CD163 Antibodies Inhibit PRRSV Infection via Receptor Blocking and Transcription Suppression
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viruses
2.2. Expression of CD163 SRCR5–9
2.3. SDS-PAGE and Western Blotting
2.4. Production and Characterization of Murine mAbs
2.5. Immunofluorescence Assays
2.6. Pre-Attachment Assay
2.7. Post-Attachment Assay
2.8. Construction of Stable Cell Lines Expressing CD163
2.9. RNA Isolation and qRT-PCR
2.10. Statistical Analysis
3. Results
3.1. mAbs 6E8 and 9A10 Were Selected for Specific Recognition of CD163
3.2. 6E8 and 9A10 Significantly Block PRRSV Infection
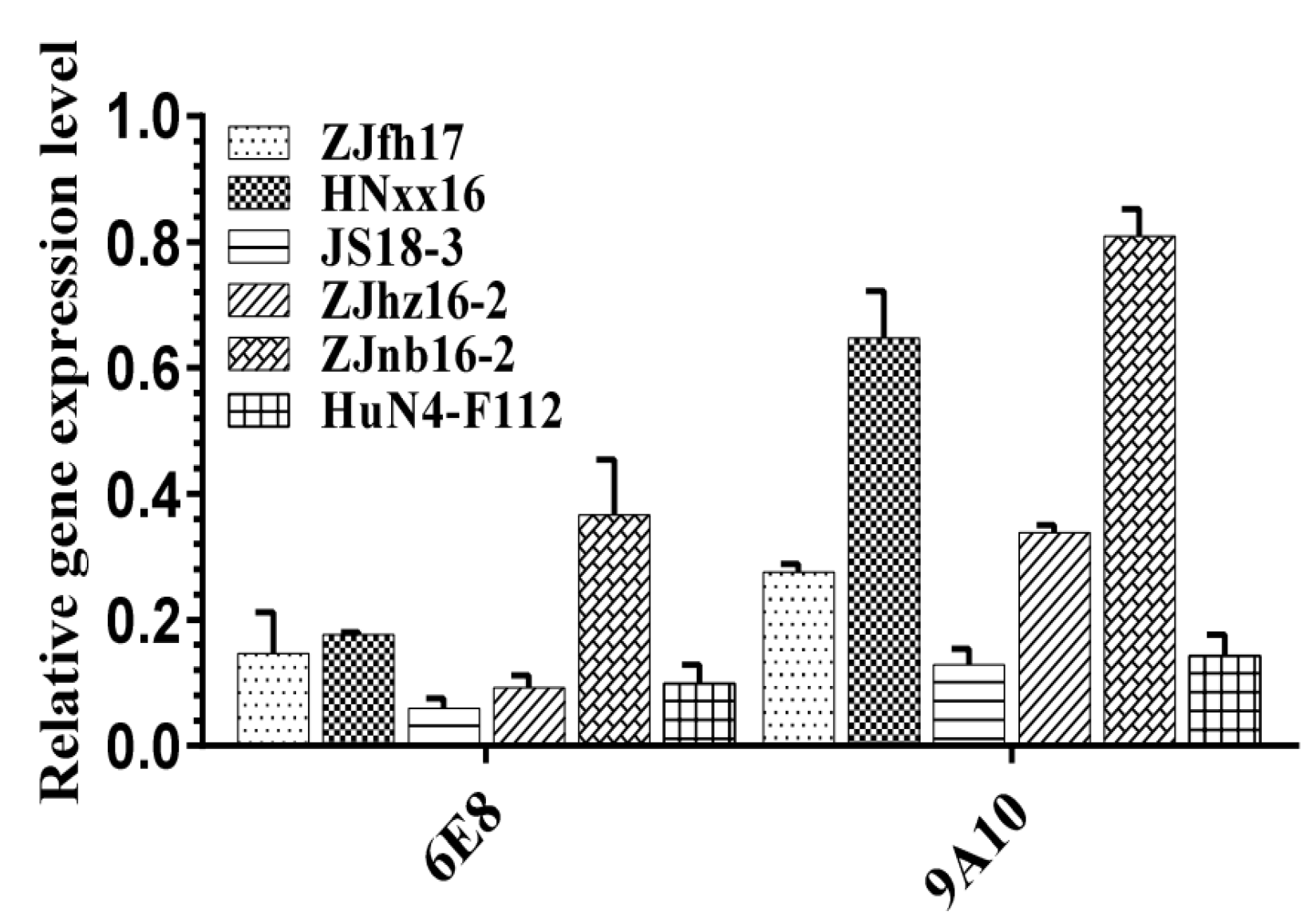
3.3. 6E8 and 9A10 Provide Broad Inhibition Against Various PRRSV Strains
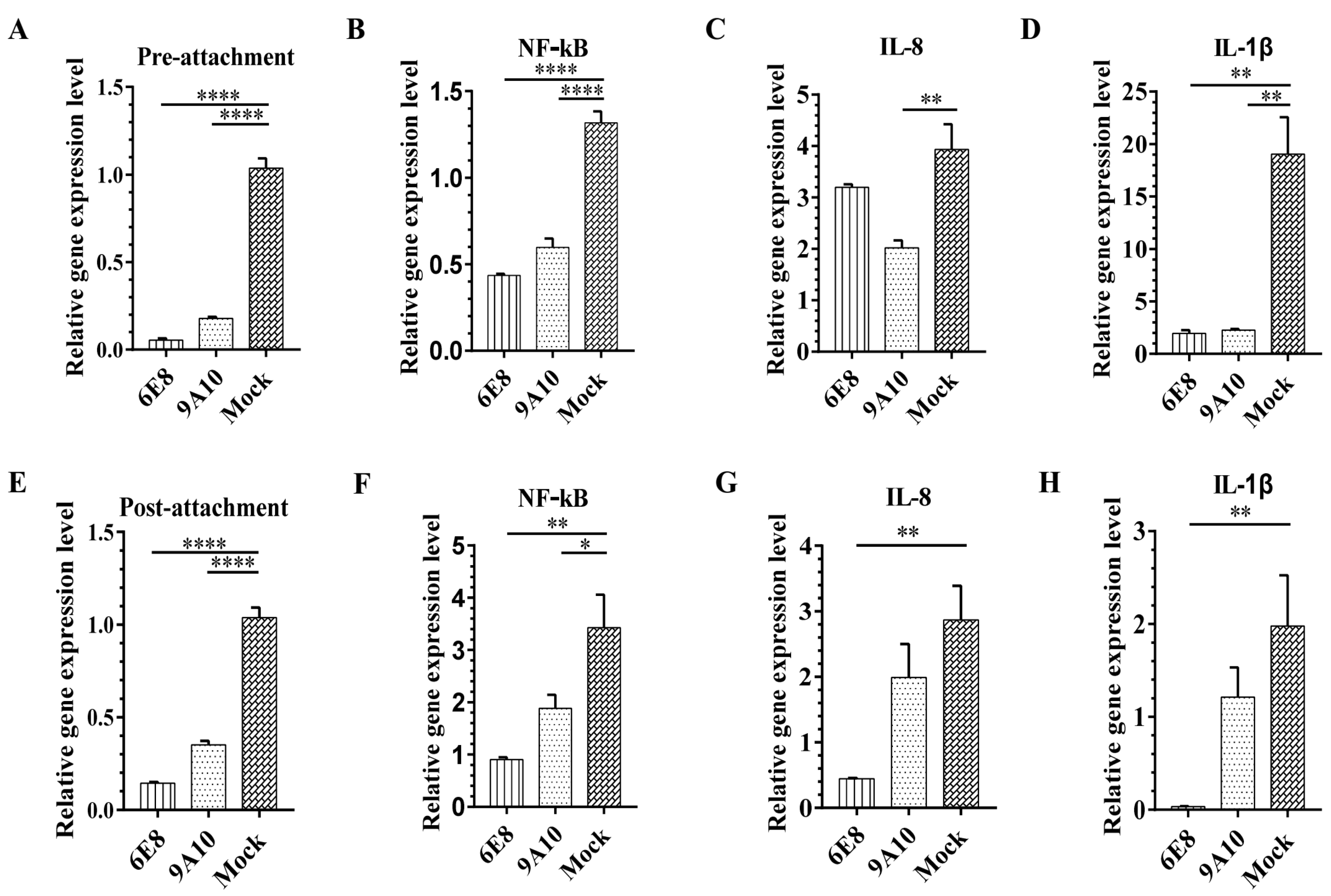
3.4. 6E8 and 9A10 Stop PRRSV Infection Either Before or After Virus Attachment
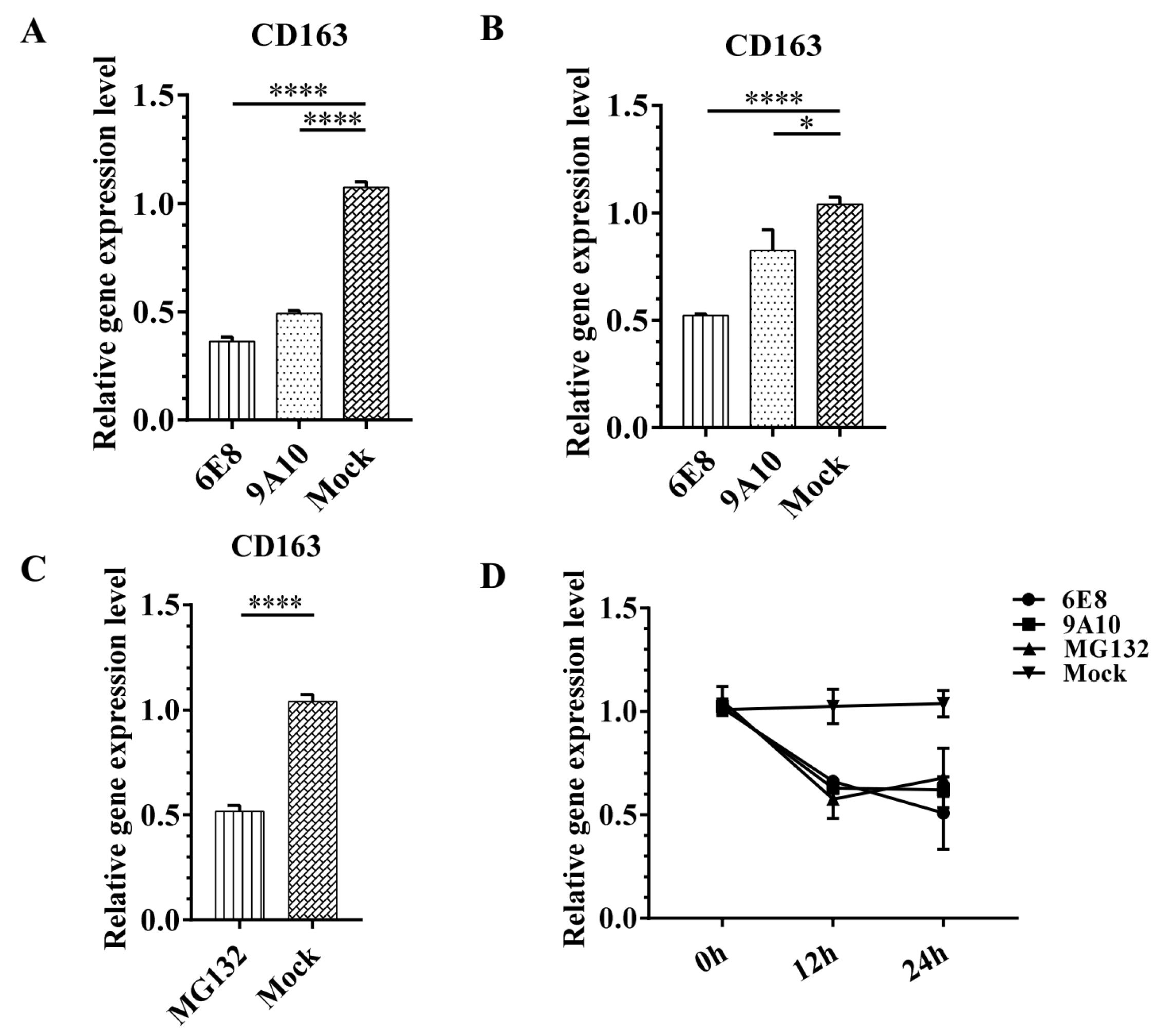
3.5. 6E8 and 9A10 Suppress CD163 Transcription in PAMs and Marc-145
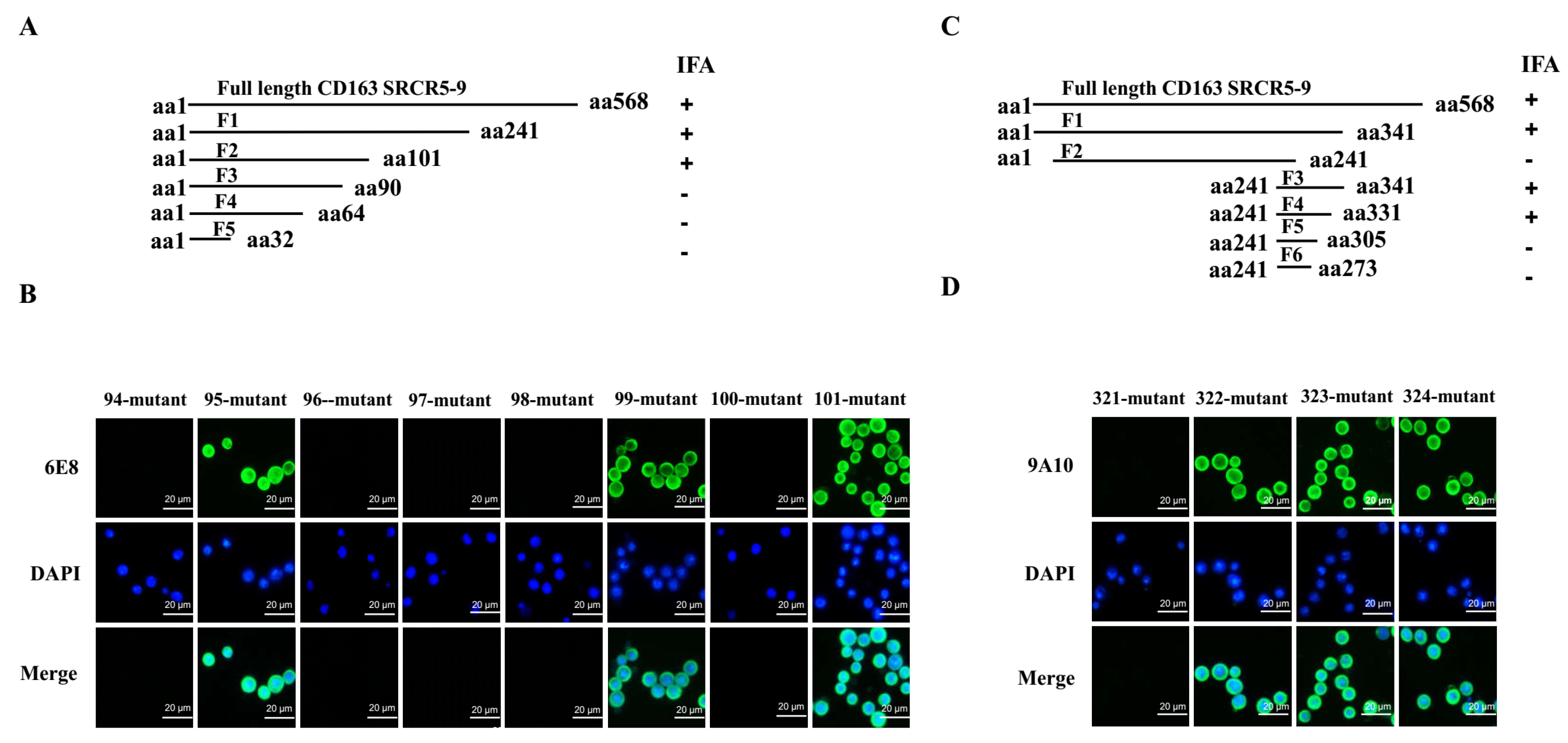
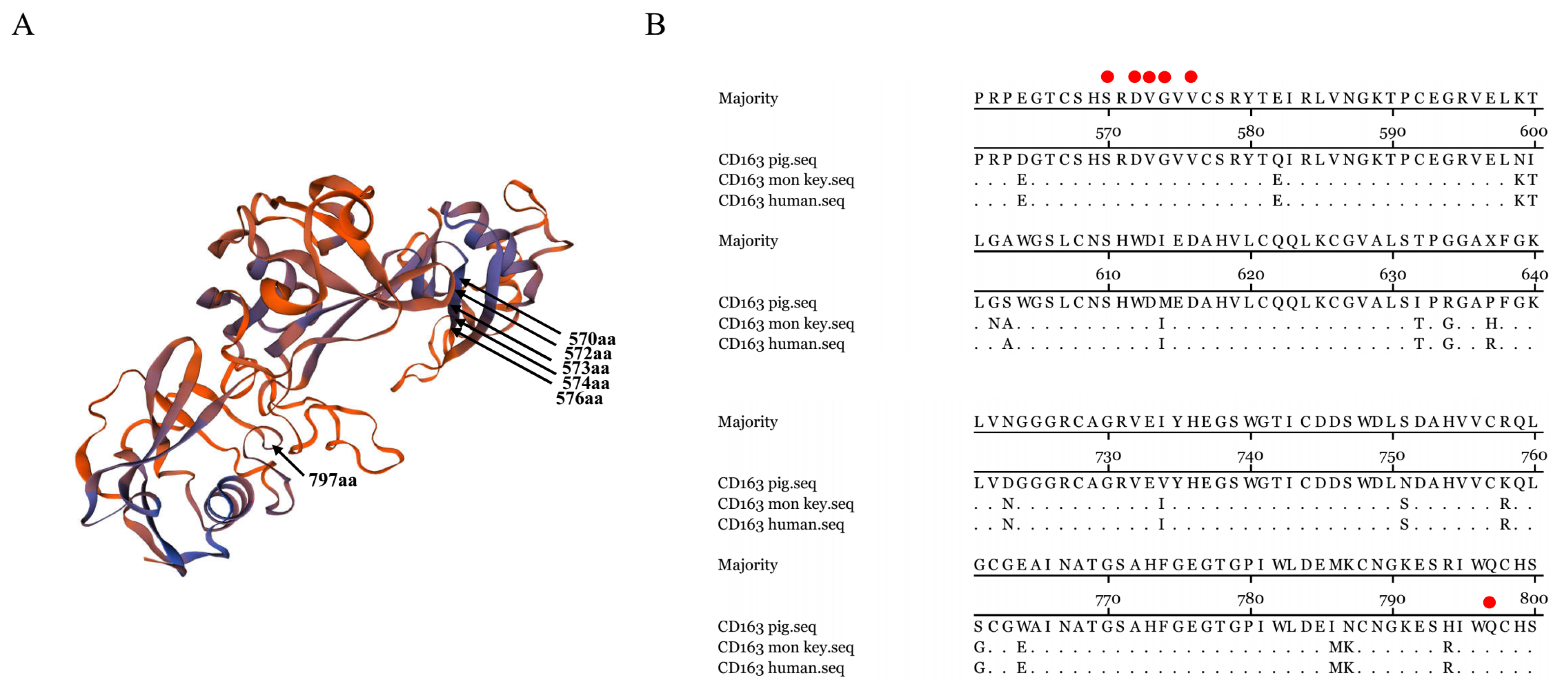
3.6. Epitope Mapping for 6E8 and 9A10 in CD163
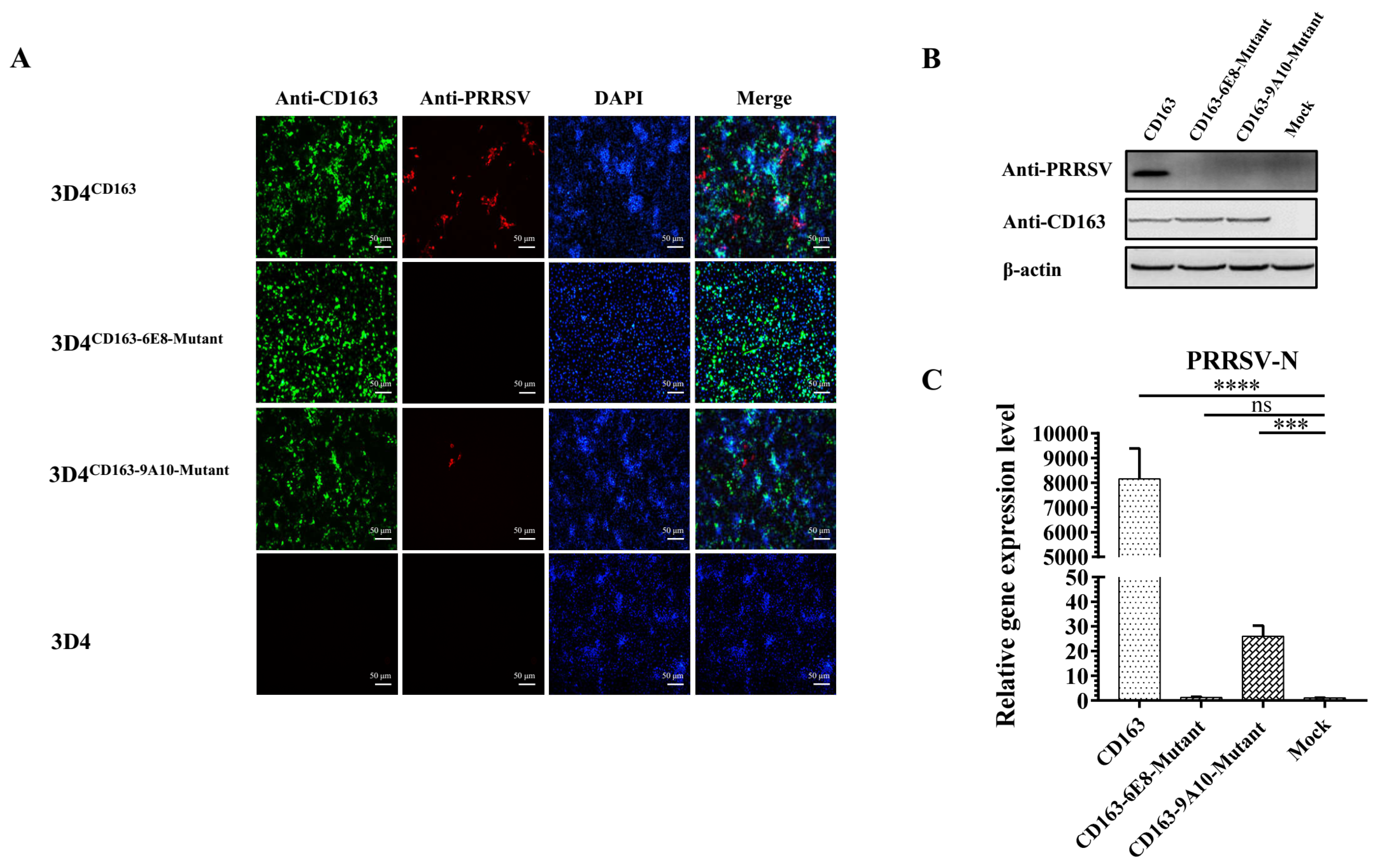
3.7. Residues 570SXDVGXV576 and Q797 of CD163 Are Required for PRRSV Infection
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Normile, D. Virology. China, Vietnam grapple with rapidly evolving pig virus. Science 2007, 317, 1017. [Google Scholar] [CrossRef] [PubMed]
- Meulenberg, J.J.; de Meijer, E.J.; Moormann, R.J. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J. Gen. Virol. 1993, 74, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G.; Terpstra, C.; Pol, J.M.; ter Laak, E.A.; Bloemraad, M.; de Kluyver, E.P.; Kragten, C.; van Buiten, L.; den Besten, A.; Wagenaar, F.; et al. Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet. Q. 1991, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.Q.; Guo, X.; Yang, H.C. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch. Virol. 2004, 149, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Yoo, D. Engineering the PRRS virus genome: Updates and perspectives. Vet. Microbiol. 2014, 174, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Zhang, S. Regulation of cell signaling and porcine reproductive and respiratory syndrome virus. Cell. Signal. 2012, 24, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Laguna, J.; Salguero, F.J.; Pallares, F.J.; Carrasco, L. Immunopathogenesis of porcine reproductive and respiratory syndrome in the respiratory tract of pigs. Vet. J. 2013, 195, 148–155. [Google Scholar] [CrossRef]
- Yoon, I.J.; Joo, H.S.; Goyal, S.M.; Molitor, T.W. A modified serum neutralization test for the detection of antibody to porcine reproductive and respiratory syndrome virus in swine sera. J. Vet. Diagn. Investig. 1994, 6, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Loving, C.L.; Osorio, F.A.; Murtaugh, M.P.; Zuckermann, F.A. Innate and adaptive immunity against Porcine Reproductive and Respiratory Syndrome Virus. Vet. Immunol. Immunopathol. 2015, 167, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Renukaradhya, G.J.; Alekseev, K.; Jung, K.; Fang, Y.; Saif, L.J. Porcine reproductive and respiratory syndrome virus-induced immunosuppression exacerbates the inflammatory response to porcine respiratory coronavirus in pigs. Viral Immunol. 2010, 23, 457–466. [Google Scholar] [CrossRef]
