Bridging the B Cell Gap: Novel Technologies to Study Antigen-Specific Human B Cell Responses
Abstract
:1. Introduction
2. Connecting Antigen Specificity to B Cell Heterogeneity
3. Connecting B Cell Repertoires to Serum Antibody Specificities
4. Animal Models to Study the Humoral Immune System
5. Ex Vivo Models to Study the Human Humoral Immune System
6. Tracking B Cell Function In Vivo
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plotkin, S.A. Correlates of Protection Induced by Vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrammert, J.; Smith, K.; Miller, J.; Langley, W.A.; Kokko, K.; Larsen, C.; Zheng, N.-Y.; Mays, I.; Garman, L.; Helms, C.; et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 2008, 453, 667–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Garman, L.; Wrammert, J.; Zheng, N.-Y.; Capra, J.D.; Ahmed, R.; Wilson, P.C. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2009, 4, 372–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guthmiller, J.J.; Dugan, H.L.; Neu, K.E.; Lan, L.Y.-L.; Wilson, P.C. An Efficient Method to Generate Monoclonal Antibodies from Human B Cells. Methods Mol. Biol. 2018, 109–145. [Google Scholar] [CrossRef]
- Shultz, L.D.; Brehm, M.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 2012, 12, 786–798. [Google Scholar] [CrossRef]
- Han, J.; Schmitz, A.J.; Richey, S.T.; Dai, Y.-N.; Turner, H.L.; Mohammed, B.M.; Fremont, D.H.; Ellebedy, A.H.; Ward, A.B. Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep. 2021, 34, 108682. [Google Scholar] [CrossRef]
- Bianchi, M.; Turner, H.L.; Nogal, B.; Cottrell, C.A.; Oyen, D.; Pauthner, M.; Bastidas, R.; Nedellec, R.; McCoy, L.E.; Wilson, I.A.; et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 2018, 49, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Setliff, I.; Shiakolas, A.; Pilewski, K.A.; Murji, A.A.; Mapengo, R.E.; Janowska, K.; Richardson, S.; Oosthuysen, C.; Raju, N.; Ronsard, L.; et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 2019, 179, 1636–1646. [Google Scholar] [CrossRef]
- Wagar, L.E.; Salahudeen, A.; Constantz, C.M.; Wendel, B.S.; Lyons, M.M.; Mallajosyula, V.; Jatt, L.P.; Adamska, J.Z.; Blum, L.K.; Gupta, N.; et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021, 27, 125–135. [Google Scholar] [CrossRef]
- Nguyen, D.C.; Garimalla, S.; Xiao, H.; Kyu, S.; Albizua, I.; Galipeau, J.; Chiang, K.-Y.; Waller, E.K.; Wu, R.; Gibson, G.; et al. Factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Garimilla, S.; Nguyen, D.C.; Halliley, J.L.; Tipton, C.; Rosenberg, A.F.; Fucile, C.F.; Saney, C.L.; Kyu, S.; Kaminsky, D.; Qian, Y.; et al. Differential transcriptome and development of human peripheral plasma cell subsets. JCI Insight 2019, 4, e126732. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.S.; Zhou, J.Q.; Han, J.; Schmitz, A.J.; Rizk, A.A.; Al Soussi, W.; Lei, T.; Amor, M.; McIntire, K.M.; Meade, P.; et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 2020, 586, 1–8. [Google Scholar] [CrossRef]
