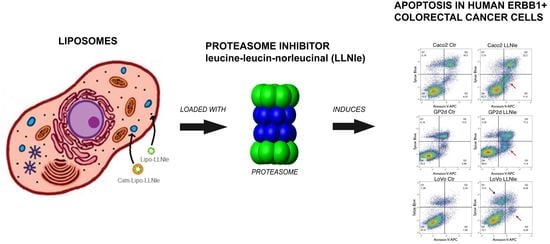
Liposomes Loaded with the Proteasome Inhibitor Z-Leucinyl-Leucinyl-Norleucinal Are Effective in Inducing Apoptosis in Colorectal Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Liposomes Preparation
2.4. Antibodies and Chemicals
2.5. Cell Cultures
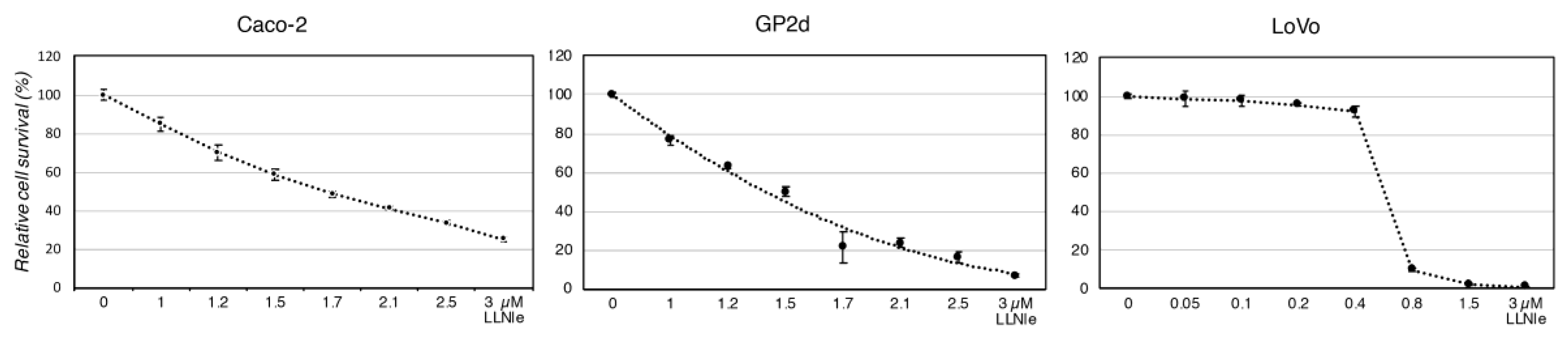
2.6. Cell Survival Assay
2.7. Immunofluorescence Analysis
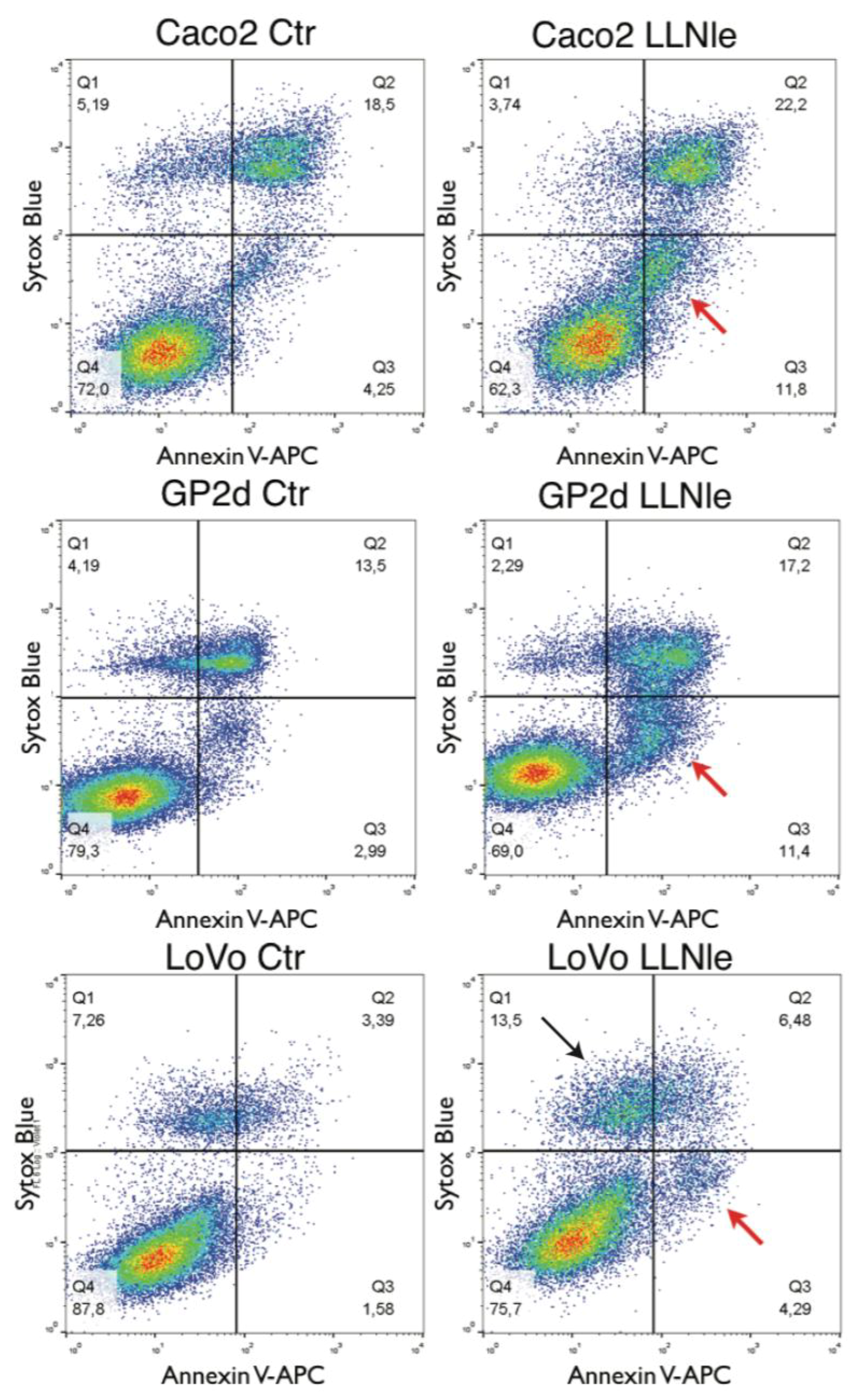
2.8. Flow Cytometry
2.9. Immunoblot Analysis
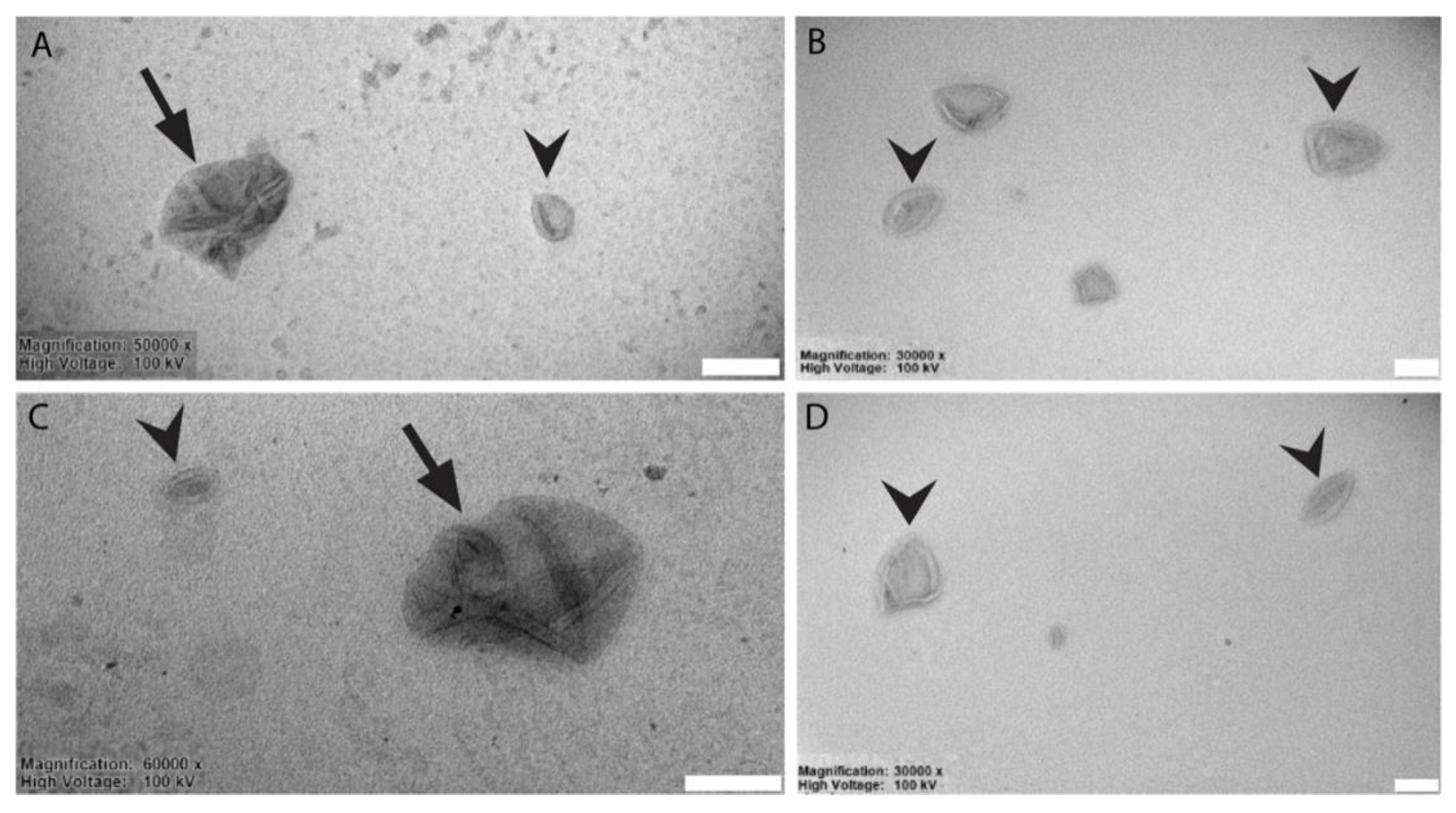
2.10. TEM Analysis
2.11. Statistical Analysis
3. Results and Discussion
3.1. LLNle Inhibits Cell Survival and Induces Apoptosis in CRC Cell Lines
3.2. Liposomes Generation and Chemical-Physical-Ultrastructural Characterization
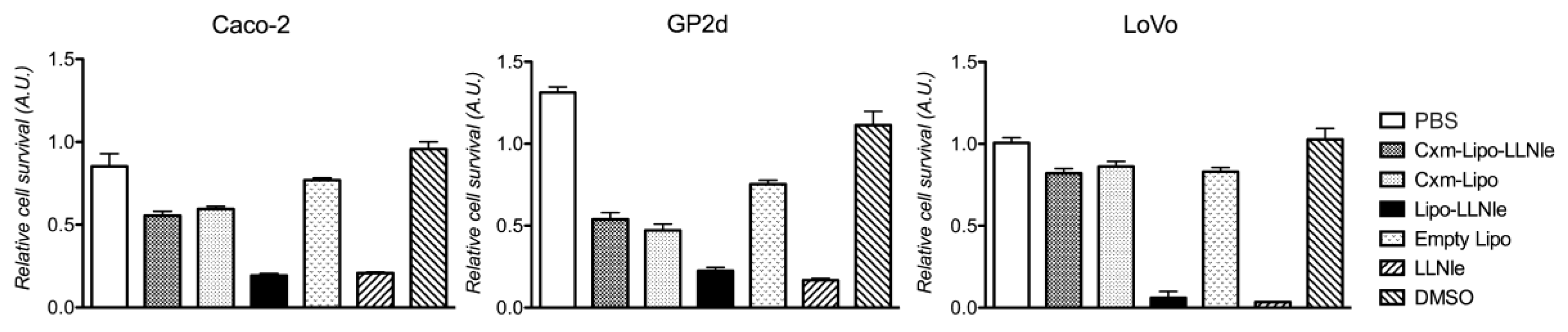
3.3. LLNle-Liposomes Reduce Cell Survival of CRC Cell Lines and Show Higher Inhibition Activity Compared to Immunoliposomes-LLNle
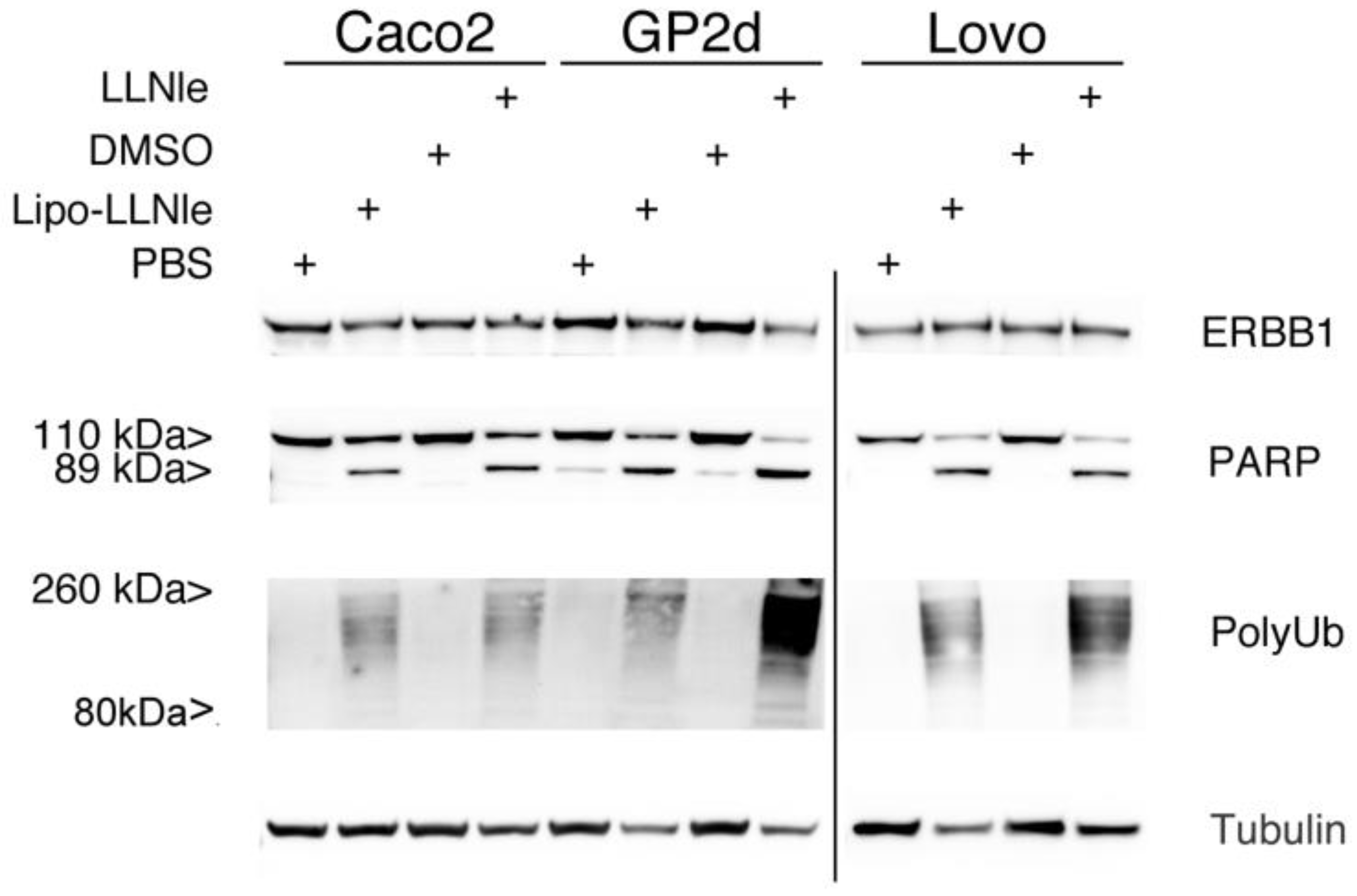
3.4. LLNle-Liposomes Inhibit Proteasomal Degradation and Induce Apoptosis in CRC Cell Lines in a Fashion Similar to Free LLNle
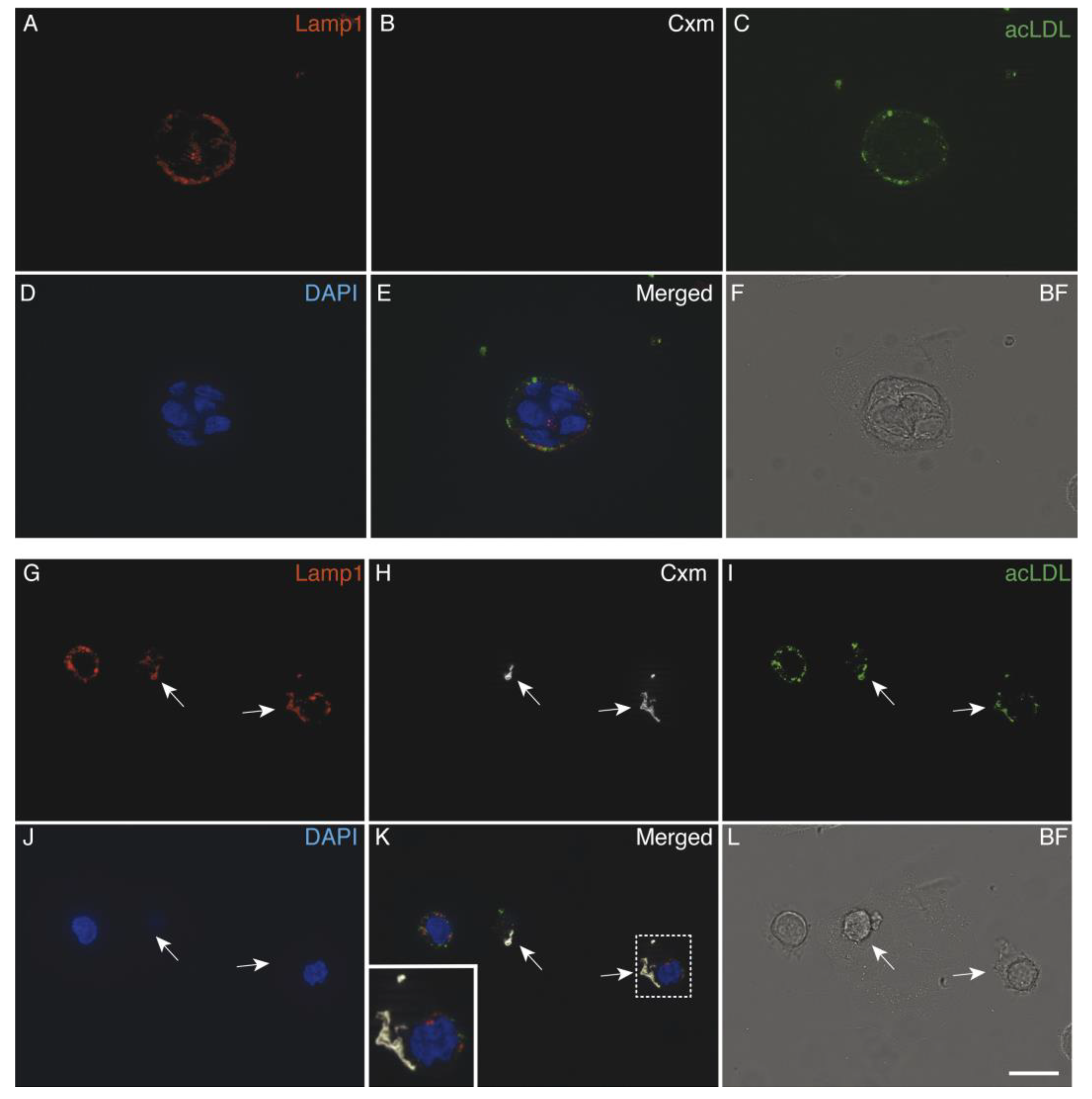