- Dwivedi, V.; Manickam, C.; Patterson, R.; Dodson, K.; Murtaugh, M.; Torrelles, J.B.; Schlesinger, L.S.; Renukaradhya, G.J. Cross-protective immunity to porcine reproductive and respiratory syndrome virus by intranasal delivery of a live virus vaccine with a potent adjuvant. Vaccine 2011, 29, 4058–4066. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, H.; Zhang, X.; He, S.; Dong, W.; Wang, X.; Chen, Y.; Liu, X.; Guo, C. Lipopolysaccharide Downregulates CD163 Expression to Inhibit PRRSV Infection via TLR4-NF-kappaB Pathway. Front. Microbiol. 2020, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, M.P.; Xiao, Z.; Zuckermann, F. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 2002, 15, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Law, S.K.; Micklem, K.J.; Shaw, J.M.; Zhang, X.P.; Dong, Y.; Willis, A.C.; Mason, D.Y. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur. J. Immunol. 1993, 23, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Buechler, C.; Langmann, T.; Schmitz, G. Genomic organization and chromosomal localization of the human CD163 (M130) gene: A member of the scavenger receptor cysteine-rich superfamily. Biochem. Biophys. Res. Commun. 1999, 260, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Delputte, P.L.; Nauwynck, H.J. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 2004, 78, 8094–8101. [Google Scholar] [CrossRef] [PubMed]
- Delputte, P.L.; Van Breedam, W.; Delrue, I.; Oetke, C.; Crocker, P.R.; Nauwynck, H.J. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 2007, 81, 9546–9550. [Google Scholar] [CrossRef]
