Single Amino Acid Deletion at N-Terminus of the Target Antigen in DNA Vaccine Induces Altered CD8+ T Cell Responses against Tumor Antigen
Abstract
1. Introduction
2. Materials and Methods
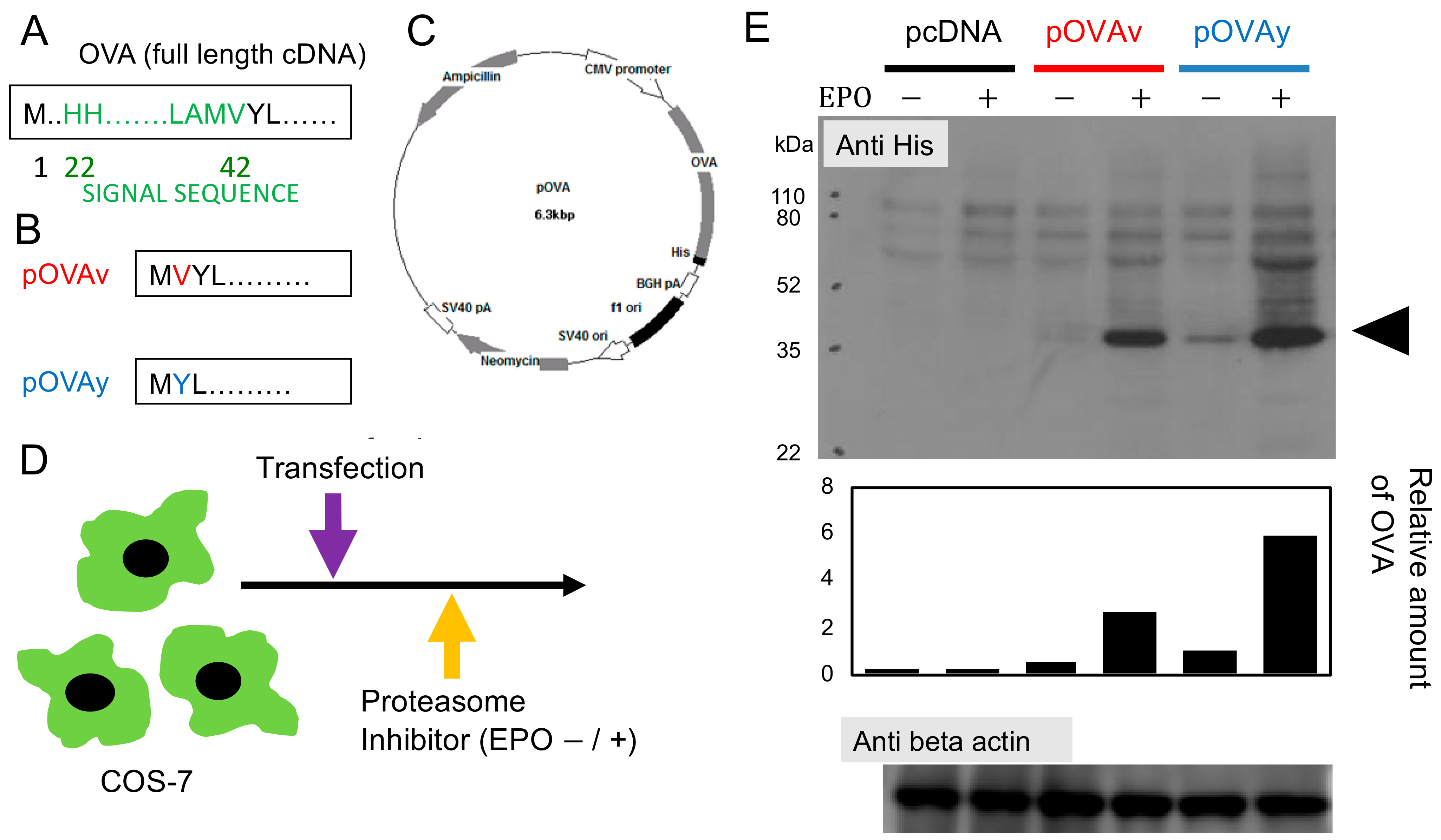
2.1. Plasmid Construction and Isolation
2.2. Transfection and Western Blotting
2.3. Mice and DNA Vaccination with a Gene Gun
2.4. Tumor Challenge, Depletion of T Cells, and In Vivo Cytotoxicity Assay
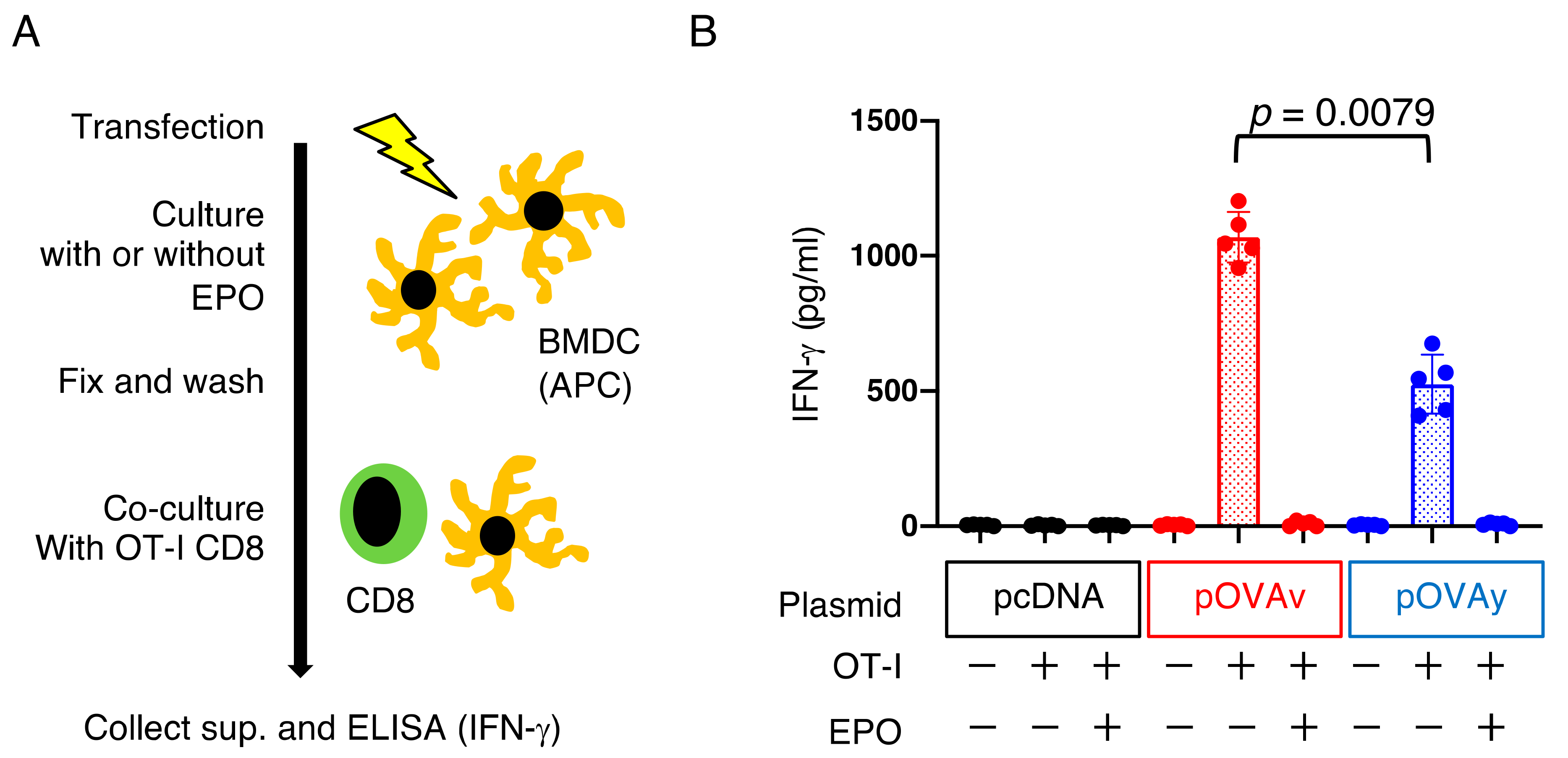
2.5. BMDC Induction, Transfection, and OVA Antigen Presentation Assay In Vitro
2.6. Statistical Analysis
3. Results
3.1. Expression of N-Terminus Modified Target Protein for DNA Vaccine
3.2. Anti-Tumor Effect of pOVAv and pOVAy with a Gene Gun
3.3. Cytotoxicity Induction by pOVAv Was Stronger Than pOVAy
3.4. Antigen Presentation by pOVAv Was Superior to pOVAy
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferraro, B.; Morrow, M.P.; Hutnick, N.A.; Shin, T.H.; Lucke, C.E.; Weiner, D.B. Clinical applications of DNA vaccines: Current progress. Clin. Infect Dis. 2011, 53, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Bergmann-Leitner, E.S.; Leitner, W.W. Vaccination Using Gene-Gun Technology. Methods Mol. Biol. 2015, 1325, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Graham, B.S.; Enama, M.E.; Nason, M.C.; Gordon, I.J.; Peel, S.A.; Ledgerwood, J.E.; Plummer, S.A.; Mascola, J.R.; Bailer, R.T.; Roederer, M.; et al. DNA vaccine delivered by a needle-free injection device improves potency of priming for antibody and CD8+ T-cell responses after rAd5 boost in a randomized clinical trial. PLoS ONE 2013, 8, e59340. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, C.J.; Assoumou, L.; Deeks, S.G.; Wilkin, T.J.; Berzins, B.; Casazza, J.P.; Lambert-Niclot, S.; Koup, R.A.; Costagliola, D.; Calvez, V.; et al. Effect of therapeutic intensification followed by HIV DNA prime and rAd5 boost vaccination on HIV-specific immunity and HIV reservoir (EraMune 02): A multicentre randomised clinical trial. Lancet HIV 2015, 2, e82–e91. [Google Scholar] [CrossRef]
- Dalmo, R.A. DNA vaccines for fish: Review and perspectives on correlates of protection. J. Fish Dis. 2018, 41, 1–9. [Google Scholar] [CrossRef]
- Chang, G.J.; Davis, B.S.; Stringfield, C.; Lutz, C. Prospective immunization of the endangered California condors (Gymnogyps californianus) protects this species from lethal West Nile virus infection. Vaccine 2007, 25, 2325–2330. [Google Scholar] [CrossRef]