3.5. Immunoliposomes-LLNle Are Directed to the Endosomal/Lysosomal Compartment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Samad, A.; Sultana, Y.; Aqil, M. Liposomal Drug Delivery Systems: An Update Review. Curr. Drug Deliv. 2007, 4, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, J. Surface Engineering of Nanomaterials with Phospholipid-Polyethylene Glycol-Derived Functional Conjugates for Molecular Imaging and Targeted Therapy. Biomaterials 2020, 230, 119646. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Liu, L.; Xiang, Y.; Lu, Y.; Deng, L.; Zhang, H.; Santos, H.A.; Cui, W. Advanced Liposome-Loaded Scaffolds for Therapeutic and Tissue Engineering Applications. Biomaterials 2019, 232, 119706. [Google Scholar] [CrossRef] [PubMed]
- El-Aneed, A. An Overview of Current Delivery Systems in Cancer Gene Therapy. J. Control. Release 2004, 94, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Matlawska-Wasowska, K.; Girodon, F.; Mazel, T.; Willman, C.L.; Atlas, S.; Chen, I.-M.; Harvey, R.C.; Hunger, S.P.; Ness, S.A.; et al. GSI-I (Z-LLNle-CHO) Inhibits γ-Secretase and the Proteosome to Trigger Cell Death in Precursor-B Acute Lymphoblastic Leukemia. Leukemia 2011, 25, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Monticone, M.; Biollo, E.; Fabiano, A.; Fabbi, M.; Daga, A.; Romeo, F.; Maffei, M.; Melotti, A.; Giaretti, W.; Corte, G.; et