- Davis, C.W.; Jackson, K.J.L.; McCausland, M.M.; Darce, J.; Chang, C.; Linderman, S.L.; Chennareddy, C.; Gerkin, R.; Brown, S.J.; Wrammert, J.; et al. Influenza vaccine–induced human bone marrow plasma cells decline within a year after vaccination. Science 2020, 370, 237–241. [Google Scholar] [CrossRef]
- Tatovic, D.; Young, P.; Kochba, E.; Levin, Y.; Wong, F.S.; Dayan, C.M. Fine-Needle Aspiration Biopsy of the Lymph Node: A Novel Tool for the Monitoring of Immune Responses after Skin Antigen Delivery. J. Immunol. 2015, 195, 386–392. [Google Scholar] [CrossRef] [Green Version]
- Muellenbeck, M.F.; Ueberheide, B.; Amulic, B.; Epp, A.; Fenyo, D.; Busse, C.E.; Esen, M.; Theisen, M.; Mordumuller, B.; Wardemann, H. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J. Exp. Med. 2013, 210, 389–399. [Google Scholar] [CrossRef]
- Moir, S.; Ho, J.; Malaspina, A.; Wang, W.; DiPoto, A.C.; O’Shea, M.A.; Roby, G.; Kottilil, S.; Arthos, J.; Proschan, M.A.; et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 2008, 205, 1797–1805. [Google Scholar] [CrossRef]
- Sutton, H.J.; Aye, R.; Idris, A.H.; Vistein, R.; Nduati, E.; Kai, O.; Mwacharo, J.; Li, X.; Gao, X.; Andrews, T.D.; et al. Atypical B cells are part of an alternative lineage of B cells that participates in responses to vaccination and infection in humans. Cell Rep. 2021, 34, 108684. [Google Scholar] [CrossRef]
- Ehrhardt, G.R.; Hsu, J.T.; Gartland, L.; Leu, C.-M.; Zhang, S.; Davis, R.S.; Cooper, M.D. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 2005, 202, 783–791. [Google Scholar] [CrossRef]
- Weisel, N.M.; Weisel, F.J.; Farber, D.L.; Borghesi, L.A.; Shen, Y.; Ma, W.; Prak, E.T.L.; Shlomchik, M.J. Comprehensive analyses of B-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood 2020, 136, 2774–2785. [Google Scholar] [CrossRef]
- Lau, D.; Lan, L.Y.-L.; Andrews, S.F.; Henry, C.; Rojas, K.T.; Neu, K.E.; Huang, M.; Huang, Y.; DeKosky, B.; Palm, A.-K.; et al. Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation. Sci. Immunol. 2017, 2, 8153. [Google Scholar] [CrossRef] [Green Version]
