvesicles As Crosstalk Mediators in Atherothrombosis and Future Pharmacology Targets: A Link between Inflammation, Atherosclerosis, and Thrombosis. Front. Pharmacol. 2016, 7, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, R.; Tan, E.; Sharghi-Namini, S.; Asada, H.H. Exosomes in Cancer Microenvironment and Beyond: Have we Overlooked these Extracellular Messengers? Cancer Microenviron. 2012, 5, 323–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Tang, W.; Zhou, M.; Dorsey, T.H.; Prieto, D.A.; Wang, X.W.; Ruppin, E.; Veenstra, T.D.; Ambs, S. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med. 2018, 10, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karasu, E.; Eisenhardt, S.U.; Harant, J.; Huber-Lang, M. Extracellular Vesicles: Packages Sent with Complement. Front. Immunol. 2018, 9, 721. [Google Scholar] [CrossRef]
- Redig, A.J.; McAllister, S.S. Breast cancer as a systemic disease: A view of metastasis. J. Intern. Med. 2013, 274, 113–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, P.-G.; Lee, J.-E.; Cho, Y.-E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.-I.; Kim, Y.-B.; Kim, I.-S.; Park, H.Y.; et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin. Cancer Res. 2016, 22, 1757–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandy, D.; Rajam, S.M.; Dutta, D. A three layered histone epigenetics in breast cancer metastasis. Cell Biosci. 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revillion, F.; Pawlowski, V.; Hornez, L.; Peyrat, J.P. Glyceraldehyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur. J. Cancer 2000, 36, 1038–1042. [Google Scholar] [CrossRef]