al. Z-Leucinyl-Leucinyl-Norleucinal Induces Apoptosis of Human Glioblastoma Tumor-Initiating Cells by Proteasome Inhibition and Mitotic Arrest Response. Mol. Cancer Res. 2009, 7, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Nguyen, D.; Yang, S.X. Targeting Notch Signaling Pathway in Cancer: Clinical Development Advances and Challenges. Pharmacol. Ther. 2014, 141, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Gavazzoni, M.; Vizzardi, E.; Gorga, E.; Bonadei, I.; Rossi, L.; Belotti, A.; Rossi, G.; Ribolla, R.; Metra, M.; Raddino, R. Mechanism of Cardiovascular Toxicity by Proteasome Inhibitors: New Paradigm Derived from Clinical and Pre-Clinical Evidence. Eur. J. Pharmacol. 2018, 828, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global Cancer Statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Chu, E. Colorectal Cancer (CRC) Continues to Be a Major Public Health Problem in the United States and throughout the World. Cancer J. 2010, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Ciardiello, D.; Martini, G.; Troiani, T.; Cardone, C.; Vitiello, P.P.; Normanno, N.; Rachiglio, A.M.; Maiello, E.; Latiano, T.; et al. Implementing Anti-Epidermal Growth Factor Receptor (EGFR) Therapy in Metastatic Colorectal Cancer: Challenges and Future Perspectives. Ann. Oncol. 