Identification and Characterization of Immunodominant Proteins from Tick Tissue Extracts Inducing a Protective Immune Response against Ixodes ricinus in Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Ticks and Animals
2.2. Protein Extracts and Antisera
2.3. Direct Antigen Co-Immunoprecipitation
2.4. Tryptic Digestion
2.5. LC-MS/MS Analysis
2.6. Cloning and Purification
2.7. Western Blotting
2.8. Bioinformatics Analysis
2.9. RNA Isolation
2.10. Quantitative RT-PCR
2.11. RNA Interference
2.12. Statistical Analysis
3. Results
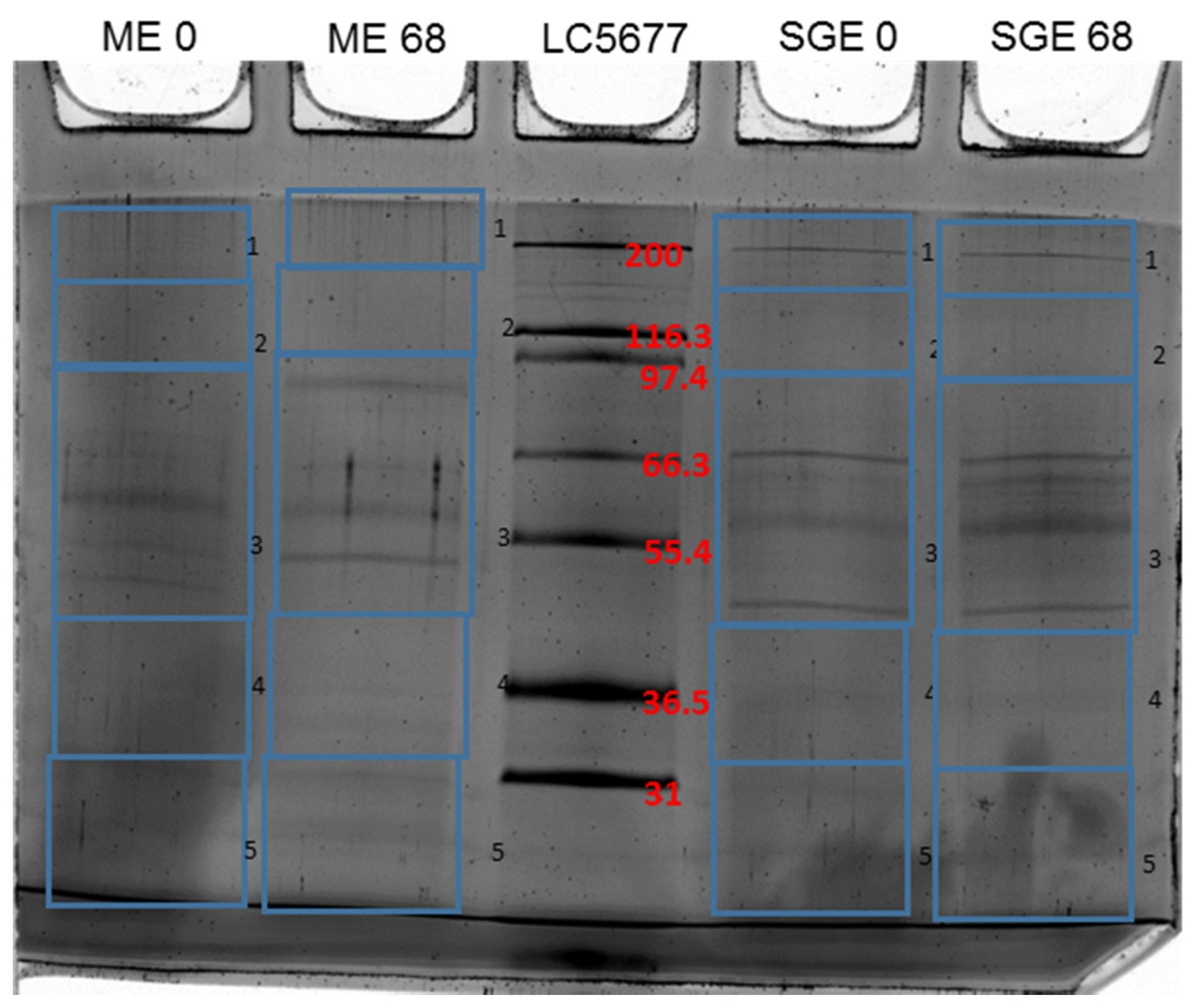
3.1. Identification of Differentially Recognized Proteins from ME and SGE by Immune Sera of Calves Immunized with ME and SGE
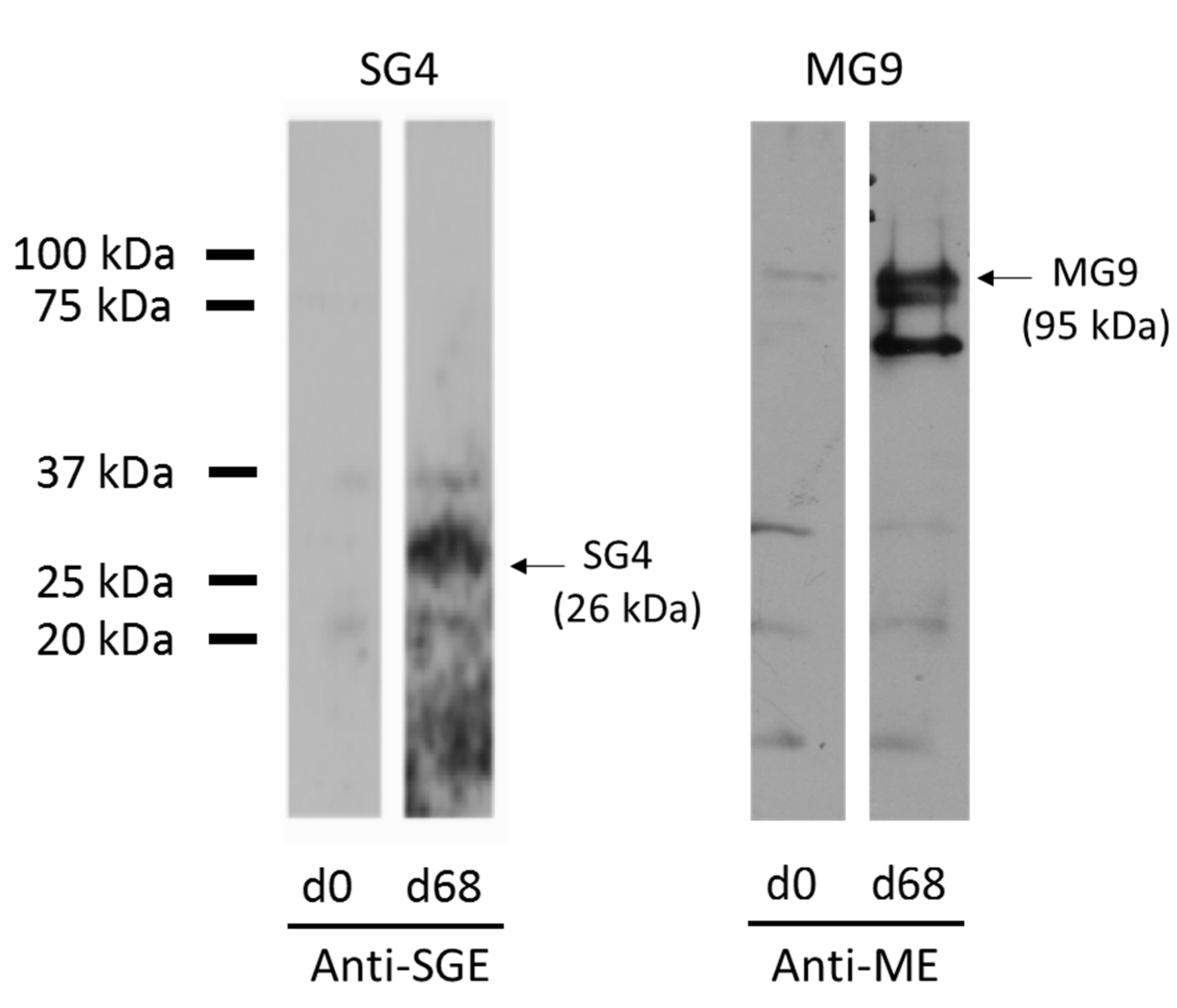
3.2. Expression of Recombinant Immunodominant Proteins and Validation by Western Blot
3.3. Expression Profile of Selected Genes in Different Tissues of Ixodes ricinus Females
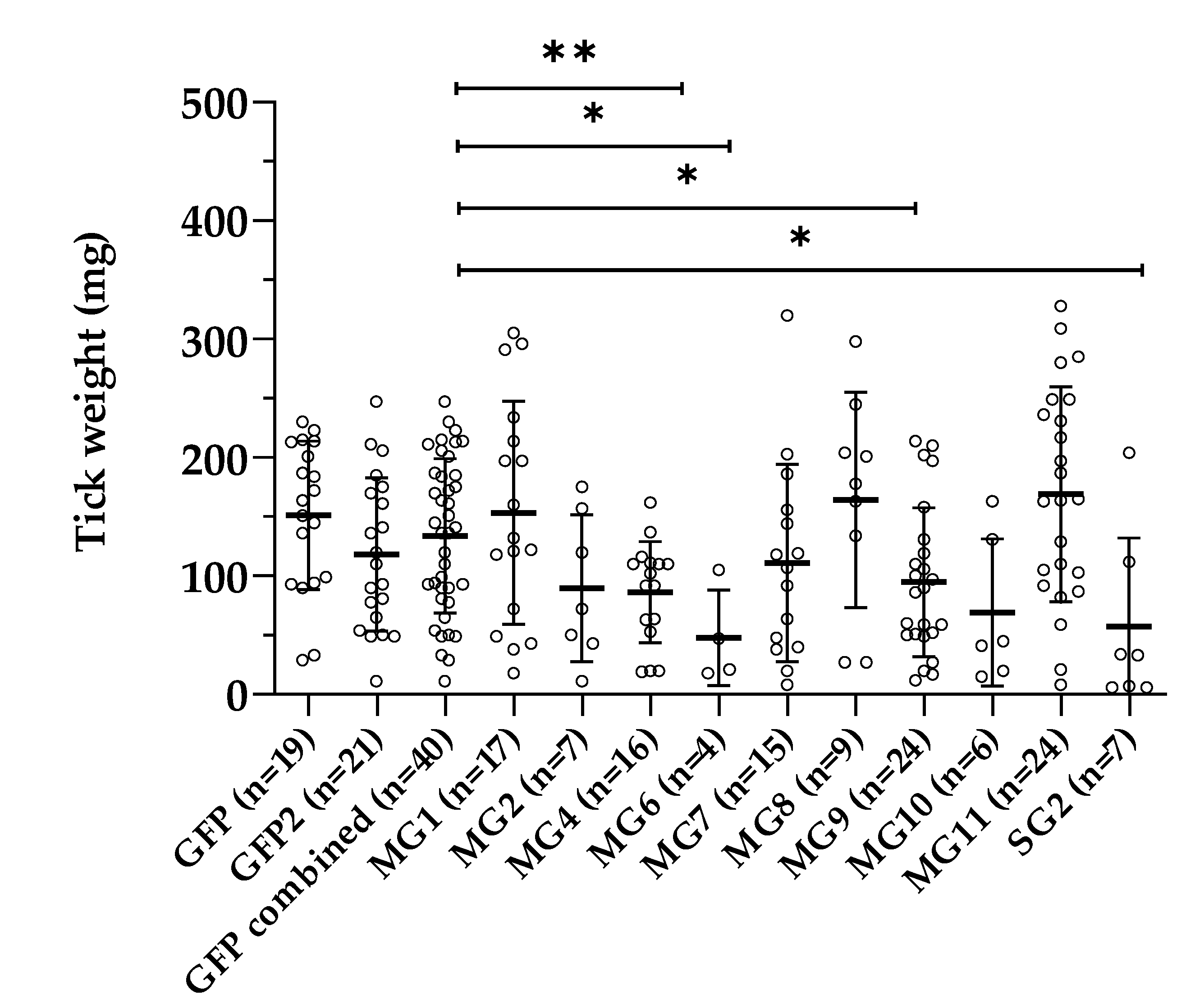
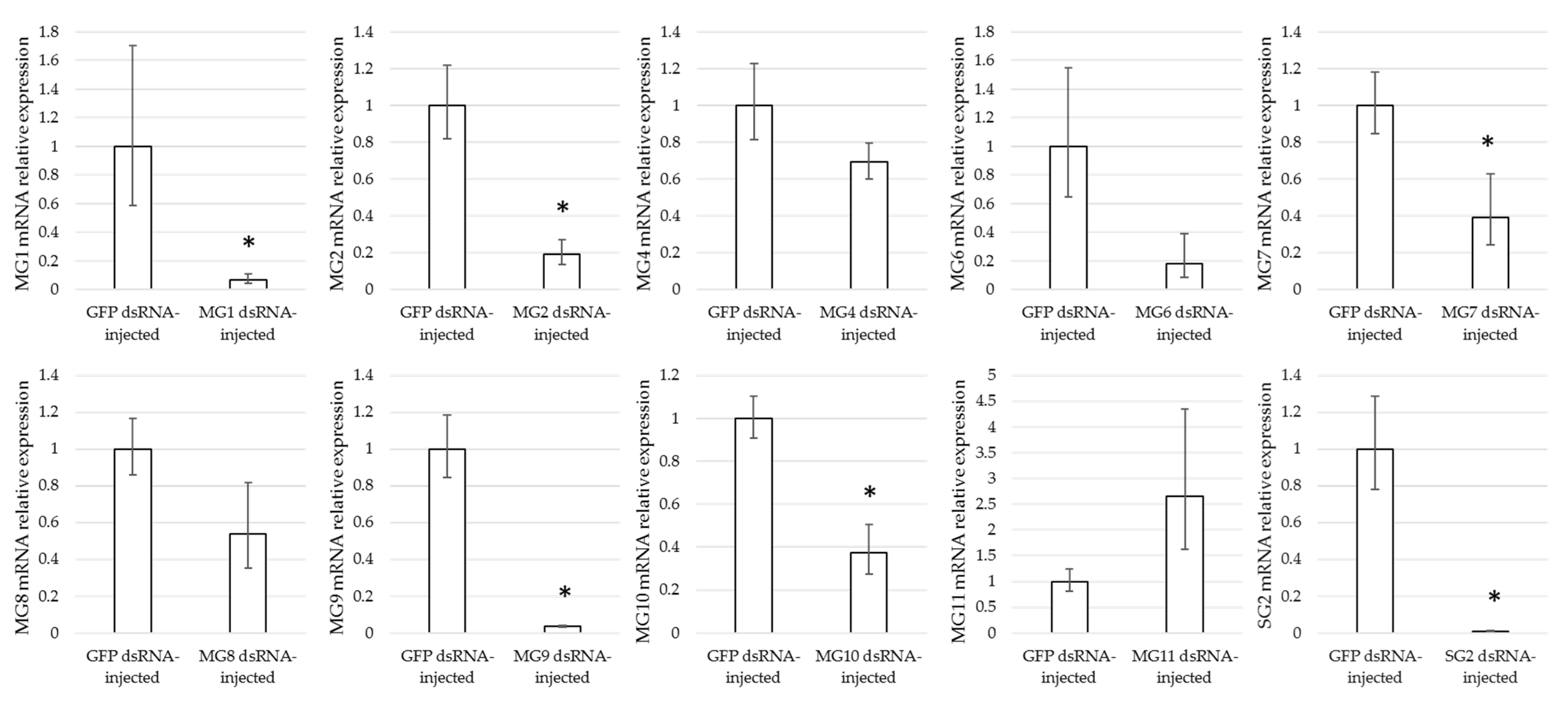
3.4. Effect of Gene Silencing of Selected Candidates on I. ricinus Adult Feeding
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heyman, P.; Cochez, C.; Hofhuis, A.; van der Giessen, J.; Sprong, H.; Porter, S.R.; Losson, B.; Saegerman, C.; Donoso-Mantke, O.; Niedrig, M.; et al. A clear and present danger: Tick-borne diseases in Europe. Expert Rev. Anti-Infect. Ther. 2010, 8, 33–50. [Google Scholar] [CrossRef]
- Apostolovic, D.; Mihailovic, J.; Commins, S.P.; Wijnveld, M.; Kazimirova, M.; Starkhammar, M.; Stockinger, H.; Platts-Mills, T.A.E.; Cirkovic Velickovic, T.; Hamsten, C.; et al. Allergenomics of the tick Ixodes ricinus reveals important alpha-Gal-carrying IgE-binding proteins in red meat allergy. Allergy 2020, 75, 217–220. [Google Scholar] [CrossRef]
- Apostolovic, D.; Tran, T.A.; Starkhammar, M.; Sanchez-Vidaurre, S.; Hamsten, C.; Van Hage, M. The red meat allergy syndrome in Sweden. Allergo J. Int. 2016, 25, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Sprong, H.; Azagi, T.; Hoornstra, D.; Nijhof, A.M.; Knorr, S.; Baarsma, M.E.; Hovius, J.W. Control of Lyme borreliosis and other Ixodes ricinus-borne diseases. Parasites Vectors 2018, 11, 145. [Google Scholar] [CrossRef]
- Embers, M.E.; Narasimhan, S. Vaccination against Lyme disease: Past, present, and future. Front. Cell. Infect. Microbiol. 2013, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Rego, R.O.M.; Trentelman, J.J.A.; Anguita, J.; Nijhof, A.M.; Sprong, H.; Klempa, B.; Hajdusek, O.; Tomas-Cortazar, J.; Azagi, T.; Strnad, M.; et al. Counterattacking the tick bite: Towards a rational design of anti-tick vaccines targeting pathogen transmission. Parasites Vectors 2019, 12, 229. [Google Scholar] [CrossRef] [PubMed]
- Koci, J.; Bista, S.; Chirania, P.; Yang, X.; Kitsou, C.; Rana, V.S.; Yas, O.B.; Sonenshine, D.E.; Pal, U. Antibodies against EGF-like domains in Ixodes scapularis BM86 orthologs impact tick feeding and survival of Borrelia burgdorferi. Sci. Rep. 2021, 11, 6095. [Google Scholar] [CrossRef]
- de la Fuente, J.; Almazan, C.; Canales, M.; de la Lastra, J.M.P.; Kocan, K.M.; Willadsen, P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 2007, 8, 23–28. [Google Scholar] [CrossRef]
