An Antigenic Thrift-Based Approach to Influenza Vaccine Design
Abstract
1. Introduction
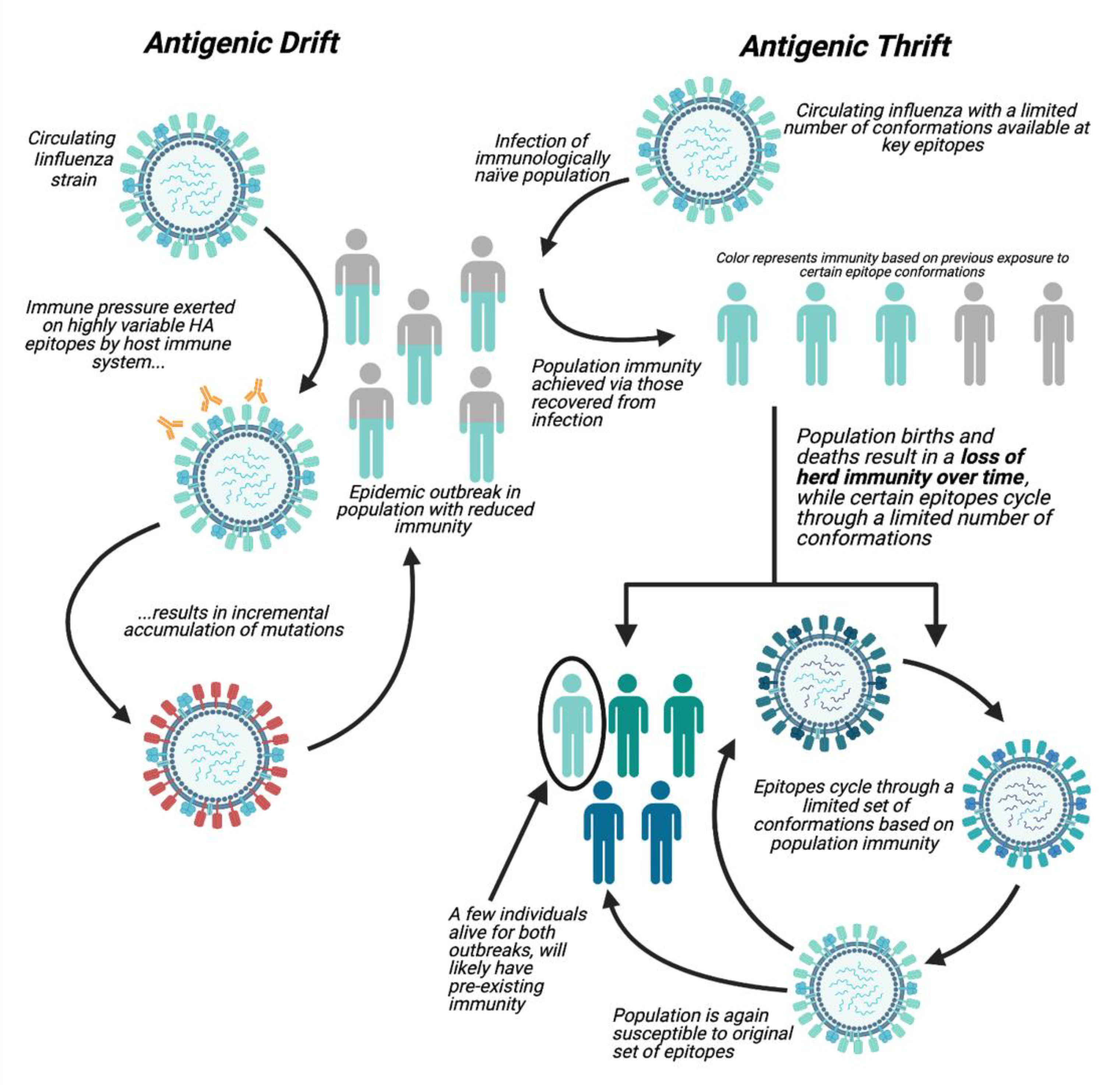
1.1. Antigenic Drift
1.2. Antigenic Thrift
1.3. Vaccine Approaches
2. Vaccines Targeting More Conserved Regions of the Influenza Virus
3. An Antigenic Thrift Approach to Universal Influenza Vaccine Design
4. Pitfalls of Effective Universal Vaccine Development
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Methods
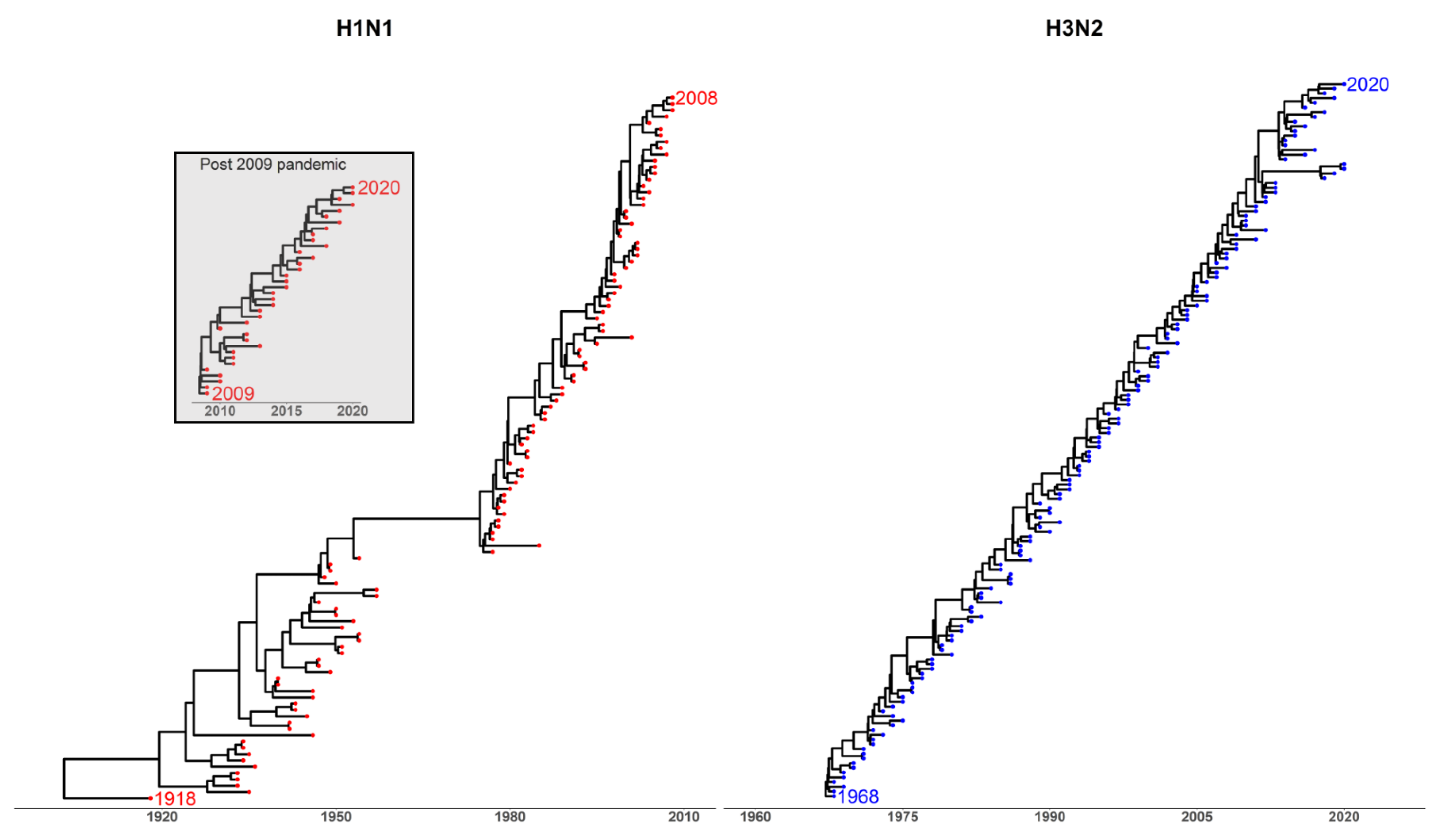
Appendix A.2. Phylogenetic Analysis
References
- Harding, A.T.; Heaton, N.S. Efforts to Improve the Seasonal Influenza Vaccine. Vaccines 2018, 6, 19. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.R.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef]
- Corder, B.N.; Bullard, B.L.; Poland, G.A.; Weaver, E.A. A Decade in Review: A Systematic Review of Universal Influenza Vaccines in Clinical Trials during the 2010 Decade. Viruses 2020, 12, 1186. [Google Scholar] [CrossRef]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Huang, S.S.H.; Lin, Z.; Banner, D.; León, A.J.; Paquette, S.G.; Rubin, B.; Rubino, S.; Guan, Y.; Kelvin, D.J.; Kelvin, A.A. Immunity toward H1N1 influenza hemagglutinin of historical and contemporary strains suggests protection and vaccine failure. Sci. Rep. 2013, 3, 1698. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Gilbertson, B.; Trifkovic, S.; Brown, L.E.; McKimm-Breschkin, J.L. Influenza Virus Neuraminidase Structure and Functions. Front. Microbiol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Pielak, R.M.; Chou, J.J. Influenza M2 proton channels. Biochim. Biophys. Acta (BBA). Biomembr. 2011, 1808, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, L.; Liu, X. Role of Post-translational Modifications in Influenza A Virus Life Cycle and Host Innate Immune Response. Front. Microbiol. 2020, 11, 2156. [Google Scholar] [CrossRef]