- Stresing, V.; Baltziskueta, E.; Rubio, N.; Blanco, J.; Arriba, M.C.; Valls, J.; Janier, M.; Clezardin, P.; Sanz-Pamplona, R.; Nieva, C.; et al. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene 2013, 32, 724–735. [Google Scholar] [CrossRef]
- Duijvesz, D.; Rodriguez-Blanco, G.; Hoogland, A.M.; Verhoef, E.I.; Dekker, L.J.; Roobol, M.J.; van Leenders, G.; Luider, T.M.; Jenster, G. Differential tissue expression of extracellular vesicle-derived proteins in prostate cancer. Prostate 2019, 79, 1032–1042. [Google Scholar] [CrossRef]
- Peng, L.; Wang, R.; Shang, J.; Xiong, Y.; Fu, Z. Peroxiredoxin 2 is associated with colorectal cancer progression and poor survival of patients. Oncotarget 2017, 8, 15057–15070. [Google Scholar] [CrossRef] [Green Version]
- Sivanand, S.; Vander Heiden, M.G. Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell 2020, 37, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Kanehira, M.; Katagiri, T.; Shimo, A.; Takata, R.; Shuin, T.; Miki, T.; Fujioka, T.; Nakamura, Y. Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 2007, 67, 3276–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Li, Y.; Zhang, X.; Liu, X.-Y.; Peng, A.; Chen, Y.; Meng, L.; Chen, H.; Zhang, Y.; Miao, X.; et al. Inhibition of kinesin family member 20B sensitizes hepatocellular carcinoma cell to microtubule-targeting agents by blocking cytokinesis. Cancer Sci. 2018, 109, 3450–3460. [Google Scholar] [CrossRef] [PubMed]
