Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Healthy Donors and PBMC Isolation
2.2. Recombinant Protein Design, Production and Purification
2.3. HA-Trimer Specific Single B Cell Sorting by Flow Cytometry
2.4. Generation of Antigen Specific Antibodies
2.5. Recombinant Antibody Production
2.6. Binding of Monoclonal Antibodies and Plasma in Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Viruses
2.8. Hemagglutination Inhibition (HAI) Assay
2.9. Micro-Neutralization (MN) Assay
2.10. Competition Assay with Monoclonal Antibodies
3. Results
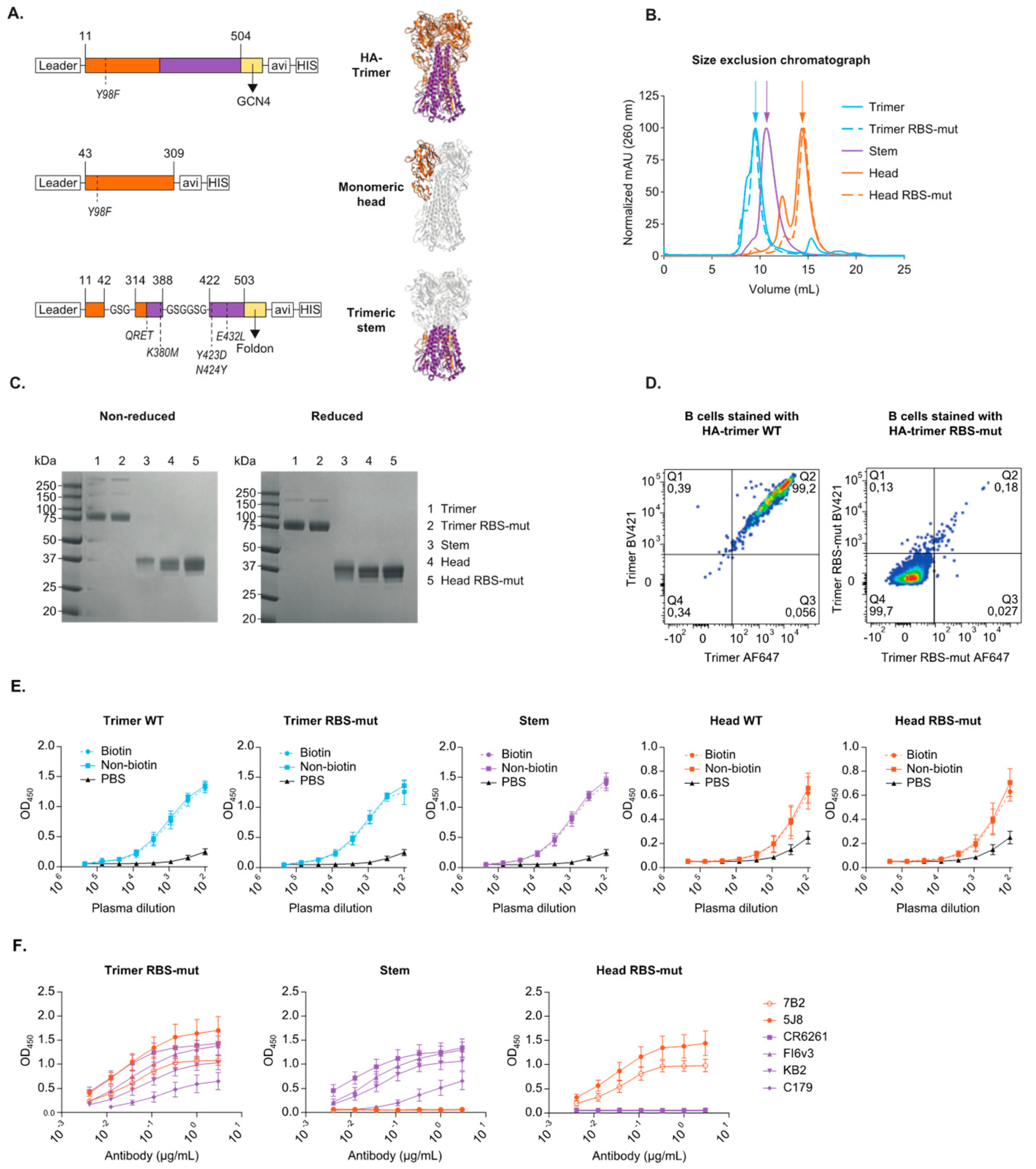
3.1. Design and Characterization of Recombinant HA-Trimer, Monomeric Head and Trimeric Stem Probes
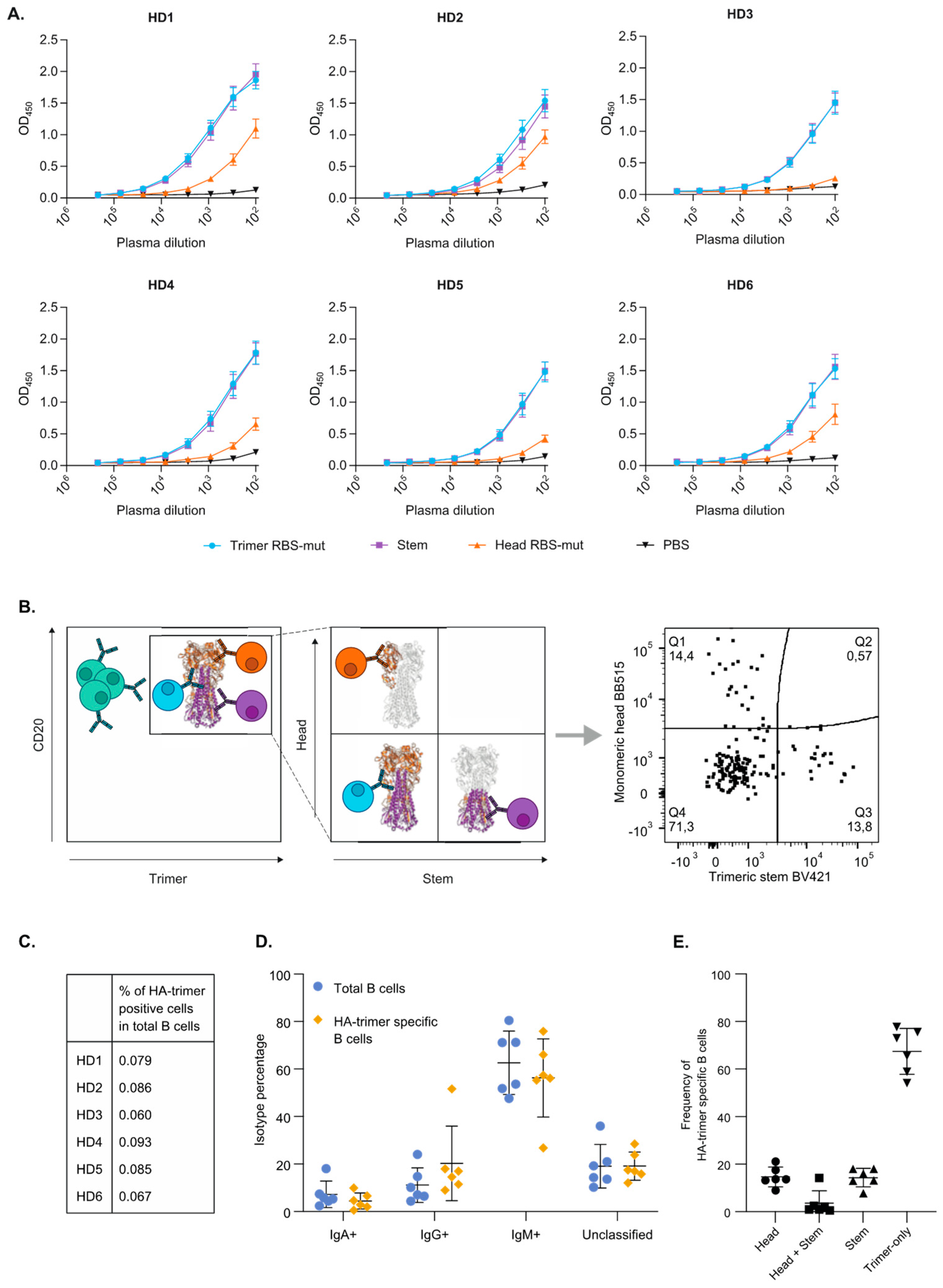
3.2. HA-Trimer, Monomeric Head and Trimeric Stem Probes Identify and Quantify Distinct HA-Specific B Cell Subsets
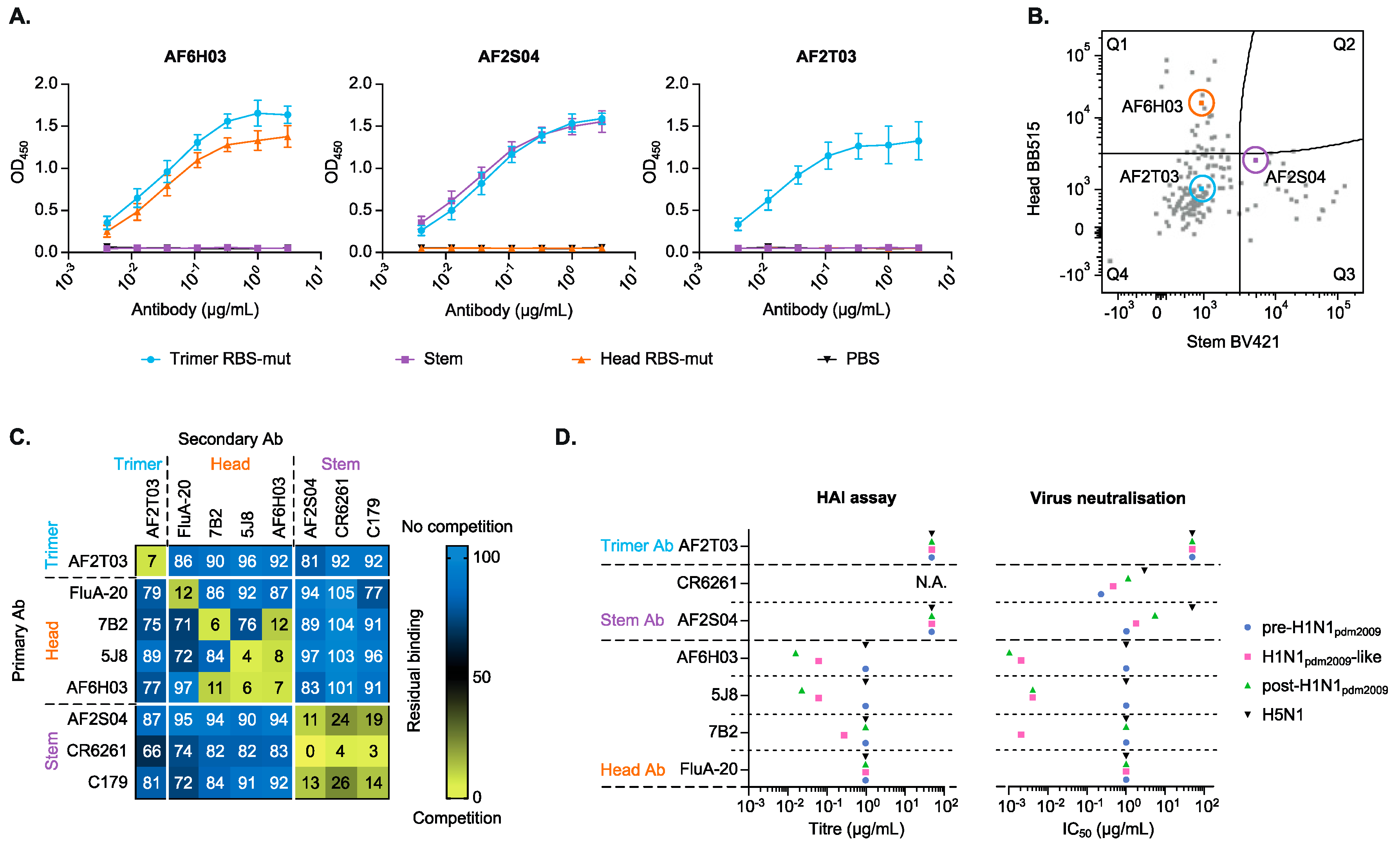
3.3. Monoclonal Antibodies Confirm the Head, Stem and Trimer-Only Specificities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Kosik, I.; Yewdell, J.W. Influenza Hemagglutinin and Neuraminidase: Yin–Yang Proteins Coevolving to Thwart Immunity. Viruses 2019, 11, 346. [Google Scholar] [CrossRef] [Green Version]
- Altman, M.O.; Angeletti, D.; Yewdell, J.W. Antibody Immunodominance: The Key to Understanding Influenza Virus Antigenic Drift. Viral Immunol. 2018, 31, 142–149. [Google Scholar] [CrossRef]
- Zhuang, Q.; Wang, S.; Liu, S.; Hou, G.; Li, J.; Jiang, W.; Wang, K.; Peng, C.; Liu, D.; Guo, A.; et al. Diversity and distribution of type A influenza viruses: An updated panorama analysis based on protein sequences. Virol. J. 2019, 16, 1–38. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.J.; Lapedes, A.S.; De Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Mapping the Antigenic and Genetic Evolution of Influenza Virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guthmiller, J.J.; Wilson, P.C. Harnessing immune history to combat influenza viruses. Curr. Opin. Immunol. 