- Luo, M. Influenza Virus Entry. Adv. Exp. Med. Biol. 2012, 726, 201–221. [Google Scholar] [CrossRef]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.K.; Meyerson, J.R.; Matsuoka, Y.; Kuybeda, O.; Moran, A.; Bliss, D.; Das, S.R.; Yewdell, J.W.; Sapiro, G.; Subbarao, K.; et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc. Natl. Acad. Sci. USA 2013, 110, 4592–4597. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Lee, J.M.; Huddleston, J.; Doud, M.B.; Hooper, K.A.; Wu, N.C.; Bedford, T.; Bloom, J.D. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc. Natl. Acad. Sci. USA 2018, 115, E8276–E8285. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Protection, Types of Influenza Viruses. Available online: https://www.cdc.gov/flu/about/viruses/types.htm (accessed on 1 February 2021).
- World Health Organization Regional Office for Europe. How Pandemic Influenza Emerges. 2021. Available online: https://www.euro.who.int/en/health-topics/communicable-diseases/influenza/pandemic-influenza/how-pandemic-influenza-emerges (accessed on 15 February 2021).
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.-L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18, 1650. [Google Scholar] [CrossRef]
- Ma, W.; Kahn, R.E.; Richt, J.A. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J. Mol. Genet. Med. 2008, 3, 158. [Google Scholar] [CrossRef]
- Landolt, G.A.; Olsen, C.W. Up to new tricks–A review of cross-species transmission of influenza A viruses. Anim. Heal. Res. Rev. 2007, 8, 1–21. [Google Scholar] [CrossRef]
- Centers for Disease Control and Protection. How Flu Viruses Can Change. 2019. Available online: https://www.cdc.gov/flu/about/viruses/change.htm. (accessed on 15 March 2021).