- Balci, H.; Genc, H.; Papila, C.; Can, G.; Papila, B.; Yanardag, H.; Uzun, H. Serum lipid hydroperoxide levels and paraoxonase activity in patients with lung, breast, and colorectal cancer. J Clin. Lab. Anal. 2012, 26, 155–160. [Google Scholar] [CrossRef]
- Bobin-Dubigeon, C.; Jaffre, I.; Joalland, M.P.; Classe, J.M.; Campone, M.; Herve, M.; Bard, J.M. Paraoxonase 1 (PON1) as a marker of short term death in breast cancer recurrence. Clin. Biochem. 2012, 45, 1503–1505. [Google Scholar] [CrossRef] [PubMed]
- Okuturlar, Y.; Gunaldi, M.; Kocoglu, H.; Hursitoglu, M.; Gedikbasi, A.; Acarer, D.; Harmankaya, O.; Kumbasar, A. Serum paraoxonase and arylesterase can be useful markers to predict neoadjuvant chemotherapy requirement in patients with breast cancer. J. Cancer Res. Ther. 2018, 14, 362–367. [Google Scholar] [CrossRef]
- Ghosh, M.; Song, X.; Mouneimne, G.; Sidani, M.; Lawrence, D.S.; Condeelis, J.S. Cofilin promotes actin polymerization and defines the direction of cell motility. Science 2004, 304, 743–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maimaiti, Y.; Tan, J.; Liu, Z.; Guo, Y.; Yan, Y.; Nie, X.; Huang, B.; Zhou, J.; Huang, T. Overexpression of cofilin correlates with poor survival in breast cancer: A tissue microarray analysis. Oncol. Lett. 2017, 14, 2288–2294. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
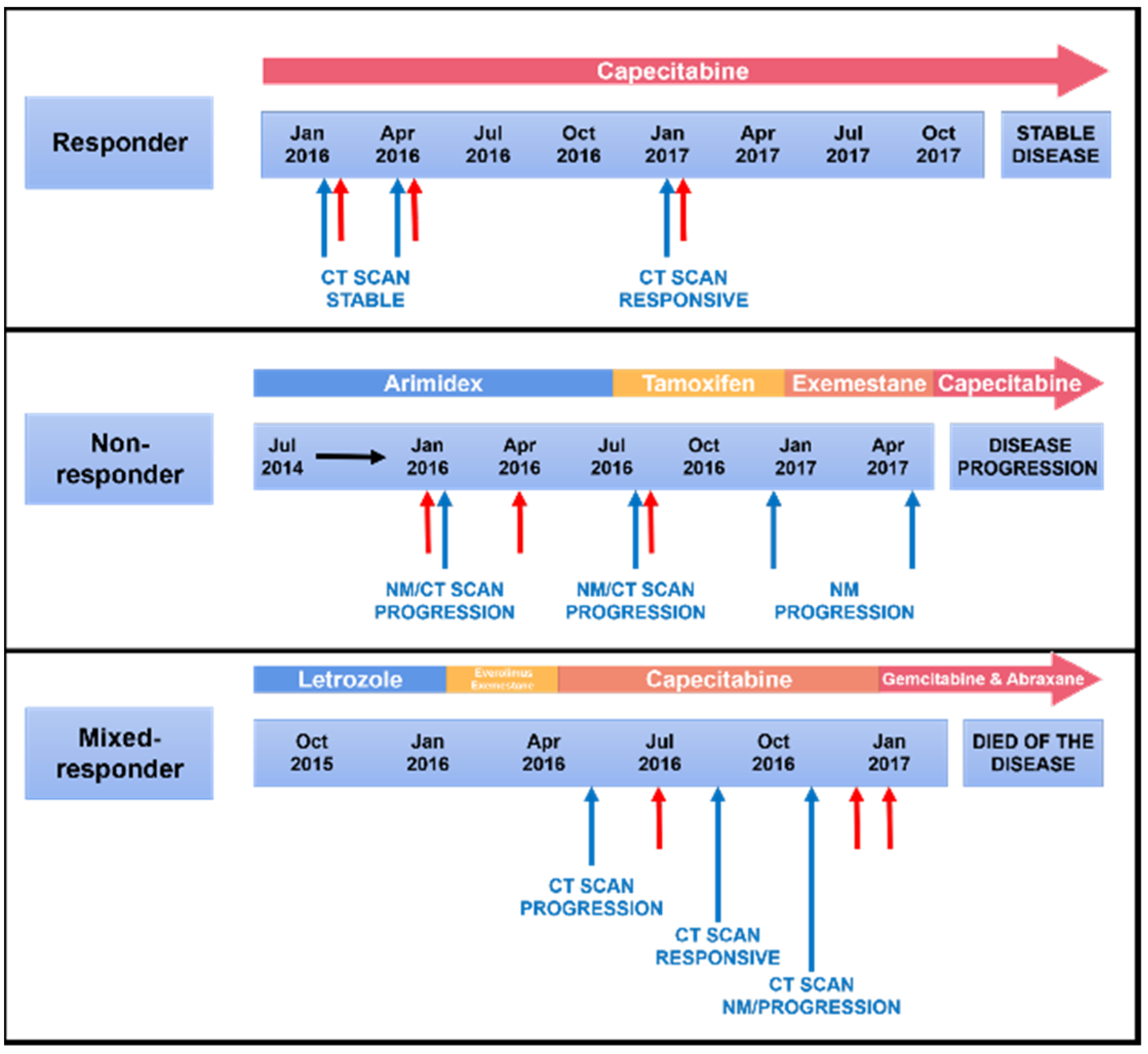
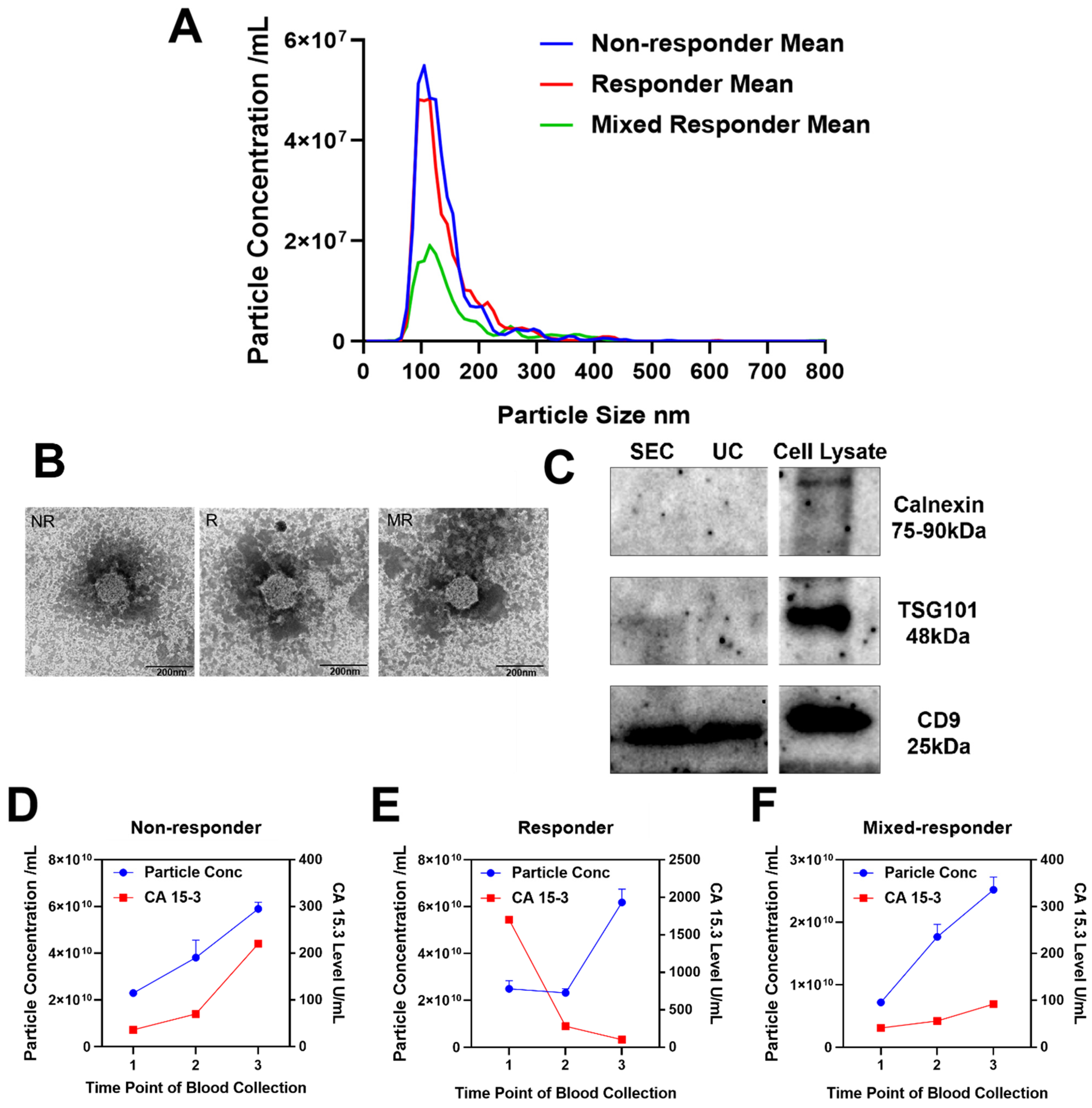


| Responder | Non-Responder | Mixed-Responder | |
|---|---|---|---|