- Delputte, P.L.; Vanderheijden, N.; Nauwynck, H.J.; Pensaert, M.B. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 2002, 76, 4312–4320. [Google Scholar] [CrossRef]
- Vanderheijden, N.; Delputte, P.; Nauwynck, H.; Pensaert, M. Effects of heparin on the entry of porcine reproductive and respiratory syndrome virus into alveolar macrophages. Adv. Exp. Med. Biol. 2001, 494, 683–689. [Google Scholar]
- Van Gorp, H.; Van Breedam, W.; Delputte, P.L.; Nauwynck, H.J. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2008, 89, 2943–2953. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Nauwynck, H.J.; Pensaert, M.B. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet. Microbiol. 1997, 56, 9–19. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; Mclaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef] [PubMed]
- Calvert, J.G.; Slade, D.E.; Shields, S.L.; Jolie, R.; Mannan, R.M.; Ankenbauer, R.G.; Welch, S.K. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007, 81, 7371–7379. [Google Scholar] [CrossRef] [PubMed]
- Backe, E.; Schwarting, R.; Gerdes, J.; Ernst, M.; Stein, H. Ber-MAC3: New monoclonal antibody that defines human monocyte/macrophage differentiation antigen. J. Clin. Pathol. 1991, 44, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Onofre, G.; Kolackova, M.; Jankovicova, K.; Krejsek, J. Scavenger receptor CD163 and its biological functions. Acta Medica (Hradec Kralove) 2009, 52, 57–61. [Google Scholar] [CrossRef]
- Subramanian, K.; Du, R.; Tan, N.S.; Ho, B.; Ding, J.L. CD163 and IgG codefend against cytotoxic hemoglobin via autocrine and paracrine mechanisms. J. Immunol. 2013, 190, 5267–5278. [Google Scholar] [CrossRef] [PubMed]
