Amphipathic Peptides Impede Lipid Domain Fusion in Phase-Separated Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Elastic Moduli
2.2. Parameterization of the System
3. Results
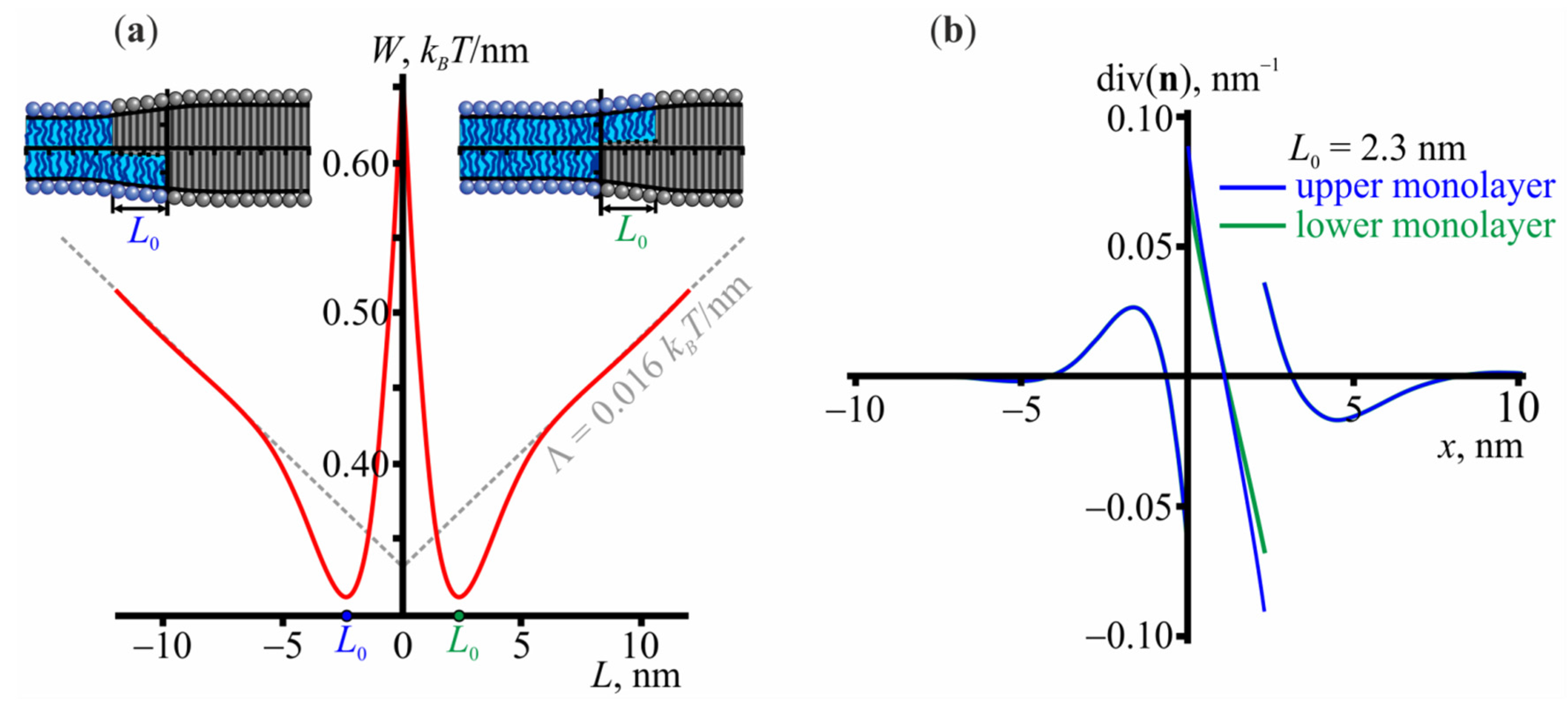
3.1. Structure of the Lo/Ld Interface
3.2. Interaction of Amphipathic Peptides with the Domain Boundary
3.3. Membrane-Mediated Interaction of Amphipathic Peptides in a Homogeneous Membrane
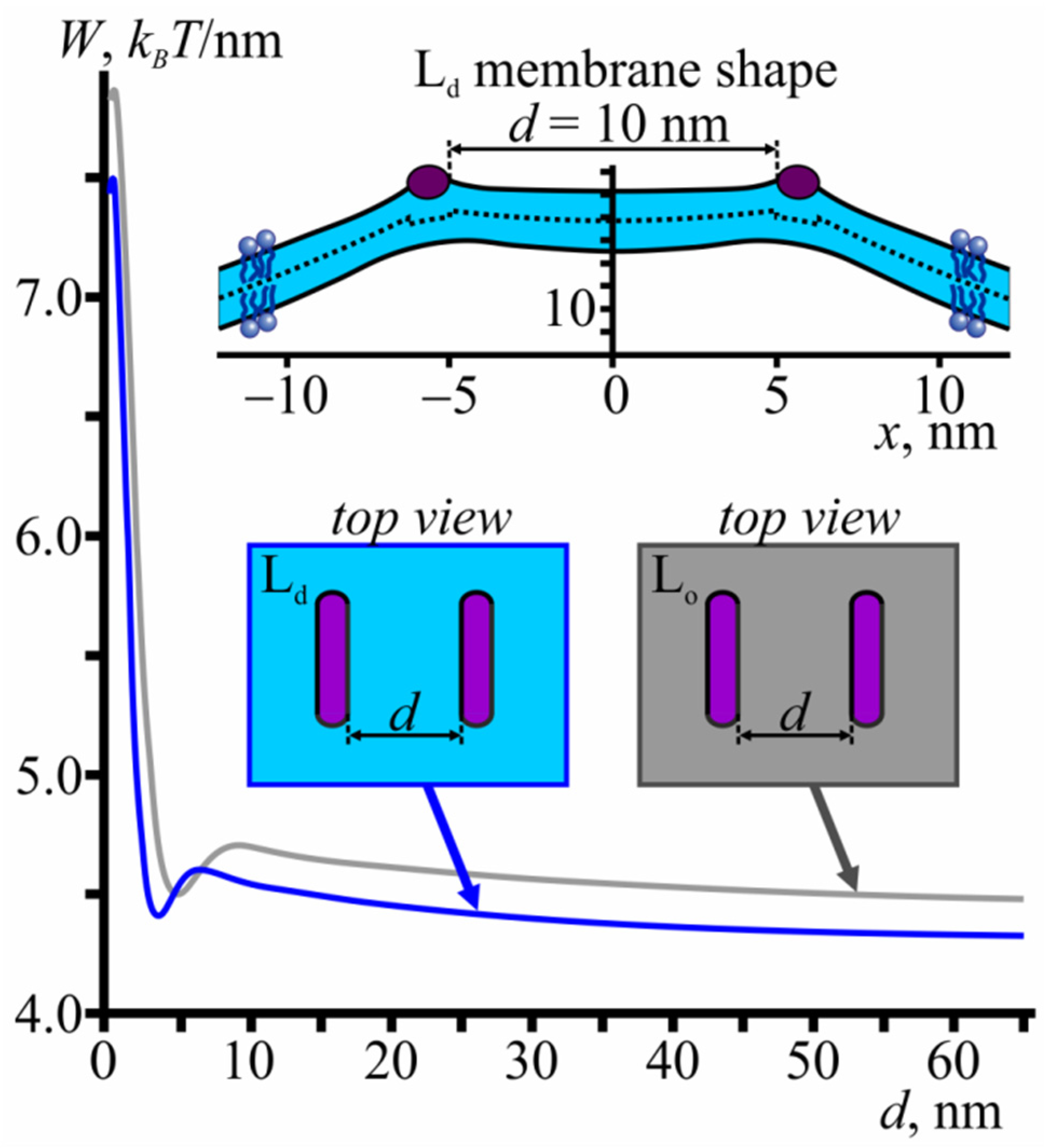
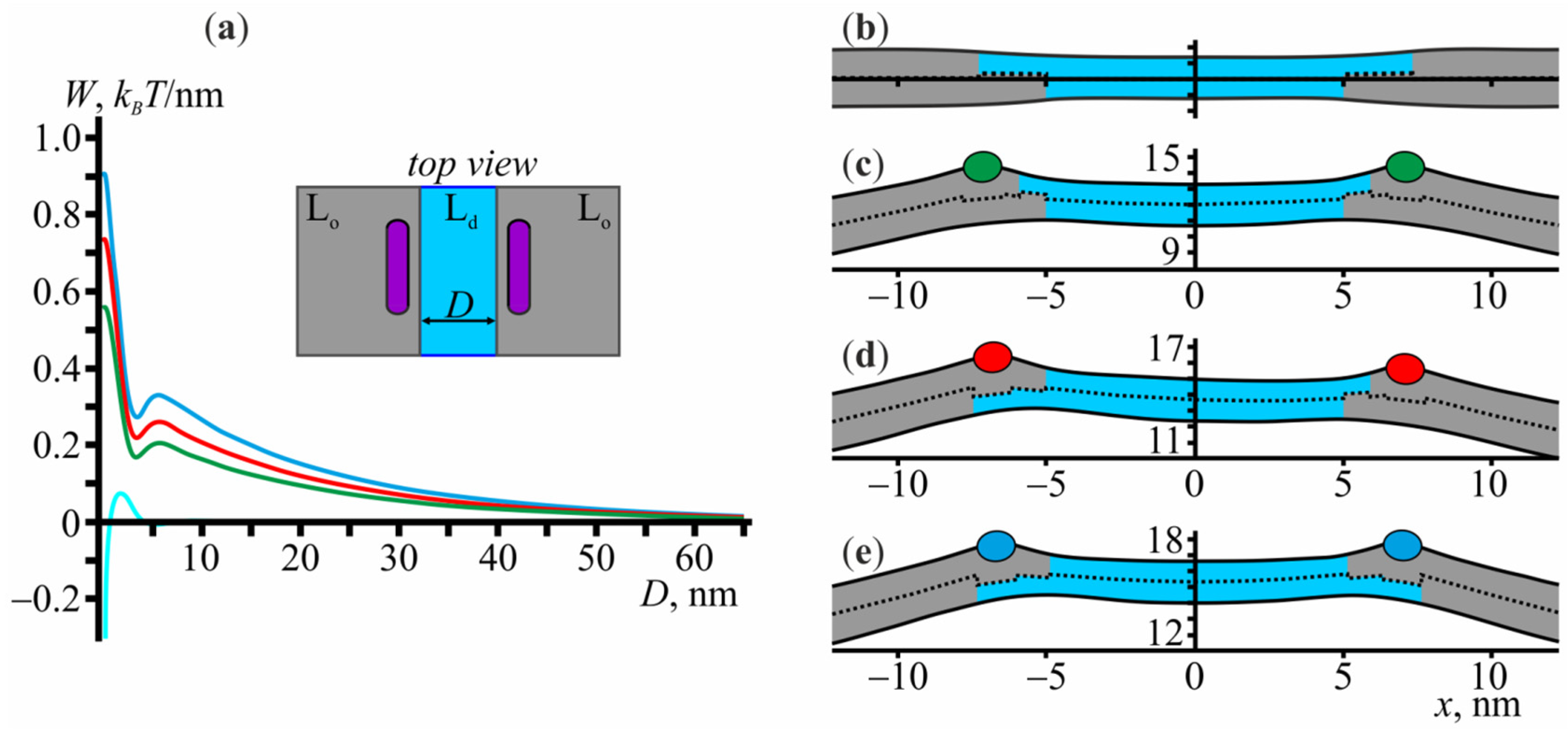
3.4. Membrane-Mediated Interaction of Lo Domains
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Lorent, J.H.; Levental, K.R.; Ganesan, L.; Rivera-Longsworth, G.; Sezgin, E.; Doktorova, M.; Lyman, E.; Levental, I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020, 16, 710. [Google Scholar] [CrossRef] [PubMed]
- Ayuyan, A.G.; Cohen, F.S. Raft Composition at physiological temperature and ph in the absence of detergents. Biophys. J. 2008, 94, 2654–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisz, J.F.; Lou, K.; Klitzing, H.A.; Hanafin, W.P.; Lizunov, V.; Wilson, R.L.; Carpenter, K.J.; Kim, R.; Hutcheon, I.D.; Zimmerberg, J.; et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. USA 2013, 110, E613–E622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisz, J.F.; Klitzing, H.A.; Lou, K.; Hutcheon, I.D.; Weber, P.K.; Zimmerberg, J.; Kraft, M.L. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J. Biol. Chem. 2013, 288, 16855–16861. [Google Scholar] [CrossRef] [Green Version]
- Owen, D.M.; Williamson, D.; Magenau, A.; Gaus, K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat. Commun. 2012, 3, 1256. [Google Scholar] [CrossRef] [Green Version]
- Nickels, J.; Chatterjee, S.; Stanley, C.; Qian, S.; Cheng, X.; Myles, D.A.A.; Standaert, R.F.; Elkins, J.G.; Katsaras, J. The in vivo structure of biological membranes and evidence for lipid domains. PLoS Biol. 2017, 15, e2002214. [Google Scholar] [CrossRef] [Green Version]
- Toulmay, A.; Prinz, W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013, 202, 35–44. [Google Scholar] [CrossRef]