- Wang, W.; Alvarado-Facundo, E.; Vassell, R.; Collins, L.; Colombo, R.E.; Ganesan, A.; Geaney, C.; Hrncir, D.; Lalani, T.; Markelz, A.E.; et al. Comparison of A(H3N2) neutralizing antibody responses elicited by 2018–2019 season quadri-valent influenza vaccines derived from eggs, cells, and recombinant hemagglutinin. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- World Health Organization, Influenza. Available online: https://www.who.int/biologicals/vaccines/influenza/en/ (accessed on 1 March 2021).
- O’Gorman, W.E.; Huang, H.; Wei, Y.-L.; Davis, K.L.; Leipold, M.D.; Bendall, S.C.; Kidd, B.; Dekker, C.L.; Maecker, H.T.; Chien, Y.-H.; et al. The Split Virus Influenza Vaccine rapidly activates immune cells through Fcγ receptors. Vaccine 2014, 32, 5989–5997. [Google Scholar] [CrossRef]
- Martínez-Sobrido, L.; Peersen, O.; Nogales, A. Temperature Sensitive Mutations in Influenza A Viral Ribonu-cleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine. Viruses 2018, 10, 560. [Google Scholar] [CrossRef]
- Soema, P.C.; Kompier, R.; Amorij, J.-P.; Kersten, G.F. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015, 94, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Osterholm, M.T.; Kelley, N.S.; Sommer, A.; Belongia, E.A. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 36–44. [Google Scholar] [CrossRef]
- Tricco, A.C.; Chit, A.; Soobiah, C.; Hallett, D.; Meier, G.; Chen, M.H.; Tashkandi, M.; Bauch, C.T.; Loeb, M. Comparing influenza vaccine efficacy against mismatched and matched strains: A systematic review and meta-analysis. BMC Med. 2013, 11, 153. [Google Scholar] [CrossRef]
- Petrie, J.G.; Ohmit, S.E.; Truscon, R.; Johnson, E.; Braun, T.M.; Levine, M.Z.; Eichelberger, M.C.; Monto, A.S. Modest Waning of Influenza Vaccine Efficacy and Antibody Titers During the 2007–2008 Influenza Season. J. Infect. Dis. 2016, 214, 1142–1149. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention, ACIP votes down use of LAIV for 2016–2017 flu season. Available online: https://www.cdc.gov/media/releases/2016/s0622-laiv-flu.html (accessed on 1 March 2021).
