Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era
Abstract
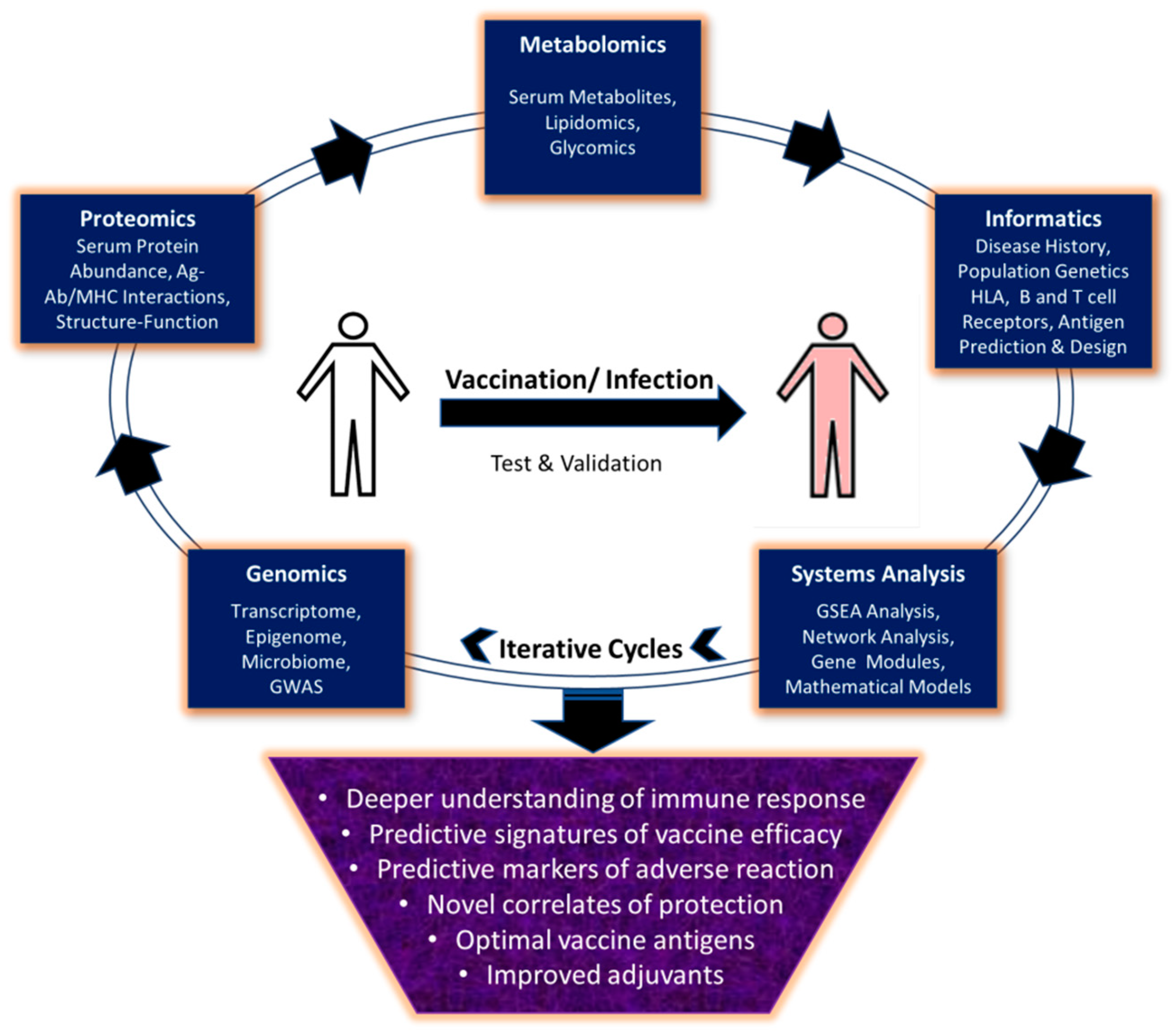
1. Introduction
2. Antigen Discovery and Development
3. Molecular Signatures of Vaccine Efficacy
4. Predictive Markers of Adverse Effects
5. Rational Use and Development of Adjuvants
6. Conclusions and Prospective
Funding
Acknowledgments
Conflicts of Interest
References
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Vaccines: The fourth century. Clin. Vaccine Immunol. 2009, 16, 1709–1719. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Garcia-Sastre, A. Antiviral innate immunity through the lens of systems biology. Virus Res. 2016, 218, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Pulendran, B. Will Systems Biology Deliver Its Promise and Contribute to the Development of New or Improved Vaccines? From Data to Understanding through Systems Biology. Cold Spring Harb. Perspect. Biol. 2018, 10, a028894. [Google Scholar] [CrossRef] [PubMed]
- Hegde, N.R.; Gauthami, S.; Sampath Kumar, H.M.; Bayry, J. The use of databases, data mining and immunoinformatics in vaccinology: Where are we? Expert Opin. Drug Discov. 2018, 13, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Meade, P.; Latorre-Margalef, N.; Stallknecht, D.E.; Krammer, F. Development of an influenza virus protein microarray to measure the humoral response to influenza virus infection in mallards. Emerg. Microbes Infect. 2017, 6, e110. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, R.; Supnet, M.; Jasinskas, A.; Jain, A.; Taghavian, O.; Obiero, J.; Milton, D.K.; Chen, W.H.; Grantham, M.; Webby, R.; et al. Protein Microarray Analysis of the Specificity and Cross-Reactivity of Influenza Virus Hemagglutinin-Specific Antibodies. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Krarup, A.; Truan, D.; Furmanova-Hollenstein, P.; Bogaert, L.; Bouchier, P.; Bisschop, I.J.M.; Widjojoatmodjo, M.N.; Zahn, R.; Schuitemaker, H.; McLellan, J.S.; et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015, 6, 8143. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef]
- Shriver, Z.; Trevejo, J.M.; Sasisekharan, R. Antibody-Based Strategies to Prevent and Treat Influenza. Front. Immunol. 2015, 6, 315. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.N.; Tharakaraman, K.; Rowley, K.J.; Costa, V.V.; Chan, K.R.; Wong, Y.H.; Ong, L.C.; Tan, H.C.; Koch, T.; Cain, D.; et al. Structure-Guided Design of an Anti-dengue Antibody Directed to a Non-immunodominant Epitope. Cell 2015, 162, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Azoitei, M.L.; Ban, Y.E.; Julien, J.P.; Bryson, S.; Schroeter, A.; Kalyuzhniy, O.; Porter, J.R.; Adachi, Y.; Baker, D.; Pai, E.F.; et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J. Mol. Biol. 2012, 415, 175–192. [Google Scholar] [CrossRef] [PubMed]