- Willadsen, P. Anti-tick vaccines. Parasitology 2004, 129, S367–S387. [Google Scholar] [CrossRef] [PubMed]
- Willadsen, P.; Riding, G.A.; McKenna, R.V.; Kemp, D.H.; Tellam, R.L.; Nielsen, J.N.; Lahnstein, J.; Cobon, G.S.; Gough, J.M. Immunologic control of a parasitic arthropod. Identification of a protective antigen from Boophilus microplus. J. Immunol. 1989, 143, 1346–1351. [Google Scholar] [PubMed]
- Kemp, D.H.; Agbede, R.I.; Johnston, L.A.; Gough, J.M. Immunization of cattle against Boophilus microplus using extracts derived from adult female ticks: Feeding and survival of the parasite on vaccinated cattle. Int. J. Parasitol. 1986, 16, 115–120. [Google Scholar] [CrossRef]
- Trentelman, J.J.A.; Teunissen, H.; Kleuskens, J.; van de Crommert, J.; de la Fuente, J.; Hovius, J.W.R.; Schetters, T.P.M. A combination of antibodies against Bm86 and Subolesin inhibits engorgement of Rhipicephalus australis (formerly Rhipicephalus microplus) larvae in vitro. Parasites Vectors 2019, 12, 362. [Google Scholar] [CrossRef] [PubMed]
- Knorr, S.; Anguita, J.; Cortazar, J.T.; Hajdusek, O.; Kopacek, P.; Trentelman, J.J.; Kershaw, O.; Hovius, J.W.; Nijhof, A.M. Preliminary evaluation of tick protein extracts and recombinant Ferritin 2 as anti-tick vaccines targeting Ixodes ricinus in cattle. Front. Physiol. 2018, 9, 1696. [Google Scholar] [CrossRef]
- Nijhof, A.M.; Balk, J.A.; Postigo, M.; Jongejan, F. Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol. Biol. 2009, 10, 112. [Google Scholar] [CrossRef]
- Trentelman, J.J.A.; Sima, R.; Krezdorn, N.; Tomas-Cortazar, J.; Barriales, D.; Takumi, K.; Butler, J.M.; Sprong, H.; Klouwens, M.J.; Urbanova, V.; et al. A combined transcriptomic approach to identify candidates for an anti-tick vaccine blocking B. afzelii transmission. Sci. Rep. 2020, 10, 20061. [Google Scholar] [CrossRef] [PubMed]
- Mans, B.J. Quantitative visions of reality at the tick-host interface: Biochemistry, genomics, proteomics, and transcriptomics as measures of complete inventories of the tick sialoverse. Front. Cell. Infect. Microbiol. 2020, 10, 574405. [Google Scholar] [CrossRef]
- Huercha; Ma, Y.; Hao, Y.; Li, M.; Hu, Z.; Song, R.; Wei, L.; Fan, S.; Chen, S.; Fan, X.; et al. Sequence identification and expression profile of seven Dermacentor marginatus glutathione S-transferase genes. Exp. Appl. Acarol. 2020, 82, 295–308. [Google Scholar] [CrossRef]
- Huercha; Song, R.; Li, M.; Fan, X.; Hu, Z.; Wu, L.; Li, Y.; Zhang, W.; Zhang, Y.; Ma, Y.; et al. Caracterization of glutathione S-transferase of Dermacantor marginatus and effect of the recombinant antigen as a potential anti-tick vaccine. Vet. Parasitol. 2020, 279, 109043. [Google Scholar] [CrossRef]
- Ndawula, C., Jr.; Sabadin, G.A.; Parizi, L.F.; da Silva Vaz, I., Jr. Constituting a glutathione S-transferase-cocktail vaccine against tick infestation. Vaccine 2019, 37, 1918–1927. [Google Scholar] [CrossRef]
- Manjunathachar, H.V.; Kumar, B.; Saravanan, B.C.; Choudhary, S.; Mohanty, A.K.; Nagar, G.; Chigure, G.; Ravi Kumar, G.; de la Fuente, J.; Ghosh, S. Identification and characterization of vaccine candidates against Hyalomma anatolicum-Vector of Crimean-Congo haemorrhagic fever virus. Transbound. Emerg. Dis. 2019, 66, 422–434. [Google Scholar] [CrossRef]