2020, 31, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Rizzuto, M.A.; Pacini, C.; Pandolfi, L.; Bonizzi, A.; Truffi, M.; Monieri, M.; Catrambone, F.; Giustra, M.; Garbujo, S.; et al. Half-Chain Cetuximab Nanoconjugates Allow Multitarget Therapy of Triple Negative Breast Cancer. Bioconjug. Chem. 2018, 29, 3817–3832. [Google Scholar] [CrossRef] [PubMed]
- Bäumer, S.; Bäumer, N.; Appel, N.; Terheyden, L.; Fremerey, J.; Schelhaas, S.; Wardelmann, E.; Buchholz, F.; Berdel, W.E.; Müller-Tidow, C. Antibody-Mediated Delivery of Anti-KRAS-SiRNA in Vivo Overcomes Therapy Resistance in Colon Cancer. Clin. Cancer Res. 2015, 21, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, E.; Barok, M.; Szöllosi, J.; Vereb, G. ErbB-Directed Immunotherapy: Antibodies in Current Practice and Promising New Agents. Immunol. Lett. 2008, 116, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Petrilli, R.; Eloy, J.O.; Lopez, R.F.V.; Lee, R.J. Cetuximab Immunoliposomes Enhance Delivery of 5-FU to Skin Squamous Carcinoma Cells. Anticancer Agents