- Baumgart, T.; Hammond, A.T.; Sengupta, P.; Hess, S.T.; Holowka, D.A.; Baird, B.A.; Webb, W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA 2007, 104, 3165–3170. [Google Scholar] [CrossRef] [Green Version]
- Lillemeier, B.F.; Pfeiffer, J.R.; Surviladze, Z.; Wilson, B.S.; Davis, M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA 2006, 103, 18992–18997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, C.; Bagatolli, L.; Volovyk, Z.; Thompson, N.; Levi, M.; Jacobson, K.; Gratton, E. Lipid Rafts Reconstituted in Model Membranes. Biophys. J. 2001, 80, 1417–1428. [Google Scholar] [CrossRef] [Green Version]
- Samsonov, A.V.; Mihalyov, I.; Cohen, F.S. Characterization of Cholesterol-Sphingomyelin Domains and Their Dynamics in Bilayer Membranes. Biophys. J. 2001, 81, 1486–1500. [Google Scholar] [CrossRef] [Green Version]
- Veatch, S.; Polozov, I.; Gawrisch, K.; Keller, S. Liquid domains in vesicles investigated by nmr and fluorescence microscopy. Biophys. J. 2004, 86, 2910–2922. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, T.; Hunt, G.; Farkas, E.R.; Webb, W.W.; Feigenson, G.W. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 2182–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staneva, G.; Puff, N.; Seigneuret, M.; Conjeaud, H.; Angelova, M.I. Segregative clustering of lo and ld membrane microdomains induced by local ph gradients in gm1-containing giant vesicles: A lipid model for cellular polarization. Langmuir 2012, 28, 16327–16337. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Molotkovsky, R.J.; Alexandrova, V.V.; Galimzyanov, T.R.; Jiménez-Munguía, I.; Pavlov, K.V.; Batishchev, O.V.; Akimov, S.A. Lateral membrane heterogeneity regulates viral-induced membrane fusion during HIV entry. Int. J. Mol. Sci. 2018, 19, 1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorent, J.; Levental, I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 2015, 192, 23–32. [Google Scholar] [CrossRef]