- Thompson, C.P.; Lourenço, J.; Walters, A.A.; Obolski, U.; Edmans, M.; Palmer, D.S.; Kooblall, K.; Carnell, G.W.; O’Connor, D.; Bowden, T.A.; et al. A naturally protective epitope of limited variability as an influenza vaccine target. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Allen, J.D.; Ray, S.; Ross, T.M. Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses. PLoS ONE 2018, 13, e0204284. [Google Scholar]
- Raymond, D.; Bajic, G.; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 2018, 115, 168–173. [Google Scholar] [CrossRef]
- Recker, M.; Pybus, O.G.; Nee, S.; Gupta, S. The generation of influenza outbreaks by a network of host immune responses against a lim-ited set of antigenic types. Proc. Natl. Acad.Sci. USA 2007, 104, 7711–7716. [Google Scholar] [CrossRef]
- Wikramaratna, P.S.; Pybus, O.G.; Gupta, S. Contact between bird species of different lifespans can promote the emergence of highly pathogenic avian influenza strains. Proc. Natl. Acad. Sci. USA 2014, 111, 10767–10772. [Google Scholar] [CrossRef] [PubMed]
- Boni, M.F. Vaccination and antigenic drift in influenza. Vaccine 2008, 26, C8–C14. [Google Scholar] [CrossRef] [PubMed]
- Fonville, J.M.; Fraaij, P.L.A.; De Mutsert, G.; Wilks, S.H.; Van Beek, R.; Fouchier, R.A.M.; Rimmelzwaan, G.F. Antigenic Maps of Influenza A(H3N2) Produced with Human Antisera Obtained After Primary Infection. J. Infect. Dis. 2016, 213, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Fonville, J.M.; Wilks, S.H.; James, S.; Fox, A.; Ventresca, M.; Aban, M.; Xue, L.; Jones, T.C.; H., L.N.M.; T., P.Q.; et al. Antibody landscapes after influenza virus infection or vaccination. Science 2014, 346, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Doud, M.B.; Hensley, S.E.; Bloom, J.D. Complete mapping of viral escape from neutralizing antibodies. PLoS Pathog. 2017, 13, e1006271. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef]
- Ghedin, E.; Sengamalay, N.A.; Shumway, M.; Zaborsky, J.; Feldblyum, T.; Subbu, V.; Spiro, D.J.; Sitz, J.; Koo, H.; Bolotov, P.; et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005, 437, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.; Reddy, K.; James, J.; Adamiak, B.; Barclay, W.; Shelton, H.; Iqbal, M. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci. Rep. 2016, 6, 18745. [Google Scholar] [CrossRef]
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31, 417–427. [Google Scholar]
- Matsuzaki, Y.; Sugawara, K.; Nakauchi, M.; Takahashi, Y.; Onodera, T.; Tsunetsugu-Yokota, Y.; Matsumura, T.; Ato, M.; Kobayashi, K.; Shimotai, Y.; et al. Epitope Mapping of the Hemagglutinin Molecule of A/(H1N1)pdm09 Influenza Virus by Using Monoclonal Antibody Escape Mutants. J. Virol. 2014, 88, 12364–12373. [Google Scholar] [CrossRef]
- Huang, K.A.; Rijal, P.; Schimanski, L.; Powell, T.J.; Lin, T.-Y.; McCauley, J.W.; Daniels, R.S.; Townsend, A.R. Focused antibody response to influenza linked to antigenic drift. J. Clin. Investig. 2015, 125, 2631–2645. [Google Scholar] [CrossRef] [PubMed]
- Zinder, D.; Bedford, T.; Gupta, S.; Pascual, M. The Roles of Competition and Mutation in Shaping Antigenic and Genetic Diversity in Influenza. PLOS Pathog. 2013, 9, e1003104. [Google Scholar] [CrossRef]
- Rambaut, A.; Pybus, O.G.; Nelson, M.I.; Viboud, C.; Taubenberger, J.K.; Holmes, E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature 2008, 453, 615–619. [Google Scholar] [CrossRef]
- Thompson, C.P.; Obolski, U. Influenza vaccination and the ‘diversity paradox’. Hum. Vaccines Immunother. 2018, 14, 3005–3009. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M.; Galvani, A.P.; Bush, R.M. Ecological and immunological determinants of influenza evolution. Nat. Cell Biol. 2003, 422, 428–433. [Google Scholar] [CrossRef]