- Tian, M.; Tian, Z.; Luo, J.; Xie, J.; Yin, H.; Zeng, Q.; Shen, H.; Chai, H.; Yuan, X.; Wang, F.; et al. Identification of the tropomyosin (HL-Tm) in Haemaphysalis longicornis. Vet. Parasitol. 2015, 207, 318–323. [Google Scholar] [CrossRef]
- Almazan, C.; Lagunes, R.; Villar, M.; Canales, M.; Rosario-Cruz, R.; Jongejan, F.; de la Fuente, J. Identification and characterization of Rhipicephalus (Boophilus) microplus candidate protective antigens for the control of cattle tick infestations. Parasitol. Res. 2010, 106, 471–479. [Google Scholar] [CrossRef]
- Almazan, C.; Moreno-Cantu, O.; Moreno-Cid, J.A.; Galindo, R.C.; Canales, M.; Villar, M.; de la Fuente, J. Control of tick infestations in cattle vaccinated with bacterial membranes containing surface-exposed tick protective antigens. Vaccine 2012, 30, 265–272. [Google Scholar] [CrossRef]
- Gao, J.; Luo, J.; Fan, R.; Guan, G.; Ren, Q.; Ma, M.; Sugimoto, C.; Bai, Q.; Yin, H. Molecular characterization of a myosin alkali light chain-like protein, a “concealed” antigen from the hard tick Haemaphysalis qinghaiensis. Vet. Parasitol. 2007, 147, 140–149. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, G.; Zhang, L.; Yin, H.; Wang, H.; Xie, J.; Zhang, P.; Luo, J. Identification of the heat shock protein 70 (HLHsp70) in Haemaphysalis longicornis. Vet. Parasitol. 2011, 181, 282–290. [Google Scholar] [CrossRef]
- Kumar, B.; Manjunathachar, H.V.; Nagar, G.; Ravikumar, G.; de la Fuente, J.; Saravanan, B.C.; Ghosh, S. Functional characterization of candidate antigens of Hyalomma anatolicum and evaluation of its cross-protective efficacy against Rhipicephalus microplus. Vaccine 2017, 35, 5682–5692. [Google Scholar] [CrossRef]
- Diaz-Martin, V.; Manzano-Roman, R.; Oleaga, A.; Encinas-Grandes, A.; Perez-Sanchez, R. Cloning and characterization of a plasminogen-binding enolase from the saliva of the argasid tick Ornithodoros moubata. Vet. Parasitol. 2013, 191, 301–314. [Google Scholar] [CrossRef]
- Willadsen, P. The Development of a New or Improved Vaccine against Boophilus microplus: Opportunities for R&D Investment; Meat & Livestock Australia Limited: Sydney, Australia, 2008. [Google Scholar]
- Xu, L.; Liu, L.; Cheng, T.Y. Cloning and expression profile of glyceraldehyde-3-phosphate dehydrogenase in Haemaphysalis flava (Acari: Ixodidae). J. Med. Entomol. 2019, 56, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Decrem, Y.; Beaufays, J.; Blasioli, V.; Lahaye, K.; Brossard, M.; Vanhamme, L.; Godfroid, E. A family of putative metalloproteases in the salivary glands of the tick Ixodes ricinus. FEBS J. 2008, 275, 1485–1499. [Google Scholar] [CrossRef] [PubMed]
- Decrem, Y.; Mariller, M.; Lahaye, K.; Blasioli, V.; Beaufays, J.; Boudjeltia, K.Z.; Vanhaeverbeek, M.; Cerutti, M.; Brossard, M.; Vanhamme, L.; et al. The impact of gene knock-down and vaccination against salivary metalloproteases on blood feeding and egg laying by Ixodes ricinus. Int. J. Parasitol. 2008, 38, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Duscher, G.G.; Galindo, R.C.; Tichy, A.; Hummel, K.; Kocan, K.M.; de la Fuente, J. Glutathione S-transferase affects permethrin detoxification in the brown dog tick, Rhipicephalus sanguineus. Ticks Tick Borne Dis. 2014, 5, 225–233. [Google Scholar] [CrossRef]