| Age | 65 | 68 | 69 |
| Sex | Female | Female | Female |
| Cancer Staging | IV | IV | IV |
| Histopathology | IDC, grade 2 | IDC, grade 3 | IDC, grade 3 |
| Receptor status | ER positive PR positive HER2 non-amplified | ER positive PR positive HER2 non-amplified | ER 100% positive PR 10% positive HER2 non-amplified |
| Treatment | Endocrine treatment Chemotherapy | Endocrine treatment | Chemotherapy Radiotherapy |
| Response | Response at all time points | Progression at all time points | Response then progression |
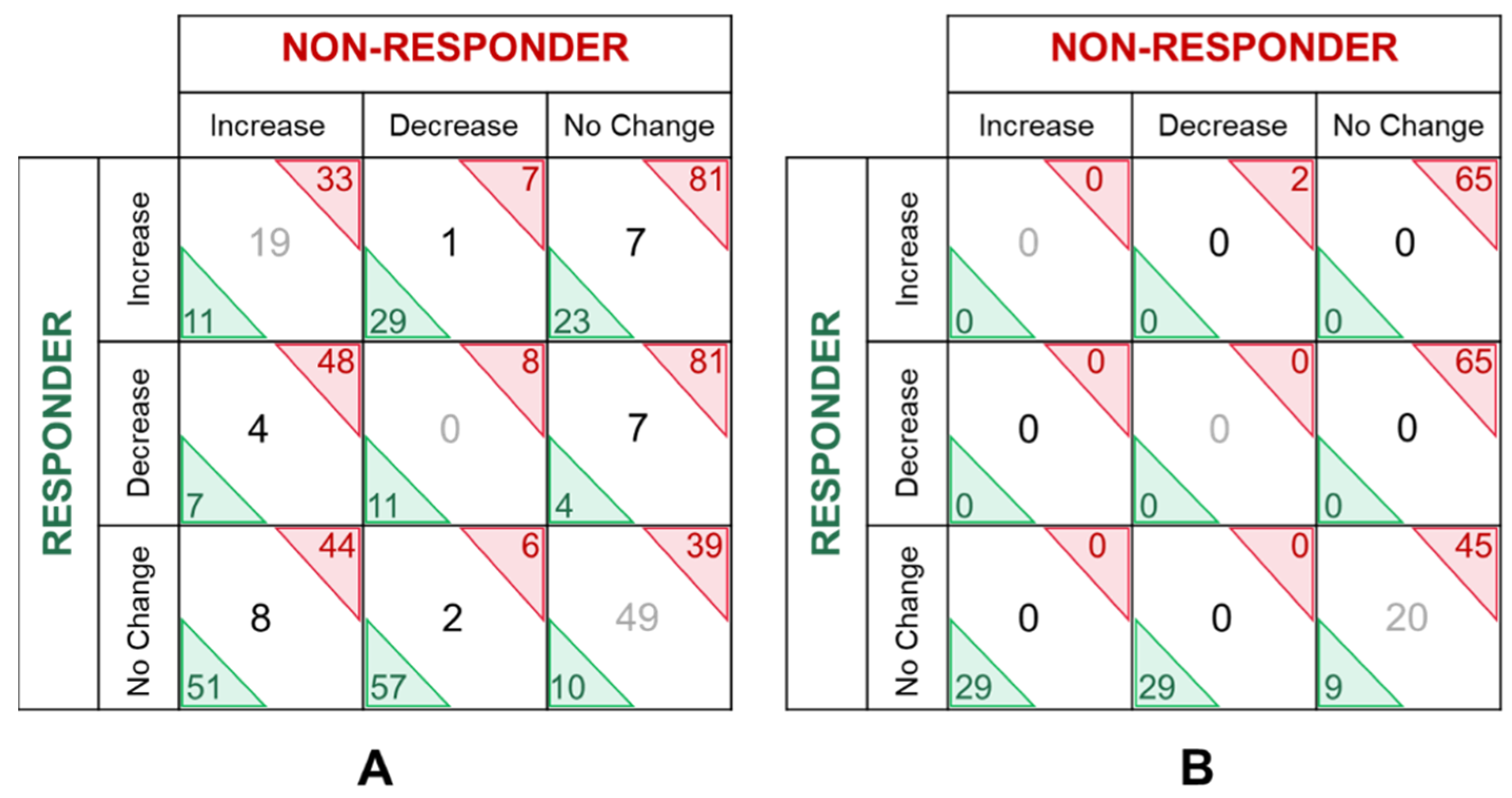
| Increase | Decrease | No Change | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical Response | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points |
| Non-Responder | 2 | 55 | 64 | 52 | 12 | 12 | 5 | 8 | 135 | 82 | 80 | 88 |
| Responder | 61 | 33 | 34 | 30 | 24 | 14 | 54 | 11 | 64 | 102 | 61 | 59 |
| Mixed Responder | 45 | 38 | 15 | 35 | 60 | 53 | 30 | 48 | 44 | 58 | 104 | 61 |
| Non-Responder | Responder | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw Abundance | Fold Change | Raw Abundance | Fold Change | ||||||||||
| Accession | Description | NR1 | NR2 | NR3 | NR1:2 | NR1:3 | NR2:3 | R1 | R2 | R3 | R1:2 | R1:3 | R2:3 |
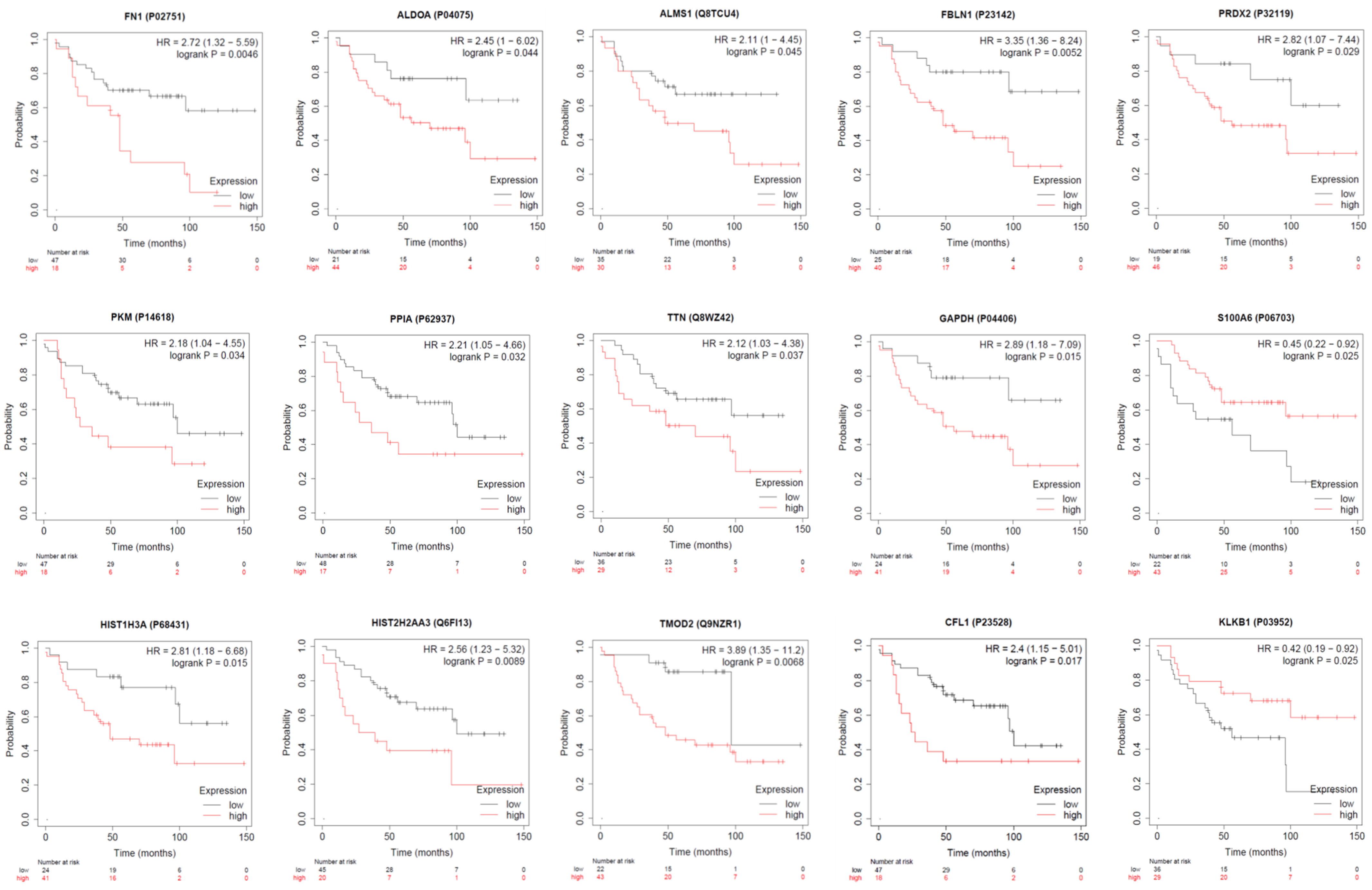
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 226.3 | 88.3 | 184.8 | 0.390 | 0.817 | 2.093 | 31.6 | 147.9 | 48.9 | 4.680 | 1.547 | 0.331 |
| Q6FI13 | Histone H2A type 2-A | 153.6 | 48.9 | 187 | 0.318 | 1.217 | 3.824 | 30.2 | 140.4 | 83.4 | 4.649 | 2.762 | 0.594 |