- Kowal, K.; Silver, R.; Slawinska, E.; Bielecki, M.; Chyczewski, L.; Kowal-Bielecka, O. CD163 and its role in inflammation. Folia Histochem. Cytobiol. 2011, 49, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Das, P.B.; Dinh, P.X.; Ansari, I.H.; de Lima, M.; Osorio, F.A.; Pattnaik, A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010, 84, 1731–1740. [Google Scholar] [CrossRef]
- Van Gorp, H.; Van Breedam, W.; Van Doorsselaere, J.; Delputte, P.L.; Nauwynck, H.J. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J. Virol. 2010, 84, 3101–3105. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, X.; Ge, X.; Chen, Y.; Sun, Q.; Yang, H. Changes in the cellular proteins of pulmonary alveolar macrophage infected with porcine reproductive and respiratory syndrome virus by proteomics analysis. J. Proteome Res. 2009, 8, 3091–3097. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, Y.; Han, G.; Fang, W.; He, F. Identification of E2 with improved secretion and immunogenicity against CSFV in piglets. BMC Microbiol. 2020, 20, 26. [Google Scholar] [CrossRef]
- Chen, N.; Tong, C.; Li, D.; Wan, J.; Yuan, X.; Li, X.; Peng, J.; Fang, W. Antigenic analysis of classical swine fever virus E2 glycoprotein using pig antibodies identifies residues contributing to antigenic variation of the vaccine C-strain and group 2 strains circulating in China. Virol. J. 2010, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Du, Q.; Ho, Y.; Kwang, J. Immunohistochemical detection of Influenza virus infection in formalin-fixed tissues with anti-H5 monoclonal antibody recognizing FFWTILKP. J. Virol. Methods 2009, 155, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5, 13875. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, J.; Shi, P.; Lu, D.; Zhao, C.; Su, Y.; Zhang, L.; Huang, J. The Immunological Regulation Roles of Porcine beta-1, 4 Galactosyltransferase V (B4GALT5) in PRRSV Infection. Front. Cell Infect. Microbiol. 2018, 8, 48. [Google Scholar] [CrossRef]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular Innate Immunity against PRRSV and Swine Influenza Viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Gu, G.; Sun, W.; Zhang, Y.J.; Zhou, E.M. Improved Vaccine against PRRSV: Current Progress and Future Perspective. Front. Microbiol. 2017, 8, 1635. [Google Scholar] [CrossRef]
- Vanderlinden, E.; Naesens, L. Emerging antiviral strategies to interfere with influenza virus entry. Med. Res. Rev. 2014, 34, 301–339. [Google Scholar] [CrossRef]
- Liu, M.; Wang, T.; Zhou, Y.; Zhao, Y.; Zhang, Y.; Li, J. Potential Role of ACE2 in Coronavirus Disease 2019 (COVID-19) Prevention and Management. J. Transl. Int. Med. 2020, 8, 9–19. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, J.; Zhang, X.; Shi, J.; Pan, Y.; Zhou, R.; Li, G.; Li, Z.; Cai, G.; Wu, Z. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir. Res. 2018, 151, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, R.; He, S.; Zhang, X.; Xia, X.; Sun, H. Additive inhibition of porcine reproductive and respiratory syndrome virus infection with the soluble sialoadhesin and CD163 receptors. Virus Res. 2014, 179, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Sulahian, T.H.; Pioli, P.A.; Wardwell, K.; Guyre, P.M. Cross-linking