- Hernandez, E.P.; Kusakisako, K.; Talactac, M.R.; Galay, R.L.; Hatta, T.; Fujisaki, K.; Tsuji, N.; Tanaka, T. Glutathione S-transferases play a role in the detoxification of flumethrin and chlorpyrifos in Haemaphysalis longicornis. Parasites Vectors 2018, 11, 460. [Google Scholar] [CrossRef]
- Henderson, B.; Nair, S.; Pallas, J.; Williams, M.A. Fibronectin: A multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 2011, 35, 147–200. [Google Scholar] [CrossRef] [PubMed]
- Vaca, D.J.; Thibau, A.; Schutz, M.; Kraiczy, P.; Happonen, L.; Malmstrom, J.; Kempf, V.A.J. Interaction with the host: The role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med. Microbiol. Immunol. 2020, 209, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Coumou, J.; Schuijt, T.J.; Boder, E.; Hovius, J.W.; Fikrig, E. A tick gut protein with fibronectin III domains aids Borrelia burgdorferi congregation to the gut during transmission. PLoS Pathog. 2014, 10, e1004278. [Google Scholar] [CrossRef] [PubMed]
- Debalke, S.; Habtewold, T.; Duchateau, L.; Christophides, G.K. The effect of silencing immunity related genes on longevity in a naturally occurring Anopheles arabiensis mosquito population from southwest Ethiopia. Parasites Vectors 2019, 12, 174. [Google Scholar] [CrossRef]
- Cooper, A.M.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef]
- Barry, G.; Alberdi, P.; Schnettler, E.; Weisheit, S.; Kohl, A.; Fazakerley, J.K.; Bell-Sakyi, L. Gene silencing in tick cell lines using small interfering or long double-stranded RNA. Exp. Appl. Acarol. 2013, 59, 319–338. [Google Scholar] [CrossRef]
- Sugahara, R.; Jouraku, A.; Nakakura, T.; Kusakabe, T.; Yamamoto, T.; Shinohara, Y.; Miyoshi, H.; Shiotsuki, T. Two adenine nucleotide translocase paralogues involved in cell proliferation and spermatogenesis in the silkworm Bombyx mori. PLoS ONE 2015, 10, e0119429. [Google Scholar] [CrossRef]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Renne, T.; Dedio, J.; Meijers, J.C.; Chung, D.; Muller-Esterl, W. Mapping of the discontinuous H-kininogen binding site of plasma prekallikrein. Evidence for a critical role of apple domain-2. J. Biol. Chem. 1999, 274, 25777–25784. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.H.; Badellino, K.; Baglia, F.A.; Walsh, P.N. A binding site for heparin in the apple 3 domain of factor XI. J. Biol. Chem. 1998, 273, 16382–16390. [Google Scholar] [CrossRef] [PubMed]
- Surakasi, V.P.; Mohamed, A.A.; Kim, Y. RNA interference of beta1 integrin subunit impairs development and immune responses of the beet armyworm, Spodoptera exigua. J. Insect. Physiol. 2011, 57, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Ahn, S.J.; Vogel, H.; Kim, Y. Integrin beta subunit and its RNA interference in immune and developmental processes of the Oriental tobacco budworm, Helicoverpa assulta. Dev. Comp. Immunol. 2014, 47, 59–67. [Google Scholar] [CrossRef]