2018, 53, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, K.; Viboud, C.; Simonsen, L. Antibody response to influenza vaccination in the elderly: A quantitative review. Vaccine 2006, 24, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Lewnard, J.A.; Cobey, S. Immune History and Influenza Vaccine Effectiveness. Vaccines 2018, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.F.; Huang, Y.; Kaur, K.; Popova, L.I.; Ho, I.Y.; Pauli, N.T.; Dunand, C.J.H.; Taylor, W.M.; Lim, S.; Huang, M.; et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015, 7, 316ra192. [Google Scholar] [CrossRef] [Green Version]
- Henry, C.; Palm, A.-K.E.; Utset, H.A.; Huang, M.; Ho, I.Y.; Zheng, N.-Y.; Fitzgerald, T.; Neu, K.E.; Chen, Y.-Q.; Krammer, F.; et al. Monoclonal Antibody Responses after Recombinant Hemagglutinin Vaccine versus Subunit Inactivated Influenza Virus Vaccine: A Comparative Study. J. Virol. 2019, 93, e01150-19. [Google Scholar] [CrossRef] [Green Version]
- Dugan, H.L.; Guthmiller, J.J.; Arevalo, P.; Huang, M.; Chen, Y.-Q.; Neu, K.E.; Henry, C.; Zheng, N.-Y.; Lan, L.Y.-L.; Tepora, M.E.; et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med. 2020, 12, 573. [Google Scholar] [CrossRef]
- Gerhard, W.; Yewdell, J.W.; Frankel, M.E.; Webster, R.G. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nat. Cell Biol. 1981, 290, 713–717. [Google Scholar] [CrossRef]
- Altman, M.; Bennink, J.R.; Yewdell, J.W.; Herrin, B.R. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. eLife 2015, 4, e07467. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, B.; Koudstaal, W.; Goudsmit, J.; Klaren, V.; Tang, C.; Bujny, M.V.; Korse, H.J.W.M.; Kwaks, T.; Otterstrom, J.J.; Juraszek, J.; et al. Mechanisms of Hemagglutinin Targeted Influenza Virus Neutralization. PLoS ONE 2013, 8, e80034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- de Jong, N.M.; Aartse, A.; van Gils, M.J.; Eggink, D. Development of broadly reactive influenza vaccines by targeting the conserved regions of the hemagglutinin stem and head domains. Expert Rev. Vaccines 2020, 19, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, D.; Kosik, I.; Santos, J.; Yewdell, W.T.; Boudreau, C.M.; Mallajosyula, V.V.A.; Mankowski, M.C.; Chambers, M.; Prabhakaran, M.; Hickman, H.D.; et al. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc. Natl. Acad. Sci. USA 2019, 116, 13474–13479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burke, D.F.; Smith, D.J. A Recommended Numbering Scheme for Influenza A HA Subtypes. PLoS ONE 2014, 9, e112302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.-J.; Kanekiyo, M.; Kong, W.-P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Vanderven, H.A.; Wragg, K.; Ana-Sosa-Batiz, F.; Kristensen, A.B.; Jegaskanda, S.; Wheatley, A.K.; Wentworth, D.; Wines, B.D.; Hogarth, P.M.; Rockman, S.; et al. Anti-Influenza Hyperimmune Immunoglobulin Enhances Fc-Functional Antibody Immunity During Human Influenza Infection. J. Infect. Dis. 2018, 218, 1383–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martína, J.; Wharton, S.A.; Lin, Y.P.; Takemoto, D.K.; Skehel, J.J.; Wiley, D.C.; Steinhauer, D.A. Studies of the Binding Properties of Influenza Hemagglutinin Receptor-Site Mutants. Virology 1998, 241, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Binley, J.M.; Sanders, R.W.; Clas, B.; Schuelke, N.; Master, A.; Guo, Y.; Kajumo, F.; Anselma, D.J.; Maddon, P.J.; Olson, W.C.; et al. A Recombinant Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Complex Stabilized by an Intermolecular Disulfide Bond between the gp120 and gp41 Subunits Is an Antigenic Mimic of the Trimeric Virion-Associated Structure. J. Virol. 2000, 74, 627–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Caniels, T.G.; Van Der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; Han, B.W.; Bontjer, I.; Mooij, P.; Garces, F.; Behrens, A.-J.; Rantalainen, K.; Kumar, S.; Sarkar, A.; Brouwer, P.J.M.; et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019, 10, 2355. [Google Scholar] [CrossRef] [Green Version]
- Tiller, T.; Meffre, E.; Yurasov, S.; Tsuiji, M.; Nussenzweig, M.C.; Wardemann, H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 2008, 329, 112–124. [Google Scholar] [CrossRef] [Green Version]
- Ho, I.Y.; Bunker, J.J.; Erickson, S.A.; Neu, K.E.; Huang, M.; Cortese, M.; Pulendran, B.; Wilson, P.C. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J. Immunol. Methods 2016, 438, 67–70. [Google Scholar] [CrossRef] [Green Version]
- Benckert, J.; Schmolka, N.; Kreschel, C.; Zoller, M.J.; Sturm, A.; Wiedenmann, B.; Wardemann, H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Investig. 2011, 121, 1946–1955. [Google Scholar] [CrossRef] [Green Version]