- DeFalco, J.; Harbell, M.; Manning-Bog, A.; Baia, G.; Scholz, A.; Millare, B.; Sumi, M.; Zhang, D.; Chu, F.; Dowd, C.; et al. Non-progressing cancer patients have persistent B cell responses expressing shared antibody paratopes that target public tumor antigens. Clin. Immunol. 2018, 187, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.; Tanner, S.D. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.R.; Tsai, A.G.; Oliveria, J.P.; Hartmann, F.J.; Kimmey, S.C.; Calderon, A.A.; Borges, L.; Glass, M.C.; Wagar, L.E.; Davis, M.M.; et al. An Integrated Multi-omic Single-Cell Atlas of Human B Cell Identity. Immunity 2020, 53, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Dzangué-Tchoupou, G.; Corneau, A.; Blanc, C.; Benveniste, O.; Allenbach, Y. Analysis of cell surface and intranuclear markers on non-stimulated human PBMC using mass cytometry. PLoS ONE 2018, 13, e0194593. [Google Scholar] [CrossRef]
- Nolan, J.P.; Condello, D. Spectral Flow Cytometry. Curr. Protoc. Cytom. 2013, 63, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Brummelman, J.; Haftmann, C.; Núñez, N.G.; Alvisi, G.; Mazza, E.M.C.; Becher, B.; Lugli, E. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat. Protoc. 2019, 14, 1946–1969. [Google Scholar] [CrossRef]
- Ferrer-Font, L.; Mayer, J.U.; Old, S.; Hermans, I.F.; Irish, J.; Price, K.M. High-Dimensional Data Analysis Algorithms Yield Comparable Results for Mass Cytometry and Spectral Flow Cytometry Data. Cytom. Part A 2020, 97, 824–831. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [Green Version]
- Neu, K.E.; Guthmiller, J.J.; Huang, M.; La, J.; Vieira, M.C.; Kim, K.; Zheng, N.-Y.; Cortese, M.; Tepora, M.E.; Hamel, N.J.; et al. Spec-seq unveils transcriptional subpopulations of antibody-secreting cells following influenza vaccination. J. Clin. Investig. 2018, 129, 93–105. [Google Scholar] [CrossRef]
- DeKosky, B.J.; Ippolito, G.C.; Deschner, R.P.; Lavinder, J.J.; Wine, Y.; Rawlings, B.M.; Varadarajan, N.; Giesecke, C.; Dörner, T.; Andrews, S.F.; et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 2013, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R.S. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Wilson, P.; Dugan, H.L.; Stamper, C.T.; Li, L.; Changrob, S.; Asby, N.W.; Halfmann, P.J.; Zheng, N.Y.; Huang, M.; Shaw, D.G.; et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity 2021, 54, 1290–1303. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Eng, C.-H.L.; Lawson, M.; Zhu, Q.; Dries, R.; Koulena, N.; Takei, Y.; Yun, J.; Cronin, C.; Karp, C.; Yuan, G.-C.; et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 2019, 568, 235–239. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Baccin, C.; Al-Sabah, J.; Velten, L.; Helbling, P.M.; Grünschläger, F.; Hernández-Malmierca, P.; Nombela-Arrieta, C.; Steinmetz, L.M.; Trumpp, A.; Haas, S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020, 22, 38–48. [Google Scholar] [CrossRef]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vazquez, G.; Black, S.; Nolan, G.P. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968–981. [Google Scholar] [CrossRef] [Green Version]