| Target | UNIPROT ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| ELF1A | CAAGATTGGTGGTATCGGCA | GACCTCAGTGGTGATGTTGGC | 106 | |
| ATP5L | CGAAAGCCATCACTACCCTG | TCTCCACCTTGGCATACTTGAC | 85 | |
| MG1 | A0A131XWI1 | GGATGTGGTTTTACCCCGTC | CCCTCTTGAGCGTTGGATG | 115 |
| MG2 | V5IFB6 | CTCTGGAAACCTGGACAACG | GCAGCGTGAAAGATAGAGTCC | 131 |
| MG3 | V5HWP4 | AATCGCCAGTTGTCAGAAGC | TCAAGCCGACAGCAAATATG | 76 |
| MG4 | V5I2L3 | CACTGGTCATCTCCTGGCTC | CGTGCTCTTGTACATAAGGTCTG | 137 |
| MG5 | A0A0K8R8I3 | GTCTCTGCTTCGGTGTCTCC | GGCGACTTGAGGTTGTAGG | 83 |
| MG6 | A0A147BXB7 | GAGACTCCCAAGGACAAGAACC | TGTAGAGATATTTTTGCCACCAGG | 78 |
| MG7 | V5IJN2 | CCGAAGTCTCCAAGGGTCC | ACCGACTCCATCGTCAAAAAG | 116 |
| MG8 | A0A147BMG4 | GACAACACCACGGCACAGG | GGTGTAGGGCTTGAAGTTGTAGAA | 92 |
| MG9 | A0A131YAQ2 | GGGGATTTCCGAAGCCAC | CTGAAGATATTGTTGACGGGGTC | 146 |
| MG10 | V5H492 | AAACGGGCATCAGCAAAGC | TTGTTGAGATCGCCAGCAGAC | 97 |
| MG11 | A0A131XS30 | CATTCGTAGATCACACCCTGC | CGGCGATTCGTAGCGTG | 107 |
| SG1 | V5HWD5 | CCACTACGAAGGCTACCACAA | CCTATTCAGCCCTGTCCATC | 56 |
| SG2 | A0A0K8RKT7 | TTGCCTACGAGATGCTGTCC | TGAACTTGTCCGACTTGAGGT | 135 |
| SG3 | A0A0K8RPW5 | AGTTTACGAGCTTCTCTTGCC | TCCGTCGTGAACACTACCG | 102 |
| SG4 | A0A0K8RQF1 | CTTCCGAAGAGTGTCAGGGTGA | GTGCCGAATGCCGACTGC | 108 |
| Target | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|
| MG1 | T7-CCCTTCCATCTTGCGGTAGC | T7- CGAACGAAGAGCGGAACG | 393 |
| MG2 | T7-GAATCCCCAGTCCAAGATGATC | T7- CTTTCGTGACCGCTCGTTC | 538 |
| MG4 | T7-CAGACATCGGCAAGGGTG | T7-GAGCCAGGAGATGACCAGTG | 229 |
| MG6 | T7-GCGGACGAAGAGGAATACG | T7-GCTAAGAGTAACATTGGTGTATCC | 493 |
| MG7 | T7-ACCACATCTGCCAACGGAG | T7-ATCCCAAGTAGGAAGCCGTT | 257 |
| MG8 | T7-ACTTTGCTTTCTTGGCATCGG | T7-GTCGTATGTGTTGCCTTTGTCG | 429 |
| MG9 | T7-GGTGGCATTGACAACGCTCTC | T7-GAACTTCTTCGTCGCTTCCTTG | 400 |
| MG10 | T7-GGCTCCAGAAAACACAATCCTC | T7-CCTTTTCCGTGGTAGAATGGG | 666 |
| MG11 | T7- CCAGGATGGGAAAGTGCGAC | T7- GAACGCCAGCGAACCAGG | 243 |
| SG2 | T7-CCAAACCTGCCCTACTACCTG | T7-GGACAGCATCTCGTAGGCAAT | 324 |
| GFP | T7-GGCCACAAGTTCAGCGTGTC | T7-GCTTGATGCCGTTCTTCTGC | 415 |
| Uniprot Accession Code | Protein Name | # of Unique Peptides | Mascot Score d0 | Mascot Score d68 | MW (kDa) | pI | Signal Peptide |
|---|---|---|---|---|---|---|---|
| V5H4T2 | Trifunctional purine biosynthetic protein adenosine-3 * | 1 | 41.20 | 71.91 | 7.3 | 8.19 | No |
| V5HMC9 | Small nuclear ribonucleoprotein G, putative | 1 | 54.37 | 8.4 | 8.54 | No | |
| V5IF42 | Myosin-2 essential light chain | 1 | 40.41 | 9.7 | 6.51 | No | |
| V5HG94 | 60S ribosomal protein L22 | 2 | 76.25 | 88.50 | 9.9 | 10.01 | No |
| A0A0K8RQM9 | Small nuclear ribonucleoprotein Sm D3 | 1 | 58.85 | 13.2 | 10.13 | No | |
| V5IJC3 | 60s ribosomal protein L11 | 1 | 128.84 | 79.57 | 13.2 | 10.62 | No |
| A0A0K8RIJ1 | Histone H2A | 1 | 50.33 | 13.4 | 10.73 | No | |
| A0A131Y512 | 40S ribosomal protein S16 | 1 | 32.98 | 14.9 | 10.04 | No | |
| V5HG43 | Stromal cell-derived factor 2, putative | 1 | 34.15 | 15.2 | 10.32 | No | |
| A0A090XEK9 | Myosin, essential light chain | 2 | 41.67 | 44.63 | 15.5 | 4.94 | No |