of FcgammaR triggers shedding of the hemoglobin-haptoglobin scavenger receptor CD163. J. Leukoc. Biol. 2004, 76, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Wells, K.D.; Bardot, R.; Whitworth, K.M.; Trible, B.R.; Fang, Y.; Mileham, A.; Kerrigan, M.A.; Samuel, M.S.; Prather, R.S.; Rowland, R. Replacement of Porcine CD163 Scavenger Receptor Cysteine-Rich Domain 5 with a CD163-Like Homolog Confers Resistance of Pigs to Genotype 1 but Not Genotype 2 Porcine Reproductive and Respiratory Syndrome Virus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Das, P.B.; Vu, H.L.; Dinh, P.X.; Cooney, J.L.; Kwon, B.; Osorio, F.A.; Pattnaik, A.K. Glycosylation of minor envelope glycoproteins of porcine reproductive and respiratory syndrome virus in infectious virus recovery, receptor interaction, and immune response. Virology 2011, 410, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Kleiboeker, S.B. Porcine arterivirus activates the NF-kappaB pathway through IkappaB degradation. Virology 2005, 342, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Song, S.; Fang, L.; Wang, D.; Jing, H.; Gao, L.; Cai, Y.; Luo, R.; Chen, H.; Xiao, S. Porcine reproductive and respiratory syndrome virus induces IL-1beta production depending on TLR4/MyD88 pathway and NLRP3 inflammasome in primary porcine alveolar macrophages. Mediators Inflamm. 2014, 2014, 403515. [Google Scholar] [CrossRef] [PubMed]
- Hintz, K.A.; Rassias, A.J.; Wardwell, K.; Moss, M.L.; Morganelli, P.M.; Pioli, P.A.; Givan, A.L.; Wallace, P.K.; Yeager, M.P.; Guyre, P.M. Endotoxin induces rapid metalloproteinase-mediated shedding followed by up-regulation of the monocyte hemoglobin scavenger receptor CD163. J. Leukoc. Biol. 2002, 72, 711–717. [Google Scholar]