- Hong, M.; Lee, P.S.; Hoffman, R.M.B.; Zhu, X.; Krause, J.C.; Laursen, N.S.; Yoon, S.-I.; Song, L.; Tussey, L.; Crowe, J.; et al. Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site. J. Virol. 2013, 87, 12471–12480. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Basler, C.; Crowe, J.E. A Broadly Neutralizing Human Monoclonal Antibody That Recognizes a Conserved, Novel Epitope on the Globular Head of the Influenza H1N1 Virus Hemagglutinin. J. Virol. 2011, 85, 10905–10908. [Google Scholar] [CrossRef] [Green Version]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.M.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; Van Deventer, E.; Guan, Y.; et al. Heterosubtypic Neutralizing Monoclonal Antibodies Cross-Protective against H5N1 and H1N1 Recovered from Human IgM+ Memory B Cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekiert, D.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bangaru, S.; Lang, S.; Schotsaert, M.; VanderVen, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 2019, 177, 1136–1152. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Friesen, R.H.; Lee, P.S.; Stoop, E.J.; Hoffman, R.M.; Ekiert, D.C.; Bhabha, G.; Yu, W.; Juraszek, J.; Koudstaal, W.; Jongeneelen, M.; et al. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. USA 2014, 111, 445–450. [Google Scholar] [CrossRef] [Green Version]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Van Gils, M.J.; Van Den Kerkhof, T.L.; Ozorowski, G.; Cottrell, C.A.; Sok, D.; Pauthner, M.; Pallesen, J.; De Val, N.; Yasmeen, A.; De Taeye, S.W. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.S.; Krammer, F.; Eggink, D.; Kongchanagul, A.; Moran, T.M.; Palese, P. A Pan-H1 Anti-Hemagglutinin Monoclonal Antibody with Potent Broad-Spectrum Efficacy In Vivo. J. Virol. 2012, 86, 6179–6188. [Google Scholar] [CrossRef] [Green Version]
- Heaton, N.S.; Leyva-Grado, V.H.; Tan, G.S.; Eggink, D.; Hai, R.; Palese, P. In Vivo Bioluminescent Imaging of Influenza A Virus Infection and Characterization of Novel Cross-Protective Monoclonal Antibodies. J. Virol. 2013, 87, 8272–8281. [Google Scholar] [CrossRef] [Green Version]