| Q13201 | Multimerin-1 | 132 | 57.1 | 95.3 | 0.433 | 0.722 | 1.669 | 92 | 38.4 | 215.9 | 0.417 | 2.347 | 5.622 |
| P32119 | Peroxiredoxin-2 | 122.4 | 93.9 | 89.9 | 0.767 | 0.734 | 0.957 | 10.4 | 150.5 | 18.6 | 14.471 | 1.788 | 0.124 |
| P11277 | Spectrin beta chain | 198.9 | 108.9 | 142.8 | 0.548 | 0.718 | 1.311 | 34.2 | 123.2 | 143.2 | 3.602 | 4.187 | 1.162 |
| Non-Responder | Responder | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw Abundance | Fold Change | Raw Abundance | Fold Change | ||||||||||
| Accession | Description | NR1 | NR2 | NR3 | NR1:2 | NR1:3 | NR2:3 | R1 | R2 | R3 | R1:2 | R1:3 | R2:3 |
| Q96H78 | Solute carrier family 25 member 44 | 299.4 | 90.8 | 115 | 0.303 | 0.384 | 1.267 | 82.7 | 44.9 | 78.7 | 0.543 | 0.952 | 1.753 |
| P27169 | Paraoxonase/arylesterase 1 | 141.7 | 141.3 | 87 | 0.997 | 0.614 | 0.616 | 95.5 | 58 | 96.1 | 0.607 | 1.006 | 1.657 |
| Q96Q89 | Kinesin-like protein KIF20B | 101.5 | 232 | 61.5 | 2.286 | 0.606 | 0.265 | 205 | 65.8 | 155.2 | 0.321 | 0.757 | 2.359 |
| P20851 | C4b-binding protein beta chain | 136.3 | 130.5 | 54.2 | 0.957 | 0.398 | 0.415 | 150.7 | 53.4 | 101.5 | 0.354 | 0.674 | 1.901 |
| Q8WZ42 | Titin | 151.8 | 66.7 | 94.1 | 0.439 | 0.620 | 1.411 | 54.1 | 113.5 | 141.4 | 2.098 | 2.614 | 1.246 |
| P02730 | Band 3 anion transport protein | 175.5 | 90.6 | 76.3 | 0.516 | 0.435 | 0.842 | 59.6 | 88.1 | 109.9 | 1.478 | 1.844 | 1.247 |
| P08697 | Alpha-2-antiplasmin | 135.4 | 114.5 | 71.3 | 0.846 | 0.527 | 0.623 | 119.8 | 125.4 | 93.5 | 1.047 | 0.780 | 0.746 |