- Salonius, K.; Simard, N.; Harland, R.; Ulmer, J.B. The road to licensure of a DNA vaccine. Curr. Opin. Investig. Drugs 2007, 8, 635–641. [Google Scholar]
- McLean, J.L.; Lobetti, R.G. Use of the melanoma vaccine in 38 dogs: The South African experience. J. S. Afr. Vet. Assoc. 2015, 86, 1246. [Google Scholar] [CrossRef]
- Kloetzel, P.M. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta 2004, 1695, 225–233. [Google Scholar] [CrossRef]
- Thomas, M.C.; Chiang, C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef]
- Li, L.; Petrovsky, N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines 2016, 15, 313–329. [Google Scholar] [CrossRef]
- Schwartz, A.L.; Ciechanover, A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 1999, 50, 57–74. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Chou, B.; Hiromatsu, K.; Hisaeda, H.; Duan, X.; Imai, T.; Murata, S.; Tanaka, K.; Himeno, K. Genetic immunization based on the ubiquitin-fusion degradation pathway against Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 2010, 392, 277–282. [Google Scholar] [CrossRef]
- Duan, X.; Hisaeda, H.; Shen, J.; Tu, L.; Imai, T.; Chou, B.; Murata, S.; Chiba, T.; Tanaka, K.; Fehling, H.J.; et al. The ubiquitin-proteasome system plays essential roles in presenting an 8-mer CTL epitope expressed in APC to corresponding CD8+ T cells. Int. Immunol. 2006, 18, 679–687. [Google Scholar] [CrossRef]
- Ishii, K.; Hisaeda, H.; Duan, X.; Imai, T.; Sakai, T.; Fehling, H.J.; Murata, S.; Chiba, T.; Tanaka, K.; Hamano, S.; et al. The involvement of immunoproteasomes in induction of MHC class I-restricted immunity targeting Toxoplasma SAG1. Microbes Infect. 2006, 8, 1045–1053. [Google Scholar] [CrossRef]
- Zhang, M.; Obata, C.; Hisaeda, H.; Ishii, K.; Murata, S.; Chiba, T.; Tanaka, K.; Li, Y.; Furue, M.; Chou, B.; et al. A novel DNA vaccine based on ubiquitin-proteasome pathway targeting ‘self’-antigens expressed in melanoma/melanocyte. Gene Ther. 2005, 12, 1049–1057. [Google Scholar] [CrossRef]
- Tobery, T.; Siliciano, R.F. Cutting edge: Induction of enhanced CTL-dependent protective immunity in vivo by N-end rule targeting of a model tumor antigen. J. Immunol. 1999, 162, 639–642. [Google Scholar]