- Hai, R.; Krammer, F.; Tan, G.S.; Pica, N.; Eggink, D.; Maamary, J.; Margine, I.; Albrecht, R.; Palese, P. Influenza Viruses Expressing Chimeric Hemagglutinins: Globular Head and Stalk Domains Derived from Different Subtypes. J. Virol. 2012, 86, 5774–5781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggink, D.; Spronken, M.; van der Woude, R.; Buzink, J.; Broszeit, F.; McBride, R.; Pawestri, H.A.; Setiawaty, V.; Paulson, J.C.; Boons, G.-J.; et al. Phenotypic Effects of Substitutions within the Receptor Binding Site of Highly Pathogenic Avian Influenza H5N1 Virus Observed during Human Infection. J. Virol. 2020, 94, e00195-20. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- WHO Global Influenza Surveillance Network: Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. Available online: https://apps.who.int/iris/bitstream/handle/10665/44518/9789241548090_eng.pdf?sequence=1 (accessed on 1 September 2020).
- Thompson, A.J.; Cao, L.; Ma, Y.; Wang, X.; Diedrich, J.K.; Kikuchi, C.; Willis, S.; Worth, C.; McBride, R.; Yates, J.R., III. Human Influenza Virus Hemagglutinins Contain Conserved Oligomannose N-Linked Glycans Allowing Potent Neutralization by Lectins. Cell Host Microbe 2020, 27, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Diedrich, J.K.; Ma, Y.; Wang, N.; Pauthner, M.; Park, S.-K.R.; Delahunty, C.M.; McLellan, J.; Burton, D.R.; Yates, J.R.; et al. Global site-specific analysis of glycoprotein N-glycan processing. Nat. Protoc. 2018, 13, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor Binding and Membrane Fusion in Virus Entry: The Influenza Hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Whittle, J.R.R.; Wheatley, A.; Wu, L.; Lingwood, D.; Kanekiyo, M.; Ma, S.S.; Narpala, S.R.; Yassine, H.M.; Frank, G.M.; Yewdell, J.W.; et al. Flow Cytometry Reveals that H5N1 Vaccination Elicits Cross-Reactive Stem-Directed Antibodies from Multiple Ig Heavy-Chain Lineages. J. Virol. 2014, 88, 4047–4057. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.C.; Crowe, J.E., Jr. Committing the oldest sins in the newest kind of ways—Antibodies targeting the influenza virus type A hemagglutinin globular head. Antibodies Infect. Dis. 2015, 2, 209–219. [Google Scholar]