| Non-Responder | Responder | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw Abundance | Fold Change | Raw Abundance | Fold Change | ||||||||||
| Accession | Description | NR1 | NR2 | NR3 | NR1:2 | NR1:3 | NR2:3 | R1 | R2 | R3 | R1:2 | R1:3 | R2:3 |
| O43866 | CD5 antigen-like | 69.1 | 79.9 | 184.8 | 1.156 | 2.674 | 2.313 | 72.5 | 231.2 | 94.6 | 3.189 | 1.305 | 0.409 |
| O60814 | Histone H2B type 1-K | 125.8 | 63 | 323.2 | 0.501 | 2.569 | 5.130 | 34.5 | 172.4 | 47.7 | 4.997 | 1.383 | 0.277 |
| P02751 | Fibronectin | 64.3 | 80.5 | 126.2 | 1.252 | 1.963 | 1.568 | 60.9 | 139.6 | 78.4 | 2.292 | 1.287 | 0.562 |
| P04075 | Fructose-bisphosphate aldolase A | 64.6 | 81.3 | 266.4 | 1.259 | 4.124 | 3.277 | 85.2 | 259.5 | 74.5 | 3.046 | 0.874 | 0.287 |
| P06681 | Complement C2 | 95.8 | 78.9 | 189.8 | 0.824 | 1.981 | 2.406 | 88.5 | 200.1 | 64.6 | 2.261 | 0.730 | 0.323 |
| P06703 | Protein S100-A6 | 82.8 | 111 | 291.1 | 1.341 | 3.516 | 2.623 | 58.3 | 183.3 | 63.5 | 3.144 | 1.089 | 0.346 |
| P07358 | Complement component C8 beta chain | 58.4 | 54.4 | 128.9 | 0.932 | 2.207 | 2.369 | 122.9 | 280.1 | 154.8 | 2.279 | 1.260 | 0.553 |
| P12109 | Collagen alpha-1(VI) chain | 64 | 61.1 | 104.5 | 0.955 | 1.633 | 1.710 | 95.6 | 217.3 | 114.1 | 2.273 | 1.194 | 0.525 |
| P14618 | Pyruvate kinase PKM | 60.9 | 72.4 | 212.1 | 1.189 | 3.483 | 2.930 | 64.5 | 156.1 | 91 | 2.420 | 1.411 | 0.583 |
| P14625 | Endoplasmin | 85.8 | 89.8 | 223.6 | 1.047 | 2.606 | 2.490 | 53.7 | 189.7 | 56.1 | 3.533 | 1.045 | 0.296 |