- Allen, J.A.; Halverson-Tamboli, R.A.; Rasenick, M.M. Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 2006, 8, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-T.; Kiessling, V.; Simmons, J.A.; White, J.M.; Tamm, L.K. HIV gp41–mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat. Chem. Biol. 2015, 11, 424–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.W.; Lemli, L.; Opitz, J.M. A newly recognized syndromeof multiple congenital anomalies. J. Pediatr. 1964, 64, 210–217. [Google Scholar] [CrossRef]
- Porter, F.D. Smith–Lemli–Opitz syndrome: Pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 2008, 16, 535–541. [Google Scholar] [CrossRef] [Green Version]
- Staneva, G.; Osipenko, D.S.; Galimzyanov, T.; Pavlov, K.V.; Akimov, S.A. Metabolic precursor of cholesterol causes formation of chained aggregates of liquid-ordered domains. Langmuir 2016, 32, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Varshney, P.; Yadav, V.; Saini, N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Mollinedo, F.; Gajate, C. Lipid rafts as major platforms for signaling regulation in cancer. Adv. Biol. Regul. 2015, 57, 130–146. [Google Scholar] [CrossRef]
- Schuck, S.; Simons, K. Polarized sorting in epithelial cells: Raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 2004, 117, 5955–5964. [Google Scholar] [CrossRef] [Green Version]
- Levental, I.; Grzybek, M.; Simons, K. Greasing their way: Lipid modifications determine protein association with membrane rafts. Biochemistry 2010, 49, 6305–6316. [Google Scholar] [CrossRef] [PubMed]
- Honda, A.; Ito, Y.; Takahashi-Niki, K.; Matsushita, N.; Nozumi, M.; Tabata, H.; Takeuchi, K.; Igarashi, M. Extracellular signals induce glycoprotein M6a clustering of lipid rafts and associated signaling molecules. J. Neurosci. 2017, 37, 4046–4064. [Google Scholar] [CrossRef] [PubMed]
- Altrock, E.; Muth, C.A.; Klein, G.; Spatz, J.P.; Lee-Thedieck, C. The significance of integrin ligand nanopatterning on lipid raft clustering in hematopoietic stem cells. Biomaterials 2012, 33, 3107–3118. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [Green Version]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic α helical antimicrobial peptides. Biopolymers 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Guha, S.; Ghimire, J.; Wu, E.; Wimley, W.C. Mechanistic landscape of membrane-permeabilizing peptides. Chem. Rev. 2019, 119, 6040–6085. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, B.; Gorr, S.-U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2016, 96, 254–260. [Google Scholar] [CrossRef] [Green Version]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Pept. Sci. 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Epand, R.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta (BBA)-Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef] [Green Version]
- Pinigin, K.V.; Kondrashov, O.; Jiménez-Munguía, I.; Alexandrova, V.V.; Batishchev, O.; Galimzyanov, T.R.; Akimov, S.A. Elastic deformations mediate interaction of the raft boundary with membrane inclusions leading to their effective lateral sorting. Sci. Rep. 2020, 10, 4087. [Google Scholar] [CrossRef]
- Rinia, H.A.; Snel, M.M.; van der Eerden, J.P.; de Kruijff, B. Visualizing detergent resistant domains in model membranes with atomic force microscopy. FEBS Lett. 2001, 501, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Saslowsky, D.E.; Lawrence, J.; Ren, X.; Brown, D.A.; Henderson, R.M.; Edwardson, J.M. Placental alkaline phosphatase is efficiently targeted to rafts in supported lipid bilayers. J. Biol. Chem. 2002, 277, 26966–26970. [Google Scholar] [CrossRef] [Green Version]
- Risselada, H.J.; Marrink, S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 17367–17372. [Google Scholar] [CrossRef] [Green Version]
- Kuzmin, P.I.; Akimov, S.A.; Chizmadzhev, Y.A.; Zimmerberg, J.; Cohen, F.S. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys. J. 2005, 88, 1120–1133. [Google Scholar] [CrossRef] [Green Version]
- Galimzyanov, T.R.; Molotkovsky, R.J.; Bozdaganyan, M.E.; Cohen, F.S.; Pohl, P.; Akimov, S.A. Elastic membrane deformations govern interleaflet coupling of lipid-ordered domains. Phys. Rev. Lett. 2015, 115, 088101. [Google Scholar] [CrossRef] [Green Version]