Med. Chem. 2017, 17, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Zalba, S.; Contreras, A.M.; Haeri, A.; Ten Hagen, T.L.M.; Navarro, I.; Koning, G.; Garrido, M.J. Cetuximab-Oxaliplatin-Liposomes for Epidermal Growth Factor Receptor Targeted Chemotherapy of Colorectal Cancer. J. Control. Release 2015, 210, 26–38. [Google Scholar] [CrossRef] [PubMed]
- De Maria, P.; Filippone, P.; Fontana, A.; Gasbarri, C.; Siani, G.; Velluto, D. Cardanol as a Replacement for Cholesterol into the Lipid Bilayer of POPC Liposomes. Colloids Surf. B Biointerfaces 2005, 40, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Kirpotin, D.; Park, J.W.; Hong, K.; Zalipsky, S.; Li, W.L.; Carter, P.; Benz, C.C.; Papahadjopoulos, D. Sterically Stabilized Anti-HER2 Immunoliposomes: Design and Targeting to Human Breast Cancer Cells in Vitro. Biochemistry 1997, 36, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Zappacosta, R.; Aschi, M.; Ammazzalorso, A.; Di Profio, P.; Fontana, A.; Siani, G. Embedding calix[4]resorcinarenes in liposomes: Experimental and computational investigation of the effect of resorcinarene inclusion on liposome properties and stability. Biochim. Biophys. Acta-Biomembr. 2019, 1861, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