| V5HWD5 | Metalloproteinase (=SG1) a | 1 | 44.26 | 16.9 | 9.92 | No | |
| A0A0K8RC23 | 40S ribosomal protein S13 | 1 | 33.57 | 17.2 | 10.68 | No | |
| A0A0K8RL33 | Superoxide-dismutase | 1 | 35.85 | 18.1 | 6.64 | Yes | |
| V5HD78 | 60S ribosomal protein L6 | 4 | 62.85 | 57.09 | 18.6 | 10.33 | No |
| V5I150 | 60S ribosomal protein L5-A | 3 | 94.73 | 50.00 | 19.0 | 7.84 | No |
| V5I135 | Alpha-crystallin A chain * | 1 | 54.14 | 20.3 | 7.64 | No | |
| V5HXA8 | 60S ribosomal protein L18 | 2 | 60.03 | 131.89 | 21.5 | 11.62 | No |
| V5H3S3 | 60S ribosomal L23 | 2 | 67.50 | 21.6 | 11.39 | No | |
| A0A0K8RQ35 | 40S ribosomal protein S8 | 1 | 40.95 | 21.9 | 10.30 | No | |
| A0A0K8RKT7 | Glutathione S-transferase (=SG2) | 1 | 39.95 | 25.5 | 7.88 | No | |
| A0A0K8RPW5 | Metalloproteinase (=SG3) a | 1 | 45.77 | 26.6 | 9.45 | Yes | |
| V5I164 | Tropomyosin, isoform close to X4 | 3 | 145.00 | 57.96 | 26.6 | 5.34 | No |
| A0A0K8RHG9 | Tubulin alpha chain | 1 | 62.27 | 27.1 | 5.57 | No | |
| A0A0K8RQF1 | Toll-like receptor, putative (=SG4) | 1 | 41.27 | 27.6 | 8.31 | Yes | |
| A0A131XW65 | 60S ribosomal protein L7 | 1 | 79.15 | 70.36 | 29.2 | 10.98 | No |
| A0A0K8RG40 | 40S ribosomal protein S4 | 3 | 74.40 | 81.92 | 29.6 | 10.29 | No |
| A0A0K8RG01 | Glyceraldehyde-3-phosphate dehydrogenase 2, isoform X1 * | 1 | 51.70 | 36.0 | 7.84 | No | |
| V5HG89 | ATP synthase subunit beta | 2 | 87.40 | 36.3 | 5.03 | No | |
| E3SS18 | Translation elongation factor EF1-alpha * | 4 | 63.01 | 184.99 | 36.7 | 8.27 | No |
| A0A147BVX5 | Venom metalloproteinase antarease-like TtrivMP_A | 1 | 34.45 | 38.8 | 5.83 | Yes | |
| Q5D579 | Actin * | 9 | 401.51 | 335.42 | 41.5 | 5.85 | No |
| A0A0K8RDN7 | Protein N-myc downstream-regulated gene 3 (NDRG3) isoform X1 | 2 | 46.54 | 88.18 | 44.6 | 6.54 | No |
| A0A0K8RCY6 | Tubulin beta chain | 3 | 40.59 | 127.53 | 45.1 | 5.97 | No |
| A0A131XPA0 | Eukaryotic translation initiation factor 3 subunit M | 1 | 36.48 | 45.2 | 5.97 | No | |
| A0A0K8RMJ6 | 60S ribosomal protein L4 | 1 | 37.81 | 46.6 | 11.19 | No | |
| V5I085 | Microsomal triglyceride transfer protein large subunit b | 1 | 36.01 | 58.33 | 48.0 | 6.89 | No |
| A0A0K8R4C2 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase 48 kDa subunit | 1 | 37.19 | 48.8 | 6.05 | Yes | |
| A0A0K8R4D7 | Cytochrome b-c1 complex subunit 2 | 3 | 105.77 | 115.65 | 48.9 | 8.62 | No |
| A0A131Y1S2 | Microsomal triglyceride transfer protein large subunit b | 2 | 36.65 | 51.53 | 49.7 | 8.97 | Yes |
| V5I095 | S-adenosylhomocysteine hydrolase-like protein | 4 | 93.01 | 50.5 | 6.54 | No | |
| A0A131XNF3 | Processing peptidase beta subunit, putative * | 2 | 105.08 | 87.93 | 53.4 | 6.15 | No |
| A0A0K8RCY2 | Metis1 | 4 | 149.35 | 176.93 | 55.5 | 7.58 | Yes |
| A0A131XPM3 | Heat shock protein 60 * | 5 | 151.93 | 59.3 | 5.62 | No | |
| A0A0K8RCE8 | Heat shock 70 kDa protein cognate 4 * | 2 | 60.97 | 115.44 | 59.8 | 7.43 | No |
| A0A0K8RP16 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 | 3 | 115.81 | 67.5 | 7.53 | Yes | |
| A0A090XC63 | Moesin/ezrin/radixin homolog 1 isoform X1 * | 7 | 81.17 | 206.20 | 70.1 | 5.66 | No |
| A0A0K8RIU3 | Heat shock protein, putative * | 3 | 66.94 | 102.00 | 72.6 | 5.41 | Yes |