- Tria, F.; Lässig, M.; Peliti, L.; Franz, S. A minimal stochastic model for influenza evolution. J. Stat. Mech. Theory Exp. 2005, 2005, P07008. [Google Scholar] [CrossRef]
- Koelle, K.; Cobey, S.; Grenfell, B.; Pascual, M. Epochal Evolution Shapes the Phylodynamics of Interpandemic Influenza A (H3N2) in Humans. Science 2006, 314, 1898–1903. [Google Scholar] [CrossRef] [PubMed]
- Bedford, T.; Rambaut, A.; Pascual, M. Canalization of the evolutionary trajectory of the human influenza virus. BMC Biol. 2012, 10, 38. [Google Scholar] [CrossRef]
- Obolski, U.; Ram, Y.; Hadany, L. Key issues review: Evolution on rugged adaptive landscapes. Rep. Prog. Phys. 2017, 81, 012602. [Google Scholar] [CrossRef]
- Koelle, K.; A Rasmussen, D. The effects of a deleterious mutation load on patterns of influenza A/H3N2’s antigenic evolution in humans. eLife 2015, 4, e07361. [Google Scholar] [CrossRef]
- Luo, S.; Reed, M.; Mattingly, J.C.; Koelle, K. The impact of host immune status on the within-host and population dynamics of antigenic immune escape. J. R. Soc. Interface 2012, 9, 2603–2613. [Google Scholar] [CrossRef]
- Volkov, I.; Pepin, K.M.; Lloyd-Smith, J.O.; Banavar, J.R.; Grenfell, B.T. Synthesizing within-host and population-level selective pressures on viral populations: The im-pact of adaptive immunity on viral immune escape. J. R. Soc. Interface 2010, 7, 1311–1318. [Google Scholar] [CrossRef]
- Yuan, H.-Y.; Koelle, K. The evolutionary dynamics of receptor binding avidity in influenza A: A mathematical model for a new antigenic drift hypothesis. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120204. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Myers, J.L.; Bostick, D.L.; Sullivan, C.B.; Madara, J.; Linderman, S.L.; Liu, Q.; Carter, D.M.; Wrammert, J.; Esposito, S.; et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 2013, 210, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Bush, R.M. Predicting the Evolution of Human Influenza A. Science 1999, 286, 1921–1925. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Genet. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Han, A.X.; Maurer-Stroh, S.; Russell, C.A. Individual immune selection pressure has limited impact on sea-sonal influenza virus evolution. Nat. Ecol. Evol. 2019, 3, 302–311. [Google Scholar] [CrossRef]
- Lumby, C.K.; Zhao, L.; Breuer, J.; Illingworth, C.J. A large effective population size for established within-host influenza virus infection. eLife 2020, 9, 9. [Google Scholar] [CrossRef]
- Morris, D.H.; Petrova, V.; Rossine, F.W.; Parker, E.; Grenfell, B.T.; A Neher, R.; A Levin, S.; A Russell, C. Asynchrony between virus diversity and antibody selection limits influenza virus evolution. eLife 2020, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Wikramaratna, P.S.; Sandeman, M.; Recker, M.; Gupta, S. The antigenic evolution of influenza: Drift or thrift? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120200. [Google Scholar] [CrossRef]
- Carter, D.M.; Bloom, C.E.; Nascimento, E.J.M.; Marques, E.; Craigo, J.K.; Cherry, J.L.; Lipman, D.J.; Ross, T.M. Sequential Seasonal H1N1 Influenza Virus Infections Protect Ferrets against Novel 2009 H1N1 Influenza Virus. J. Virol. 2012, 87, 1400–1410. [Google Scholar] [CrossRef]
- Andrews, S.F.; Huang, Y.; Kaur, K.; Popova, L.I.; Ho, I.Y.; Pauli, N.T.; Dunand, C.J.H.; Taylor, W.M.; Lim, S.; Huang, M.; et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015, 7, 316ra192. [Google Scholar] [CrossRef]