- Galimzyanov, T.R.; Molotkovsky, R.J.; Cohen, F.S.; Pohl, P.; Akimov, S.A. Galimzyanov et al. Reply. Phys. Rev. Lett. 2016, 116, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimov, S.A.; Aleksandrova, V.V.; Galimzyanov, T.R.; Batishchev, F.V.; Batishchev, O.V. Interaction of amphipathic peptides mediated by elastic membrane deformations. Biol. Membr. 2017, 34, 162–173. [Google Scholar] [CrossRef]
- Kondrashov, O.V.; Galimzyanov, T.R.; Jiménez-Munguía, I.; Batishchev, O.V.; Akimov, S.A. Membrane-mediated interaction of amphipathic peptides can be described by a one-dimensional approach. Phys. Rev. E 2019, 99, 022401. [Google Scholar] [CrossRef] [PubMed]
- Pinigin, K.V.; Kuzmin, P.I.; Akimov, S.A.; Galimzyanov, T.R. Additional contributions to elastic energy of lipid membranes: Tilt-curvature coupling and curvature gradient. Phys. Rev. E 2020, 102, 042406. [Google Scholar] [CrossRef] [PubMed]
- Galimzyanov, T.R.; Molotkovsky, R.J.; Kheyfets, B.B.; Akimov, S.A. Energy of the interaction between membrane lipid domains calculated from splay and tilt deformations. JETP Lett. 2013, 96, 681–686. [Google Scholar] [CrossRef]
- Landau, L.D.; Lifshitz, E.M. Course of Theoretical Physics Vol 7: Theory and Elasticity; Pergamon Press: Oxford, UK, 1975. [Google Scholar]
- Hamm, M.; Kozlov, M.M. Elastic energy of tilt and bending of fluid membranes. Eur. Phys. J. E 2000, 3, 323–335. [Google Scholar] [CrossRef]
- Terzi, M.M.; Deserno, M. Novel tilt-curvature coupling in lipid membranes. J. Chem. Phys. 2017, 147, 084702. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Rawicz, W. Entropy-driven tension and bending elasticity in condensed-fluid membranes. Phys. Rev. Lett. 1990, 64, 2094–2097. [Google Scholar] [CrossRef]
- Pan, J.; Tristram-Nagle, S.; Nagle, J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E 2009, 80, 021931. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, T.; Das, S.; Webb, W.W.; Jenkins, J.T. Membrane elasticity in giant vesicles with fluid phase coexistence. Biophys. J. 2005, 89, 1067–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawicz, W.; Olbrich, K.; McIntosh, T.; Needham, D.; Evans, E. Effect of chain length and unsaturation on elasticity of lipid Bilayers. Biophys. J. 2000, 79, 328–339. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.; Homann, U. Cell surface area regulation and membrane tension. J. Membr. Biol. 2001, 179, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Braganza, L.F.; Worcester, D.L. Structural changes in lipid bilayers and biological membranes caused by hydrostatic pressure. Biochemistry 1986, 25, 7484–7488. [Google Scholar] [CrossRef]
- Scarlata, S. Compression of lipid membranes as observed at varying membrane positions. Biophys. J. 1991, 60, 334–340. [Google Scholar] [CrossRef] [Green Version]
- Terzi, M.M.; Deserno, M.; Nagle, J.F. Mechanical properties of lipid bilayers: A note on the Poisson ratio. Soft Matter 2019, 15, 9085–9092. [Google Scholar] [CrossRef]
- Galimzyanov, T.R.; Kuzmin, P.I.; Pohl, P.; Akimov, S.A. Undulations drive domain registration from the two membrane leaflets. Biophys. J. 2017, 112, 339–345. [Google Scholar] [CrossRef] [Green Version]
- Galimzyanov, T.R.; Kalutsky, M.A.; Kondrashov, O.V.; Pinigin, K.V.; Molotkovsky, R.J.; Kuzmin, P.I.; Batishchev, O.V.; Akimov, S.A. Normal Fluctuations of Biological Membrane Shape as a Coupling Factor for Ordered Monolayer Domains. Biol. Membr 2019, 36, 184–191. [Google Scholar] [CrossRef]
- Blosser, M.C.; Honerkamp-Smith, A.; Han, T.; Haataja, M.; Keller, S.L. Transbilayer colocalization of lipid domains explained via measurement of strong coupling parameters. Biophys. J. 2015, 109, 2317–2327. [Google Scholar] [CrossRef] [Green Version]
- Israelachvili, J.N. Intermolecular and Surface Forces, 3rd ed.; Academic Press: Waltham, MA, USA, 2011. [Google Scholar]
- Pinigin, K.V.; Volovik, M.V.; Batishchev, O.V.; Akimov, S.A. Interaction of Ordered Lipid Domain Boundaries and Amphipathic Peptides Regulates Probability of Pore Formation in Membranes. Biol. Membr. 2020, 37, 337–349. [Google Scholar] [CrossRef]