- Ma, H.; Jiang, L.; Qiao, S.; Zhi, Y.; Chen, X.X.; Yang, Y.; Huang, X.; Huang, M.; Li, R.; Zhang, G.P. The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef]


| Prmier | Sequence(5′–3′) |
|---|---|
| ORF(N)-F | AAAACCAGTCCAGAGGCAAG |
| ORF(N)-R | CGGATCAGACGCACAGTATG |
| CD163-F | ATTCATCATCCTCGGACCCAT |
| CD163-R | CCCAGCACAACGACCACCT |
| NF-κB-F | TCGCTGCCAAAGAAGGACAT |
| NF-κB-R | AGCGTTCAGACCTTCACCGT |
| IL-1β-F | CCCAAAAGTTACCCGAAGAGG |
| IL-1β-R | TCTGCTTGAGAGGTGCTGATG |
| IL-8-F | GGCATCACCTTTGGCATCTT |
| IL-8-R | TGGCATCGAAGTTCTGCACT |
| mGAPDH-F | TGACAACAGCCTCAAGATCG |
| mGAPDH-R | GTCTTCTGGGTGGCAGTGAT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Liu, Z.; Zheng, S.; Han, G.; He, F. CD163 Antibodies Inhibit PRRSV Infection via Receptor Blocking and Transcription Suppression. Vaccines 2020, 8, 592. https://doi.org/10.3390/vaccines8040592
Xu H, Liu Z, Zheng S, Han G, He F. CD163 Antibodies Inhibit PRRSV Infection via Receptor Blocking and Transcription Suppression. Vaccines. 2020; 8(4):592. https://doi.org/10.3390/vaccines8040592
Chicago/Turabian StyleXu, Huiling, Zehui Liu, Suya Zheng, Guangwei Han, and Fang He. 2020. "CD163 Antibodies Inhibit PRRSV Infection via Receptor Blocking and Transcription Suppression" Vaccines 8, no. 4: 592. https://doi.org/10.3390/vaccines8040592
APA StyleXu, H., Liu, Z., Zheng, S., Han, G., & He, F. (2020). CD163 Antibodies Inhibit PRRSV Infection via Receptor Blocking and Transcription Suppression. Vaccines, 8(4), 592. https://doi.org/10.3390/vaccines8040592