- Whittle, J.R.R.; Zhang, R.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 2011, 108, 14216–14221. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Piepenbrink, M.S.; Wang, J.; Ortega, S.; Basu, M.; Fucile, C.F.; Treanor, J.J.; Rosenberg, A.F.; Zand, M.S.; Keefer, M.C.; et al. A Highly Potent and Broadly Neutralizing H1 Influenza-Specific Human Monoclonal Antibody. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Guptill, J.; Naficy, A.; Nachbagauer, R.; Berlanda-Scorza, F.; Feser, J.; Wilson, P.C.; Solórzano, A.; Van Der Wielen, M.; Walter, E.B.; et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine can-didates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 2020, 20, 80–91. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Liu, W.-C.; Choi, A.; Wohlbold, T.J.; Atlas, T.; Rajendran, M.; Solórzano, A.; Berlanda-Scorza, F.; García-Sastre, A.; Palese, P.; et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2017, 2, 1–13. [Google Scholar] [CrossRef]
- Choi, A.; Bouzya, B.; Franco, K.-D.C.; Stadlbauer, D.; Rajabhathor, A.; Rouxel, R.N.; Mainil, R.; Van Der Wielen, M.; Palese, P.; García-Sastre, A.; et al. Chimeric Hemagglutinin-Based Influenza Virus Vaccines Induce Protective Stalk-Specific Humoral Immunity and Cellular Responses in Mice. Immunohorizons 2019, 3, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1, 16015. [Google Scholar] [CrossRef]
- Amitai, A.; Sangesland, M.; Barnes, R.M.; Rohrer, D.; Lonberg, N.; Lingwood, D.; Chakraborty, A.K. Defining and Manipulating B Cell Immunodominance Hierarchies to Elicit Broadly Neutralizing Antibody Responses against Influenza Virus. Cell Syst. 2020, 11, 573–588.e9. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.P. GSK Dumps Universal Flu Vaccine after Interim Data Readout. Available online: https://www.fiercebiotech.com/biotech/gsk-dumps-universal-flu-vaccine-after-interim-data-readout (accessed on 30 March 2021).
- Hayward, A.C.; Wang, L.; Goonetilleke, N.; Fragaszy, E.; Bermingham, A.; Copas, A.; Dukes, O.; Millett, E.; Nazareth, I.; Nguyen-Van-Tam, J.S.; et al. Natural T Cell–mediated Protection against Seasonal and Pandemic Influenza. Results of the Flu Watch Cohort Study. Am. J. Respir. Crit. Care Med. 2015, 191, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.S.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef]
- Wang, Z.; Wan, Y.; Qiu, C.; Quiñones-Parra, S.; Zhu, Z.; Loh, L.; Tian, D.; Ren, Y.; Hu, Y.; Zhang, X.; et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. Nat. Commun. 2015, 6, 6833. [Google Scholar] [CrossRef]
- Wilkinson, T.M.; Li, C.K.F.; Chui, C.S.C.; Huang, A.K.Y.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against in-fluenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef]
- Berthoud, T.K.; Hamill, M.; Lillie, P.J.; Hwenda, L.; Collins, K.A.; Ewer, K.J.; Milicic, A.; Poyntz, H.C.; Lambe, T.; Fletcher, H.A.; et al. Potent CD8+ T-Cell Immunogenicity in Humans of a Novel Heterosubtypic Influenza A Vac-cine, MVA-NP+M1. Clin. Infect. Dis. 2011, 52, 1–7. [Google Scholar] [CrossRef]
- Lillie, P.J.; Berthoud, T.K.; Powell, T.J.; Lambe, T.; E Mullarkey, C.; Spencer, A.J.; Hamill, M.M.; Peng, Y.; Blais, M.-E.; Duncan, C.; et al. Preliminary Assessment of the Efficacy of a T-Cell–Based Influenza Vaccine, MVA-NP+M1, in Humans. Clin. Infect. Dis. 2012, 55, 19–25. [Google Scholar] [CrossRef]
- Coughlan, L.; Sridhar, S.; Payne, R.; Edmans, M.; Milicic, A.; Venkatraman, N.; Lugonja, B.; Clifton, L.; Qi, C.; Folegatti, P.M.; et al. Heterologous Two-Dose Vaccination with Simian Adenovirus and Poxvirus Vectors Elicits Long-Lasting Cellular Immunity to Influenza Virus A in Healthy Adults. EBioMedicine 2018, 29, 146–154. [Google Scholar] [CrossRef]
- Vaccitech. Phase 2 Clinical Results for Vaccitech’s Universal Influenza A Vaccine. 2020. Available online: https://www.vaccitech.co.uk/phase-2-clinical-results-for-vaccitech-universal-influenza/. (accessed on 1 March 2021).