- Cirelli, K.M.; Carnathan, D.G.; Nogal, B.; Martin, J.; Rodriguez, O.L.; Upadhyay, A.; Enemuo, C.A.; Gebru, E.H.; Choe, Y.; Viviano, F.; et al. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 2019, 177, 1153–1171. [Google Scholar] [CrossRef]
- Moyer, T.J.; Kato, Y.; Abraham, W.; Chang, J.Y.H.; Kulp, D.W.; Watson, N.; Turner, H.L.; Menis, S.; Abbott, R.K.; Bhiman, J.N.; et al. Author Correction: A Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat. Med. 2020, 26, 804. [Google Scholar] [CrossRef]
- Nogal, B.; Bianchi, M.; Cottrell, C.; Kirchdoerfer, R.N.; Sewall, L.M.; Turner, H.; Zhao, F.; Sok, D.; Burton, D.R.; Hangartner, L.; et al. Mapping Polyclonal Antibody Responses in Non-human Primates Vaccinated with HIV Env Trimer Subunit Vaccines. Cell Rep. 2020, 30, 3755–3765. [Google Scholar] [CrossRef]
- Georgiou, G.; Ippolito, G.C.; Beausang, J.; Busse, C.; Wardemann, H.; Quake, S.R. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014, 32, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Chappaz, S.; Finke, D. The IL-7 Signaling Pathway Regulates Lymph Node Development Independent of Peripheral Lymphocytes. J. Immunol. 2010, 184, 3562–3569. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Masse-Ranson, G.; Garcia, Z.; Bruel, T.; Kök, A.; Strick-Marchand, H.; Jouvion, G.; Serafini, N.; Lim, A.I.; Dusseaux, M.; et al. A human immune system mouse model with robust lymph node development. Nat. Methods 2018, 15, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Jardine, J.G.; Kulp, D.W.; Havenar-Daughton, C.; Sarkar, A.; Briney, B.; Sok, D.; Sesterhenn, F.; Ereño-Orbea, J.; Kalyuzhniy, O.; Deresa, I.; et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016, 351, 1458–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangesland, M.; Ronsard, L.; Kazer, S.W.; Bals, J.; Boyoglu-Barnum, S.; Yousif, A.S.; Barnes, R.; Feldman, J.; Quirindongo-Crespo, M.; McTamney, P.M.; et al. Germline-Encoded Affinity for Cognate Antigen Enables Vaccine Amplification of a Human Broadly Neutralizing Response against Influenza Virus. Immuity 2019, 51, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Fleming, J.; Clair, E.W.S.; Katinger, H.; Stiegler, G.; Kunert, R.; Robinson, J.; Scearce, R.M.; Plonk, K.; Staats, H.; et al. Cardiolipin Polyspecific Autoreactivity in Two Broadly Neutralizing HIV-1 Antibodies. Science 2005, 308, 1906–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Yang, G.; Wiehe, K.; Nicely, N.I.; Vandergrift, N.A.; Rountree, W.; Bonsignori, M.; Alam, S.M.; Gao, J.; Haynes, B.F.; et al. Polyreactivity and Autoreactivity among HIV-1 Antibodies. J. Virol. 2014, 89, 784–798. [Google Scholar] [CrossRef] [Green Version]