- Andersson, H.A.; Barry, M.A. Maximizing antigen targeting to the proteasome for gene-based vaccines. Mol. Ther. 2004, 10, 432–446. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Teramoto, K.; Kontani, K.; Ozaki, Y.; Sawai, S.; Tezuka, N.; Nagata, T.; Fujino, S.; Itoh, Y.; Taguchi, O.; Koide, Y.; et al. Deoxyribonucleic acid (DNA) encoding a pan-major histocompatibility complex class II peptide analogue augmented antigen-specific cellular immunity and suppressive effects on tumor growth elicited by DNA vaccine immunotherapy. Cancer Res. 2003, 63, 7920–7925. [Google Scholar] [PubMed]
- Imai, T.; Duan, X.; Hisaeda, H.; Himeno, K. Antigen-specific CD8+ T cells induced by the ubiquitin fusion degradation pathway. Biochem. Biophys. Res. Commun. 2008, 365, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Hogquist, K.A.; Jameson, S.C.; Heath, W.R.; Howard, J.L.; Bevan, M.J.; Carbone, F.R. T cell receptor antagonist peptides induce positive selection. Cell 1994, 76, 17–27. [Google Scholar] [CrossRef]
- Moore, M.W.; Carbone, F.R.; Bevan, M.J. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell 1988, 54, 777–785. [Google Scholar] [CrossRef]
- Haymaker, C.L.; Hailemichael, Y.; Yang, Y.; Nurieva, R. In Vivo Assay for Detection of Antigen-specific T-cell Cytolytic Function Using a Vaccination Model. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Kato, Y.; Kajiwara, C.; Mizukami, S.; Ishige, I.; Ichiyanagi, T.; Hikida, M.; Wang, J.Y.; Udono, H. Heat shock protein 90 (HSP90) contributes to cytosolic translocation of extracellular antigen for cross-presentation by dendritic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16363–16368. [Google Scholar] [CrossRef]
- Elbahnasawy, M.A.; Farag, M.M.S.; Mansour, M.T.; El-Ghamery, A.A. Cloning, expression and nanodiscs assemble of recombinant HIV-1 gp41. Microb. Pathog. 2020, 138, 103824. [Google Scholar] [CrossRef] [PubMed]
- Varland, S.; Osberg, C.; Arnesen, T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 2015, 15, 2385–2401. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef]
- Hwang, C.S.; Shemorry, A.; Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 2010, 327, 973–977. [Google Scholar] [CrossRef]
- Huang, L.; Kuhls, M.C.; Eisenlohr, L.C. Hydrophobicity as a driver of MHC class I antigen processing. Embo J. 2011, 30, 1634–1644. [Google Scholar] [CrossRef]
- Delamarre, L.; Couture, R.; Mellman, I.; Trombetta, E.S. Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. J. Exp. Med. 2006, 203, 2049–2055. [Google Scholar] [CrossRef]