- Powell, T.J.; Silk, J.D.; Sharps, J.; Fodor, E.; Townsend, A.R.M. Pseudotyped Influenza A Virus as a Vaccine for the Induction of Heterotypic Immunity. J. Virol. 2012, 86, 13397–13406. [Google Scholar] [CrossRef]
- Morgan, S.B.; Hemmink, J.D.; Porter, E.; Harley, R.; Shelton, H.; Aramouni, M.; Everett, H.E.; Brookes, S.M.; Bailey, M.; Townsend, A.M.; et al. Aerosol Delivery of a Candidate Universal Influenza Vaccine Reduces Viral Load in Pigs Challenged with Pandemic H1N1 Virus. J. Immunol. 2016, 196, 5014–5023. [Google Scholar] [CrossRef]
- Holzer, B.; Morgan, S.B.; Matsuoka, Y.; Edmans, M.; Salguero, F.J.; Everett, H.; Brookes, S.M.; Porter, E.; Macloughlin, R.; Charleston, B.; et al. Comparison of Heterosubtypic Protection in Ferrets and Pigs Induced by a Single-Cycle Influ-enza Vaccine. J. Immunol. 2018, 200, 4068–4077. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Wohlbold, T.J.; Zheng, N.Y.; Huang, M.; Huang, Y.; Neu, K.E.; Lee, J.; Wan, H.; Rojas, K.T.; Kirkpatrick, E.; et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuramini-dase-Reactive Antibodies. Cell 2018, 173, 417–429.e10. [Google Scholar] [CrossRef] [PubMed]
- Eichelberger, M.C.; Wan, H. Influenza Neuraminidase as a Vaccine Antigen. Curr. Top. Microbiol. Immunol. 2014, 386, 275–299. [Google Scholar] [CrossRef]
- Krammer, F.; Fouchier, R.A.M.; Eichelberger, M.C.; Webby, R.J.; Shaw-Saliba, K.; Wan, H.; Wilson, P.C.; Compans, R.W.; Skountzou, I.; Monto, A.S. NAction! How Can Neuraminidase-Based Immunity Contribute to Better Influenza Virus Vaccines? MBio 2018, 9, e02332-17. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Hashem, A.M.; Li, C.; Van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013, 100, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Lee, Y.-T.; Park, S.; Jung, Y.-J.; Lee, Y.; Ko, E.-J.; Kim, Y.-J.; Li, X.; Kang, S.-M. Neuraminidase expressing virus-like particle vaccine provides effective cross protection against influenza virus. Virology 2019, 535, 179–188. [Google Scholar] [CrossRef]
- Zebedee, S.L.; Richardson, C.D.; Lamb, R.A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 1985, 56, 502–511. [Google Scholar] [CrossRef]
- Zebedee, S.L.; Lamb, R.A. Nucleotide sequences of influenza A virus RNA segment 7: A comparison of five isolates. Nucleic Acids Res. 1989, 17, 2870. [Google Scholar] [CrossRef][Green Version]
- Ito, T.; Gorman, O.T.; Kawaoka, Y.; Bean, W.J.; Webster, R.G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J. Virol. 1991, 65, 5491–5498. [Google Scholar] [CrossRef]
- El Bakkouri, K.; Descamps, F.; De Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; Van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal Vaccine Based on Ectodomain of Matrix Protein 2 of Influenza A: Fc Receptors and Alveolar Macrophages Mediate Protection. J. Immunol. 2010, 186, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef] [PubMed]
- Mezhenskaya, D.; Isakova-Sivak, I.; Rudenko, L. M2e-based universal influenza vaccines: A historical overview and new approaches to development. J. Biomed. Sci. 2019, 26, 1–15. [Google Scholar] [CrossRef]
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K.; et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214. [Google Scholar] [CrossRef]
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e–flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152. [Google Scholar] [CrossRef]
- Talbot, H.K.; Rock, M.T.; Johnson, C.; Tussey, L.; Kavita, U.; Shanker, A.; Shaw, A.R.; Taylor, D.N. Immunopotentiation of Trivalent Influenza Vaccine When Given with VAX102, a Recombinant Influenza M2e Vaccine Fused to the TLR5 Ligand Flagellin. PLoS ONE 2010, 5, e14442. [Google Scholar] [CrossRef]
- Tsybalova, L.M.; Stepanova, L.A.; Kuprianov, V.V.; Blokhina, E.A.; Potapchuk, M.V.; Korotkov, A.V.; Gorshkov, A.N.; Kasyanenko, M.A.; Ravin, N.V.; Kiselev, O.I. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant region of hepatitis B core antigen: Broad protective efficacy of particles carrying four copies of M2e. Vaccine 2015, 33, 3398–3406. [Google Scholar] [CrossRef]
- Neirynck, S.; DeRoo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Blue Water Vaccinces. Our Vaccine. 2019. Available online: https://www.bluewatervaccines.com (accessed on 1 February 2021).