- Guthmiller, J.J.; Lan, L.Y.-L.; Fernández-Quintero, M.L.; Han, J.; Utset, H.A.; Bitar, D.J.; Hamel, N.J.; Stovicek, O.; Li, L.; Tepora, M.; et al. Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses. Immunity 2020, 53, 1230–1244. [Google Scholar] [CrossRef]
- Lin, Y.; Pecetta, S.; Steichen, J.M.; Kratochvil, S.; Melzi, E.; Arnold, J.; Dougan, S.K.; Wu, L.; Kirsch, K.H.; Nair, U.; et al. One-step CRISPR /Cas9 method for the rapid generation of human antibody heavy chain knock-in mice. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Jacobsen, J.T.; Mesin, L.; Markoulaki, S.; Schiepers, A.; Cavazzoni, C.B.; Bousbaine, D.; Jaenisch, R.; Victora, G.D. One-step generation of monoclonal B cell receptor mice capable of isotype switching and somatic hypermutation. J. Exp. Med. 2018, 215, 2686–2695. [Google Scholar] [CrossRef] [Green Version]
- Dougan, S.K.; Ogata, S.; Hu, C.C.A.; Grotenbreg, G.M.; Guillen, E.; Jaenisch, R.; Ploegh, H.L. IgG1+ ovalbumin-specific B-cell transnuclear mice show class switch recombination in rare allelically included B cells. Proc. Natl. Acad. Sci. USA 2012, 109, 13739–13744. [Google Scholar] [CrossRef] [Green Version]
- Jameson, S.C.; Masopust, D. What Is the Predictive Value of Animal Models for Vaccine Efficacy in Humans? Reevaluating the Potential of Mouse Models for the Human Immune System. Cold Spring Harb. Perspect. Biol. 2018, 10, a029132. [Google Scholar] [CrossRef] [Green Version]
- Watkins, I.D.; Burton, D.R.; Kallas, E.G.; Moore, J.P.; Koff, W.C. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 2008, 14, 617–621. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.; Huber, J.; Alp, Ö.S.; Gürkov, R.; Reichel, C.A.; Herrmann, M.; Keppler, O.T.; Leeuw, T.; Baumjohann, D. Complex human adenoid tissue-based ex vivo culture systems reveal anti-inflammatory drug effects on germinal center T and B cells. EBioMedicine 2020, 53, 102684. [Google Scholar] [CrossRef]
- Slifka, M.K.; Antia, R.; Whitmire, J.; Ahmed, R. Humoral Immunity Due to Long-Lived Plasma Cells. Immunity 1998, 8, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Manz, R.A.; Thiel, A.; Radbruch, A. Lifetime of plasma cells in the bone marrow. Nature 1997, 388, 133–134. [Google Scholar] [CrossRef]
- Rappuoli, R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 2007, 25, 1361–1366. [Google Scholar] [CrossRef]
- Germain, R.N. Vaccines and the Future of Human Immunology. Immunity 2010, 33, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509–517. [Google Scholar] [CrossRef]
- Amanna, I.J.; Carlson, N.E.; Slifka, M.K. Duration of Humoral Immunity to Common Viral and Vaccine Antigens. N. Engl. J. Med. 2007, 357, 1903–1915. [Google Scholar] [CrossRef] [Green Version]