| P55201 | Peregrin | 2.5 | 2.2 | 4.8 | 0.880 | 1.920 | 2.182 | 3.5 | 6.4 | 4.0 | 1.829 | 1.143 | 0.625 |
| P62937 | Peptidyl-prolyl cis-trans isomerase A | 81 | 75.5 | 235.3 | 0.932 | 2.905 | 3.117 | 60.3 | 156.9 | 89.9 | 2.602 | 1.491 | 0.573 |
| P62979 | Ubiquitin-40S ribosomal protein S27a | 68 | 66.1 | 124.2 | 0.972 | 1.826 | 1.879 | 92.5 | 174.3 | 110.4 | 1.884 | 1.194 | 0.633 |
| P68431 | Histone H3.1 | 54.2 | 60.9 | 263.4 | 1.124 | 4.860 | 4.325 | 48.6 | 174.5 | 64.3 | 3.591 | 1.323 | 0.368 |
| Q13093 | Platelet-activating factor acetylhydrolase | 97.6 | 89.6 | 189.4 | 0.918 | 1.941 | 2.114 | 89 | 215.2 | 93.1 | 2.418 | 1.046 | 0.433 |
| Q8TCU4 | Alstrom syndrome protein 1 | 72.4 | 82.1 | 235.4 | 1.134 | 3.251 | 2.867 | 72.2 | 282.6 | 85.8 | 3.914 | 1.188 | 0.304 |
| Q9NZR1 | Tropomodulin-2 | 63.9 | 49.3 | 604.3 | 0.772 | 9.457 | 12.258 | 27.3 | 175.8 | 10.6 | 6.440 | 0.388 | 0.060 |
| P08670 | Vimentin | 101.5 | 59.9 | 316.7 | 0.590 | 3.120 | 5.287 | 31.8 | 184.9 | 19.2 | 5.814 | 0.604 | 0.104 |
| P23142 | Fibulin-1 | 115.9 | 115.9 | 204.1 | 1.000 | 1.761 | 1.761 | 52.7 | 127.1 | 35.0 | 2.412 | 0.664 | 0.275 |
| P23528 | Cofilin-1 | 41.8 | 54 | 229.9 | 1.292 | 5.500 | 4.257 | 71.7 | 101.7 | 81.9 | 1.418 | 1.142 | 0.805 |
| P68363 | Tubulin alpha-1B chain | 58.1 | 72.5 | 252.9 | 1.248 | 4.353 | 3.488 | 100.2 | 76.5 | 105.4 | 0.763 | 1.052 | 1.378 |
| Q9BWP8 | Collectin-11 | 85.2 | 82.1 | 193.5 | 0.964 | 2.271 | 2.357 | 91 | 132.9 | 97.9 | 1.460 | 1.076 | 0.737 |