| V5HP83 | Coatomer subunit alpha | 2 | 61.57 | 75.6 | 8.79 | No | |
| A0A0K8R8N9 | Heat shock protein HSP 90-alpha | 1 | 37.81 | 84.2 | 5.02 | No | |
| V5HRY6 | Sodium/potassium-transporting ATPase subunit alpha-B | 3 | 144.31 | 91.7 | 5.16 | No | |
| A0A131XXE4 | F-box only protein 11 | 1 | 37.74 | 99.1 | 7.06 | No | |
| A0A131XWG4 | Coatomer subunit beta | 2 | 59.80 | 103.2 | 5.21 | No | |
| V5GY25 | Clathrin heavy chain 1 * | 3 | 35.11 | 119.33 | 190.7 | 5.81 | No |
| V5I4B8 | Myosin heavy chain, muscle isoform X3 * | 40 | 1378.54 | 1750.99 | 222.0 | 6.09 | No |
| V5I3C9 | Myosin heavy chain, non-muscle isoform X1 * | 15 | 354.64 | 650.15 | 227.5 | 5.55 | No |
| Uniprot Accession Code | Protein Name | # of Unique Peptides | Mascot Score d0 | Mascot Score d68 | MW (kDa) | pI | Signal Peptide |
|---|---|---|---|---|---|---|---|
| V5H4T2 | Trifunctional purine biosynthetic protein adenosine-3 * | 1 | 52.26 | 7.3 | 8.19 | No | |
| V5HY31 | Histone H4-like, putative | 1 | 37.81 | 9.8 | 10.43 | No | |
| A0A0K8RK48 | Uncharacterized protein | 1 | 43.68 | 44.01 | 13.2 | 7.83 | Yes |
| A0A0K8RQA6 | Ubiquitin | 1 | 47.08 | 14.5 | 9.82 | No | |
| V5H0K4 | Pantetheinase, putative a | 1 | 161.39 | 16.3 | 6.54 | Yes | |
| A0A131Y7G4 | Uncharacterized protein b | 1 | 58.35 | 84.84 | 18.2 | 8.29 | Yes |
| V5I2L3 | ADP/ATP translocase, putative (=MG4) | 1 | 42.62 | 19.3 | 9.58 | No | |
| V5I135 | Alpha-crystallin A chain * | 1 | 110.22 | 20.3 | 7.64 | No | |
| A0A090X8W5 | Uncharacterized protein b | 1 | 73.12 | 61.59 | 21.7 | 8.94 | Yes |
| A0A131YAP7 | Tropomyosin isoform X15/X16 | 1 | 61.58 | 23.6 | 4.74 | No | |
| V5HHC0 | Uncharacterized protein | 1 | 58.35 | 95.16 | 23.8 | 9.85 | Yes |
| A0A131XX88 | 60S ribosomal protein L19 | 1 | 44.69 | 24.2 | 11.43 | No | |
| A0A131XRL8 | Cathepsin L | 1 | 72.31 | 27.7 | 5.48 | No | |
| A0A131XWI1 | Salivary secreted cytotoxin, putative (=MG1) | 1 | 66.40 | 32.0 | 9.33 | No | |
| V5HBQ2 | Lysosomal Pro-X carboxypeptidase | 1 | 40.01 | 33.9 | 5.08 | No | |
| V5HWP4 | Uncharacterized protein (=MG3) | 1 | 48.01 | 35.0 | 7.01 | No | |
| A0A0K8RNA0 | Malate dehydrogenase | 2 | 73.48 | 35.8 | 9.09 | No | |
| A0A0K8RG01 | Glyceraldehyde-3-phosphate dehydrogenase 2 isoform X1 * | 1 | 39.91 | 36.0 | 7.84 | No | |
| E3SS18 | Translation elongation factor EF1-alpha * | 3 | 54.98 | 36.81 | 36.7 | 8.27 | No |
| Q5D579 | Actin * | 7 | 248.34 | 38.91 | 41.5 | 5.85 | No |
| A0A0K8RCB1 | Enolase | 1 | 38.63 | 47.1 | 6.01 | No | |
| A0A131XPI3 | Aminopeptidase, putative W07G4.4 | 1 | 34.48 | 47.6 | 8.32 | No | |
| A0A147BSS4 | Pantetheinase a | 1 | 206.82 | 52.5 | 7.01 | Yes | |
| A0A131XNF3 | Processing peptidase beta subunit, putative * | 1 | 50.89 | 53.4 | 6.15 | No | |
| V5HEY6 | Alpha-L-fucosidase | 1 | 55.17 | 53.5 | 6.92 | Yes | |
| V5HB74 | Retinal dehydrogenase 1 | 4 | 181.55 | 90.92 | 54.6 | 6.89 | No |
| V5IJN2 | Calcium-activated chloride channel regulator (=MG7) | 1 | 39.96 | 55.1 | 4.96 | No | |
| A0A131Y0J3 | Alpha-aminoadipic semialdehyde dehydrogenase | 1 | 39.78 | 58.7 | 6.79 | No | |
| A0A131XPM3 | Heat shock protein 60 * | 1 | 89.26 | 59.3 | 5.62 | No | |