- Eggink, D.; Goff, P.H.; Palese, P. Guiding the Immune Response against Influenza Virus Hemagglutinin toward the Conserved Stalk Domain by Hyperglycosylation of the Globular Head Domain. J. Virol. 2013, 88, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.-X.; Jegaskanda, S.; Juno, J.A.; Esterbauer, R.; Wong, J.; Kelly, H.G.; Liu, Y.; Tilmanis, D.; Hurt, A.C.; Yewdell, J.W.; et al. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J. Clin. Investig. 2019, 129, 850–862. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med. 2019, 10, a038778. [Google Scholar] [CrossRef] [Green Version]
- Sangster, M.Y.; Nguyen, P.Q.T.; Topham, D.J. Role of Memory B Cells in Hemagglutinin-Specific Antibody Production Following Human Influenza A Virus Infection. Pathogens 2019, 8, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, M.; Changrob, S.; Li, L.; Wilson, P.C. Imprinting, immunodominance, and other impediments to generating broad influenza immunity. Immunol. Rev. 2020, 296, 191–204. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.R.; Watanabe, A.; Kuraoka, M.; Do, K.T.; McGee, C.E.; Sempowski, G.D.; Kepler, T.B.; Schmidt, A.G.; Kelsoe, G.; Harrison, S.C. Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 2018, 48, 174–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, A.; McCarthy, K.R.; Kuraoka, M.; Schmidt, A.G.; Adachi, Y.; Onodera, T.; Tonouchi, K.; Caradonna, T.; Bajic, G.; Song, S.; et al. Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 2019, 177, 1124–1135.e1116. [Google Scholar] [CrossRef]
- Koutsakos, M.; Sekiya, T.; Chua, B.Y.; Nguyen, T.H.O.; Wheatley, A.K.; Juno, J.A.; Ohno, M.; Nomura, N.; Ohara, Y.; Nishimura, T. Immune profiling of influenza-specific B-and T-cell responses in macaques using flow cytometry-based assays. Immunol. Cell Biol. 2021, 99, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, Z.; Sheehan, J.; Avnir, Y.; Ridenour, C.; Sachnik, T.; Sun, J.; Hossain, M.J.; Chen, L.-M.; Zhu, Q.; et al. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat. Commun. 2016, 7, 12780. [Google Scholar] [CrossRef]