| Increase | Decrease | No Change | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical Response | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points | 1–2 | 1–3 | 2–3 | Overlap ≥ 2 Time Points |
| Non-Responder | 11 | 20 | 16 | 0 | 6 | 37 | 35 | 2 | 120 | 80 | 86 | 65 |
| Responder | 47 | 30 | 40 | 0 | 32 | 17 | 48 | 0 | 58 | 90 | 49 | 29 |
| Mixed-Responder | 33 | 50 | 24 | 2 | 47 | 29 | 7 | 5 | 57 | 58 | 106 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruhen, O.; Qu, X.; Jamaluddin, M.F.B.; Salomon, C.; Gandhi, A.; Millward, M.; Nixon, B.; Dun, M.D.; Meehan, K. Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer. Membranes 2021, 11, 880. https://doi.org/10.3390/membranes11110880
Ruhen O, Qu X, Jamaluddin MFB, Salomon C, Gandhi A, Millward M, Nixon B, Dun MD, Meehan K. Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer. Membranes. 2021; 11(11):880. https://doi.org/10.3390/membranes11110880
Chicago/Turabian StyleRuhen, Olivia, Xinyu Qu, M. Fairuz B. Jamaluddin, Carlos Salomon, Aesha Gandhi, Michael Millward, Brett Nixon, Matthew D. Dun, and Katie Meehan. 2021. "Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer" Membranes 11, no. 11: 880. https://doi.org/10.3390/membranes11110880
APA StyleRuhen, O., Qu, X., Jamaluddin, M. F. B., Salomon, C., Gandhi, A., Millward, M., Nixon, B., Dun, M. D., & Meehan, K. (2021). Dynamic Landscape of Extracellular Vesicle-Associated Proteins Is Related to Treatment Response of Patients with Metastatic Breast Cancer. Membranes, 11(11), 880. https://doi.org/10.3390/membranes11110880