- A Breakthrough in the Development of a Universal Flu Vaccine. Available online: https://www.ox.ac.uk/research/research-impact/breakthrough-development-universal-flu-vaccine (accessed on 15 March 2021).
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U.; et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemaggluti-nin Vaccine for H1N1 Influenza Viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.M.; Allen, J.D.; Bebin-Blackwell, A.G.; Carter, D.M.; Alefantis, T.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Computationally Optimized Broadly Reactive Hemagglutinin Elicits Hemagglutination Inhibi-tion Antibodies against a Panel of H3N2 Influenza Virus Cocirculating Variants. J. Virol. 2017, 91, e01581-17. [Google Scholar] [CrossRef]
- Crevar, C.J.; Carter, D.M.; Lee, K.Y.J.; Ross, T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccines Immunother. 2015, 11, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.M.; Hairong, L.; Chia, B.S.; Hill, E.; Weirback, H.; Zimmer, S. Prevalence of antibodies against seasonal influenza A and B viruses during the 2009–2010 and 2010–2011 influenza seasons in residents of Pittsburgh, PA, USA. PLoS Curr. 2011, 3, RRN1265. [Google Scholar] [CrossRef] [PubMed]
- DIOSynVax. Technology. 2021. Available online: https://diosvax.com/ (accessed on 1 April 2021).
- Zhang, X.-S.; Pebody, R.; De Angelis, D.; White, P.J.; Charlett, A.; McCauley, J.W. The Possible Impact of Vaccination for Seasonal Influenza on Emergence of Pandemic Influ-enza via Reassortment. PLoS ONE 2014, 9, e114637. [Google Scholar]
- Thompson, R.N.; Thompson, C.P.; Pelerman, O.; Gupta, S.; Obolski, U. Increased frequency of travel in the presence of cross-immunity may act to decrease the chance of a global pandemic. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180274. [Google Scholar] [CrossRef]
- Subramanian, R.; Graham, A.L.; Grenfell, B.T.; Arinaminpathy, N. Universal or Specific? A Modeling-Based Comparison of Broad-Spectrum Influenza Vac-cines against Conventional, Strain-Matched Vaccines. PLoS Comput. Biol. 2016, 12, e1005204. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing Computer Program, Version 3.6.1; R Foundation for Sta-tistical Computing: Vienna, Austria, 2019; Available online: http://www.r-project.org (accessed on 2 January 2021).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Flouri, T.; Izquierdo-Carrasco, F.; Darriba, D.; Aberer, A.; Nguyen, L.-T.; Minh, B.; Von Haeseler, A.; Stamatakis, A. The Phylogenetic Likelihood Library. Syst. Biol. 2015, 64, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef]
- Gasbarra, D.; Pirinen, M.; Sillanpää, M.J.; Salmela, E.; Arjas, E. Estimating genealogies from unlinked marker data: A Bayesian approach. Theor. Popul. Biol. 2007, 72, 305–322. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian Coalescent Inference of Past Population Dynamics from Molecular Sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T.-A. Dating of the human-ape splitting by a molecular clock of mitochon-drial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Figtree, A.R. A Graphical Viewer of Phylogenetic Trees. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree. (accessed on 1 April 2021).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Yu, G.; Lam, T.T.-Y.; Zhu, H.; Guan, Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolton, J.S.; Klim, H.; Wellens, J.; Edmans, M.; Obolski, U.; Thompson, C.P. An Antigenic Thrift-Based Approach to Influenza Vaccine Design. Vaccines 2021, 9, 657. https://doi.org/10.3390/vaccines9060657
Bolton JS, Klim H, Wellens J, Edmans M, Obolski U, Thompson CP. An Antigenic Thrift-Based Approach to Influenza Vaccine Design. Vaccines. 2021; 9(6):657. https://doi.org/10.3390/vaccines9060657
Chicago/Turabian StyleBolton, Jai S., Hannah Klim, Judith Wellens, Matthew Edmans, Uri Obolski, and Craig P. Thompson. 2021. "An Antigenic Thrift-Based Approach to Influenza Vaccine Design" Vaccines 9, no. 6: 657. https://doi.org/10.3390/vaccines9060657
APA StyleBolton, J. S., Klim, H., Wellens, J., Edmans, M., Obolski, U., & Thompson, C. P. (2021). An Antigenic Thrift-Based Approach to Influenza Vaccine Design. Vaccines, 9(6), 657. https://doi.org/10.3390/vaccines9060657