| A0A0K8RCE8 | Heat shock 70 kDa protein cognate 4 * | 3 | 171.54 | 59.8 | 7.43 | No | |
| A0A131XQI6 | Moesin/ezrin/radixin homolog 1 * | 1 | 97.17 | 62.5 | 5.52 | No | |
| V5HN24 | Beta-hexosaminidase subunit beta | 1 | 38.14 | 63.4 | 5.40 | No | |
| A0A0K8RIU3 | Heat shock protein, putative * | 3 | 179.61 | 72.6 | 5.41 | Yes | |
| A0A0K8R8I3 | Uncharacterized protein (cubilin-like?) (=MG5) | 2 | 80.81 | 75.8 | 6.67 | Yes | |
| V5IFB6 | Integrin beta-PS (=MG2) | 3 | 193.69 | 83.8 | 5.17 | No | |
| V5GPX7 | Alpha-actinin isoform X2 | 1 | 38.52 | 89.4 | 6.13 | No | |
| A0A131YAQ2 | Uncharacterized protein (=MG9) | 1 | 71.18 | 94.3 | 6.62 | Yes | |
| A0A147BXB7 | Cell adhesion molecule, putative (=MG6) | 3 | 70.59 | 104.8 | 6.34 | Yes | |
| A0A147BMG4 | Uncharacterized protein (=MG8) | 1 | 34.22 | 109.1 | 5.95 | Yes | |
| A0A0K8RQE7 | Lysosomal alpha-mannosidase-like | 1 | 62.70 | 109.3 | 7.56 | No | |
| V5H492 | Integrin alpha-PS1 (=MG10) | 1 | 43.73 | 110.3 | 6.14 | No | |
| V5H7Z4 | Alpha-2-macroglobulin-like protein | 2 | 114.53 | 152.0 | 5.59 | Yes | |
| V5GY25 | Clathrin heavy chain 1 * | 1 | 47.55 | 190.7 | 5.81 | No | |
| V5I4B8 | Myosin heavy chain, muscle isoform X3 * | 6 | 178.22 | 52.04 | 222.0 | 6.09 | No |
| V5I3C9 | Myosin heavy chain, non-muscle isoform X1 * | 6 | 68.25 | 71.36 | 227.5 | 5.55 | No |
| A0A131XS30 | MAM and LDL-receptor class A domain-containing protein 1 (=MG11) | 1 | 40.27 | 420.4 | 5.49 | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knorr, S.; Reissert-Oppermann, S.; Tomás-Cortázar, J.; Barriales, D.; Azkargorta, M.; Iloro, I.; Elortza, F.; Pinecki-Socias, S.; Anguita, J.; Hovius, J.W.; et al. Identification and Characterization of Immunodominant Proteins from Tick Tissue Extracts Inducing a Protective Immune Response against Ixodes ricinus in Cattle. Vaccines 2021, 9, 636. https://doi.org/10.3390/vaccines9060636
Knorr S, Reissert-Oppermann S, Tomás-Cortázar J, Barriales D, Azkargorta M, Iloro I, Elortza F, Pinecki-Socias S, Anguita J, Hovius JW, et al. Identification and Characterization of Immunodominant Proteins from Tick Tissue Extracts Inducing a Protective Immune Response against Ixodes ricinus in Cattle. Vaccines. 2021; 9(6):636. https://doi.org/10.3390/vaccines9060636
Chicago/Turabian StyleKnorr, Sarah, Sophia Reissert-Oppermann, Julen Tomás-Cortázar, Diego Barriales, Mikel Azkargorta, Ibon Iloro, Félix Elortza, Sophia Pinecki-Socias, Juan Anguita, Joppe W. Hovius, and et al. 2021. "Identification and Characterization of Immunodominant Proteins from Tick Tissue Extracts Inducing a Protective Immune Response against Ixodes ricinus in Cattle" Vaccines 9, no. 6: 636. https://doi.org/10.3390/vaccines9060636
APA StyleKnorr, S., Reissert-Oppermann, S., Tomás-Cortázar, J., Barriales, D., Azkargorta, M., Iloro, I., Elortza, F., Pinecki-Socias, S., Anguita, J., Hovius, J. W., & Nijhof, A. M. (2021). Identification and Characterization of Immunodominant Proteins from Tick Tissue Extracts Inducing a Protective Immune Response against Ixodes ricinus in Cattle. Vaccines, 9(6), 636. https://doi.org/10.3390/vaccines9060636