| Protocol Step | Liposomes | LLNle-Liposomes | Cmx-Liposomes | Cmx-Liposomes-LLNle |
|---|---|---|---|---|
| Extrusion | 170.7 ± 26.5 | 170.7 ± 26.5 | 176.1 ± 12.1 | 176.1 ± 12.1 |
| LLNle incubation | - | 189.4 ± 13.5 | - | 189.5 ± 3.0 |
| Cxm conjugation | - | - | 187.1 ± 7.3 | 199.9 ± 8.7 |
| G-25 filtration | 143.7 ± 10.0 | 183.1 ± 16.0 | 165.6 ± 7.2 | 162.7 ± 8.8 |
| Rehydration after freeze-drying 1 | 587.5 ± 7.6 221.6 ± 26.1 | 479.6 ± 6.7 171.3 ± 27.5 | 815.4 ± 267.0 285.6 ± 28.8 | 1198 ± 63 291.9 ± 13.6 |
| Protocol Step | Liposomes | LLNle−Liposomes | Cmx−Liposomes | Cmx−Liposomes−LLNle |
|---|---|---|---|---|
| Extrusion | −23.3 ± 0.6 | −23.3 ± 0.6 | −22.9 ± 2.8 | −22.9 ± 2.8 |
| LLNle incubation | - | - | - | - |
| Cxm conjugation | - | - | −20.0 ± 1.1 | −22.2 ± 4.3 |
| G−25 filtration | −23.6 ± 1.6 | −21.4 ± 0.5 | −20.3 ± 0.2 | −20.9 ± 3.5 |
| Rehydration after freeze−drying | −26.1 ± 2.9 | −27.9 ± 2.1 | −23.1 ± 2.2 | −29.2 ± 8.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortese, K.; Marconi, S.; Aiello, C.; Gagliani, M.C.; Pilato, S.; Zappacosta, R.; Fontana, A.; Castagnola, P. Liposomes Loaded with the Proteasome Inhibitor Z-Leucinyl-Leucinyl-Norleucinal Are Effective in Inducing Apoptosis in Colorectal Cancer Cell Lines. Membranes 2020, 10, 91. https://doi.org/10.3390/membranes10050091