- Bins, A.D.; Wolkers, M.C.; van den Boom, M.D.; Haanen, J.B.; Schumacher, T.N. In vivo antigen stability affects DNA vaccine immunogenicity. J. Immunol. 2007, 179, 2126–2133. [Google Scholar] [CrossRef]
- Delamarre, L.; Pack, M.; Chang, H.; Mellman, I.; Trombetta, E.S. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 2005, 307, 1630–1634. [Google Scholar] [CrossRef]
- Eisenbarth, S.C. Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 2019, 19, 89–103. [Google Scholar] [CrossRef]
- Kumamoto, T.; Huang, E.K.; Paek, H.J.; Morita, A.; Matsue, H.; Valentini, R.F.; Takashima, A. Induction of tumor-specific protective immunity by in situ Langerhans cell vaccine. Nat. Biotechnol. 2002, 20, 64–69. [Google Scholar] [CrossRef]
- Porgador, A.; Irvine, K.R.; Iwasaki, A.; Barber, B.H.; Restifo, N.P.; Germain, R.N. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J. Exp. Med. 1998, 188, 1075–1082. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef]
- Hitzeroth, I.I.; Chabeda, A.; Whitehead, M.P.; Graf, M.; Rybicki, E.P. Optimizing a Human Papillomavirus Type 16 L1-Based Chimaeric Gene for Expression in Plants. Front. Bioeng. Biotechnol. 2018, 6, 101. [Google Scholar] [CrossRef]
- Mbewana, S.; Mortimer, E.; Pera, F.F.; Hitzeroth, I.I.; Rybicki, E.P. Production of H5N1 Influenza Virus Matrix Protein 2 Ectodomain Protein Bodies in Tobacco Plants and in Insect Cells as a Candidate Universal Influenza Vaccine. Front. Bioeng. Biotechnol. 2015, 3, 197. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imai, T. Single Amino Acid Deletion at N-Terminus of the Target Antigen in DNA Vaccine Induces Altered CD8+ T Cell Responses against Tumor Antigen. Vaccines 2021, 9, 540. https://doi.org/10.3390/vaccines9060540
Imai T. Single Amino Acid Deletion at N-Terminus of the Target Antigen in DNA Vaccine Induces Altered CD8+ T Cell Responses against Tumor Antigen. Vaccines. 2021; 9(6):540. https://doi.org/10.3390/vaccines9060540
Chicago/Turabian StyleImai, Takashi. 2021. "Single Amino Acid Deletion at N-Terminus of the Target Antigen in DNA Vaccine Induces Altered CD8+ T Cell Responses against Tumor Antigen" Vaccines 9, no. 6: 540. https://doi.org/10.3390/vaccines9060540
APA StyleImai, T. (2021). Single Amino Acid Deletion at N-Terminus of the Target Antigen in DNA Vaccine Induces Altered CD8+ T Cell Responses against Tumor Antigen. Vaccines, 9(6), 540. https://doi.org/10.3390/vaccines9060540