- Lee, J.; Boutz, D.R.; Chromikova, V.; Joyce, M.G.; Vollmers, C.; Leung, K.; Horton, A.P.; DeKosky, B.J.; Lee, C.-H.; Lavinder, J.J.; et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. 2016, 22, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, M.; Adachi, Y.; Takahashi, Y. Hide and seek: Interplay between influenza viruses and B cells. Int. Immunol. 2020, 32, 605–611. [Google Scholar] [CrossRef] [Green Version]
- Turner, H.L.; Pallesen, J.; Lang, S.; Bangaru, S.; Urata, S.; Li, S.; Cottrell, C.A.; Bowman, C.A.; Crowe, J.E., Jr.; Wilson, I.A. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol. 2019, 17, e3000139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Gilchuk, I.; Li, S.; Irving, R.; Goff, M.T.; Turner, H.L.; Ward, A.B.; Carnahan, R.H.; Crowe, J.E. Anti–influenza H7 human antibody targets antigenic site in hemagglutinin head domain interface. J. Clin. Investig. 2020, 130, 4734–4739. [Google Scholar] [CrossRef]
- Bangaru, S.; Zhang, H.; Gilchuk, I.M.; Voss, T.G.; Irving, R.P.; Gilchuk, P.; Matta, P.; Zhu, X.; Lang, S.; Nieusma, T.; et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018, 9, 2669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aartse, A.; Eggink, D.; Claireaux, M.; van Leeuwen, S.; Mooij, P.; Bogers, W.M.; Sanders, R.W.; Koopman, G.; van Gils, M.J. Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans. Vaccines 2021, 9, 717. https://doi.org/10.3390/vaccines9070717
Aartse A, Eggink D, Claireaux M, van Leeuwen S, Mooij P, Bogers WM, Sanders RW, Koopman G, van Gils MJ. Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans. Vaccines. 2021; 9(7):717. https://doi.org/10.3390/vaccines9070717
Chicago/Turabian StyleAartse, Aafke, Dirk Eggink, Mathieu Claireaux, Sarah van Leeuwen, Petra Mooij, Willy M. Bogers, Rogier W. Sanders, Gerrit Koopman, and Marit J. van Gils. 2021. "Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans" Vaccines 9, no. 7: 717. https://doi.org/10.3390/vaccines9070717
APA StyleAartse, A., Eggink, D., Claireaux, M., van Leeuwen, S., Mooij, P., Bogers, W. M., Sanders, R. W., Koopman, G., & van Gils, M. J. (2021). Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans. Vaccines, 9(7), 717. https://doi.org/10.3390/vaccines9070717