- Morel, S.; Didierlaurent, A.; Bourguignon, P.; Delhaye, S.; Baras, B.; Jacob, V.; Planty, C.; Elouahabi, A.; Harvengt, P.; Carlsen, H. Adjuvant System AS03 containing α-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine 2011, 29, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Havenar-Daughton, C.; Newton, I.G.; Zare, S.Y.; Reiss, S.M.; Schwan, B.; Suh, M.J.; Hasteh, F.; Levi, G.; Crotty, S. Normal human lymph node T follicular helper cells and germinal center B cells accessed via fine needle aspirations. J. Immunol. Methods 2020, 479, 112746. [Google Scholar] [CrossRef] [PubMed]
- Havenar-Daughton, C.; Carnathan, D.G.; de la Peña, A.T.; Pauthner, M.; Briney, B.; Reiss, S.M.; Wood, J.S.; Kaushik, K.; van Gils, M.J.; Rosales, S.L.; et al. Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep. 2016, 17, 2195–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauthner, M.; Havenar-Daughton, C.; Sok, D.; Nkolola, J.P.; Bastidas, R.; Boopathy, A.V.; Carnathan, D.G.; Chandrashekar, A.; Cirelli, K.M.; Cottrell, C.A.; et al. Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 2017, 46, 1073–1088. [Google Scholar] [CrossRef] [Green Version]
- Vickovic, S.; Eraslan, G.; Salmén, F.; Klughammer, J.; Stenbeck, L.; Schapiro, D.; Äijö, T.; Bonneau, R.; Bergenstråhle, L.; Navarro, J.F.; et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 2019, 16, 987–990. [Google Scholar] [CrossRef]
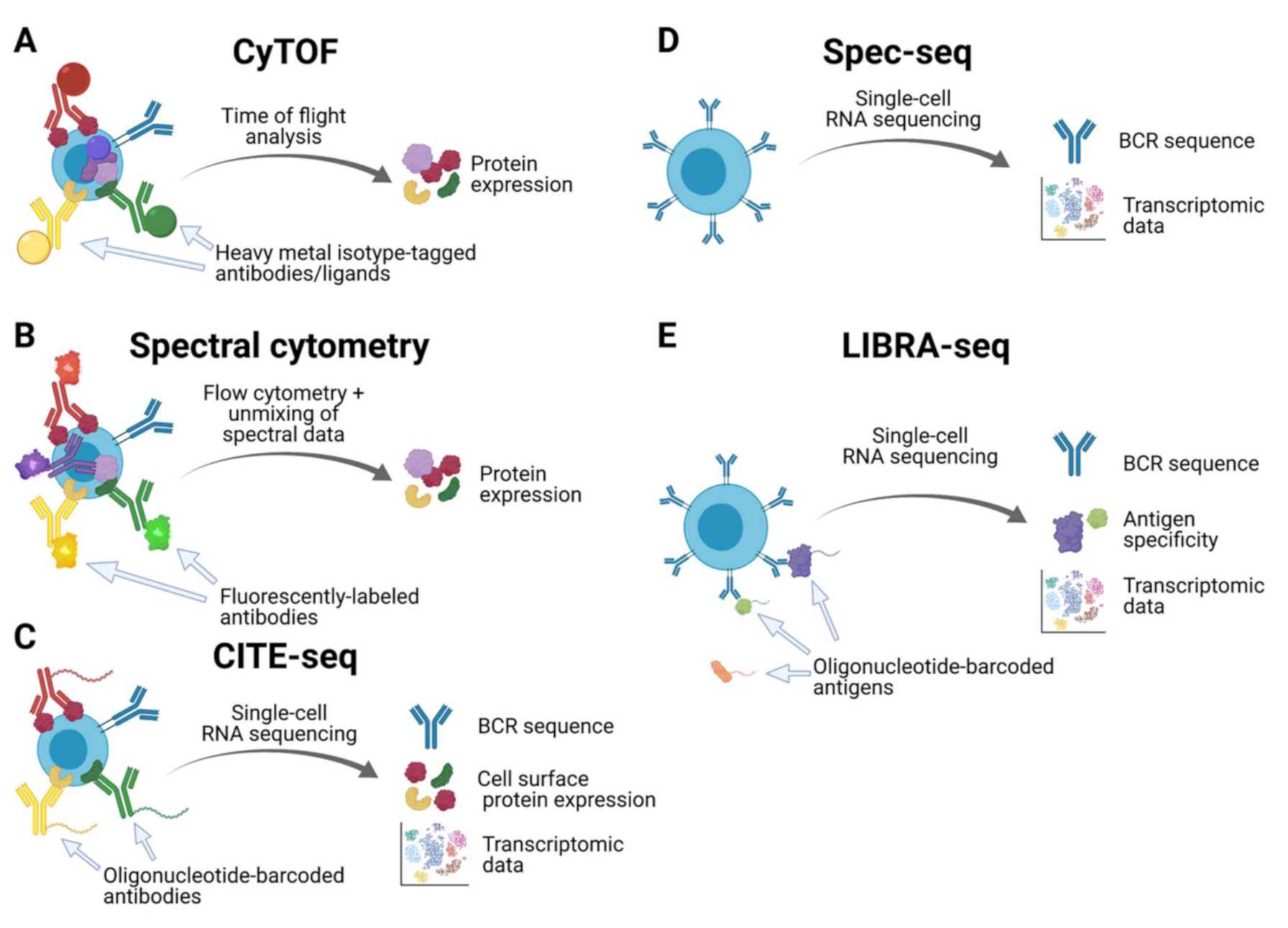

| CyTOF | Spectral Cytometry | CITE-seq | Spec-seq | LIBRA-seq | |
|---|---|---|---|---|---|
| BCR sequence | No | No | Yes * | Yes | Yes |
| Antigen specificity | No | No | Yes * | No | Yes |
| Transcriptomic data | No | No | Yes * | Yes | Yes |
| Cell surface protein expression | Yes | Yes | Yes | No | No |
| Intracellular protein expression | Yes | Yes | No | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Utset, H.A.; Guthmiller, J.J.; Wilson, P.C. Bridging the B Cell Gap: Novel Technologies to Study Antigen-Specific Human B Cell Responses. Vaccines 2021, 9, 711. https://doi.org/10.3390/vaccines9070711
Utset HA, Guthmiller JJ, Wilson PC. Bridging the B Cell Gap: Novel Technologies to Study Antigen-Specific Human B Cell Responses. Vaccines. 2021; 9(7):711. https://doi.org/10.3390/vaccines9070711
Chicago/Turabian StyleUtset, Henry A., Jenna J. Guthmiller, and Patrick C. Wilson. 2021. "Bridging the B Cell Gap: Novel Technologies to Study Antigen-Specific Human B Cell Responses" Vaccines 9, no. 7: 711. https://doi.org/10.3390/vaccines9070711
APA StyleUtset, H. A., Guthmiller, J. J., & Wilson, P. C. (2021). Bridging the B Cell Gap: Novel Technologies to Study Antigen-Specific Human B Cell Responses. Vaccines, 9(7), 711. https://doi.org/10.3390/vaccines9070711