Cortese K, Marconi S, Aiello C, Gagliani MC, Pilato S, Zappacosta R, Fontana A, Castagnola P. Liposomes Loaded with the Proteasome Inhibitor Z-Leucinyl-Leucinyl-Norleucinal Are Effective in Inducing Apoptosis in Colorectal Cancer Cell Lines. Membranes. 2020; 10(5):91. https://doi.org/10.3390/membranes10050091
Chicago/Turabian StyleCortese, Katia, Silvia Marconi, Cinzia Aiello, Maria Cristina Gagliani, Serena Pilato, Romina Zappacosta, Antonella Fontana, and Patrizio Castagnola. 2020. "Liposomes Loaded with the Proteasome Inhibitor Z-Leucinyl-Leucinyl-Norleucinal Are Effective in Inducing Apoptosis in Colorectal Cancer Cell Lines" Membranes 10, no. 5: 91. https://doi.org/10.3390/membranes10050091
APA StyleCortese, K., Marconi, S., Aiello, C., Gagliani, M. C., Pilato, S., Zappacosta, R., Fontana, A., & Castagnola, P. (2020). Liposomes Loaded with the Proteasome Inhibitor Z-Leucinyl-Leucinyl-Norleucinal Are Effective in Inducing Apoptosis in Colorectal Cancer Cell Lines. Membranes, 10(5), 91. https://doi.org/10.3390/membranes10050091