- Usery, R.D.; Enoki, T.A.; Wickramasinghe, S.; Weiner, M.; Tsai, W.-C.; Kim, M.B.; Wang, S.; Torng, T.; Ackerman, D.G.; Heberle, F.; et al. Line tension controls liquid-disordered + liquid-ordered domain size transition in lipid bilayers. Biophys. J. 2017, 112, 1431–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitov, A.; Akimov, S.A.; Galimzyanov, T.R.; Glasnov, T.; Pohl, P. Ordered lipid domains assemble via concerted recruitment of constituents from both membrane leaflets. Phys. Rev. Lett. 2020, 124, 108102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frolov, V.A.J.; Chizmadzhev, Y.A.; Cohen, F.S.; Zimmerberg, J. “Entropic traps” in the kinetics of phase separation in multicomponent membranes stabilize nanodomains. Biophys. J. 2006, 91, 189–205. [Google Scholar] [CrossRef] [Green Version]
- Puff, N.; Watanabe, C.; Seigneuret, M.; Angelova, M.I.; Staneva, G. Lo/Ld phase coexistence modulation induced by GM1. Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2105–2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galimzyanov, T.R.; Lyushnyak, A.S.; Aleksandrova, V.V.; Shilova, L.A.; Mikhalyov, I.I.; Molotkovskaya, I.M.; Akimov, S.A.; Batishchev, O.V. Line activity of ganglioside gm1 regulates the raft size distribution in a cholesterol-dependent manner. Langmuir 2017, 33, 3517–3524. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinigin, K.V.; Galimzyanov, T.R.; Akimov, S.A. Amphipathic Peptides Impede Lipid Domain Fusion in Phase-Separated Membranes. Membranes 2021, 11, 797. https://doi.org/10.3390/membranes11110797
Pinigin KV, Galimzyanov TR, Akimov SA. Amphipathic Peptides Impede Lipid Domain Fusion in Phase-Separated Membranes. Membranes. 2021; 11(11):797. https://doi.org/10.3390/membranes11110797
Chicago/Turabian StylePinigin, Konstantin V., Timur R. Galimzyanov, and Sergey A. Akimov. 2021. "Amphipathic Peptides Impede Lipid Domain Fusion in Phase-Separated Membranes" Membranes 11, no. 11: 797. https://doi.org/10.3390/membranes11110797
APA StylePinigin, K. V., Galimzyanov, T. R., & Akimov, S. A. (2021). Amphipathic Peptides Impede Lipid Domain Fusion in Phase-Separated Membranes. Membranes, 11(11), 797. https://doi.org/10.3390/membranes11110797