- Azoitei, M.L.; Correia, B.E.; Ban, Y.E.; Carrico, C.; Kalyuzhniy, O.; Chen, L.; Schroeter, A.; Huang, P.S.; McLellan, J.S.; Kwong, P.D.; et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 2011, 334, 373–376. [Google Scholar] [CrossRef]
- Correia, B.E.; Bates, J.T.; Loomis, R.J.; Baneyx, G.; Carrico, C.; Jardine, J.G.; Rupert, P.; Correnti, C.; Kalyuzhniy, O.; Vittal, V.; et al. Proof of principle for epitope-focused vaccine design. Nature 2014, 507, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, X.; Zhang, B.; McKee, K.; O’Dell, S.; Soto, C.; Zhou, T.; Casazza, J.P.; Program, N.C.S.; Mullikin, J.C.; et al. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc. Natl. Acad. Sci. USA 2013, 110, E4088–E4097. [Google Scholar] [CrossRef]
- Lee, J.; Boutz, D.R.; Chromikova, V.; Joyce, M.G.; Vollmers, C.; Leung, K.; Horton, A.P.; DeKosky, B.J.; Lee, C.H.; Lavinder, J.J.; et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. 2016, 22, 1456–1464. [Google Scholar] [CrossRef]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Gaucher, D.; Therrien, R.; Kettaf, N.; Angermann, B.R.; Boucher, G.; Filali-Mouhim, A.; Moser, J.M.; Mehta, R.S.; Drake, D.R., 3rd; Castro, E.; et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 2008, 205, 3119–3131. [Google Scholar] [CrossRef]
- Pulendran, B. Learning immunology from the yellow fever vaccine: Innate immunity to systems vaccinology. Nat. Rev. Immunol. 2009, 9, 741–747. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef]
- Reif, D.M.; Motsinger-Reif, A.A.; McKinney, B.A.; Rock, M.T.; Crowe, J.E., Jr.; Moore, J.H. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009, 10, 112–119. [Google Scholar] [CrossRef]
- Zak, D.E.; Andersen-Nissen, E.; Peterson, E.R.; Sato, A.; Hamilton, M.K.; Borgerding, J.; Krishnamurty, A.T.; Chang, J.T.; Adams, D.J.; Hensley, T.R.; et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. USA 2012, 109, E3503–E3512. [Google Scholar] [CrossRef]
- Li, S.; Rouphael, N.; Duraisingham, S.; Romero-Steiner, S.; Presnell, S.; Davis, C.; Schmidt, D.S.; Johnson, S.E.; Milton, A.; Rajam, G.; et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014, 15, 195–204. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Franco, L.M.; Bucasas, K.L.; Wells, J.M.; Nino, D.; Wang, X.; Zapata, G.E.; Arden, N.; Renwick, A.; Yu, P.; Quarles, J.M.; et al. Integrative genomic analysis of the human immune response to influenza vaccination. Elife 2013, 2, e00299. [Google Scholar] [CrossRef]
- Li, S.; Sullivan, N.L.; Rouphael, N.; Yu, T.; Banton, S.; Maddur, M.S.; McCausland, M.; Chiu, C.; Canniff, J.; Dubey, S.; et al. Metabolic Phenotypes of Response to Vaccination in Humans. Cell 2017, 169, 862–877. [Google Scholar] [CrossRef]
- Pulendran, B.; Miller, J.; Querec, T.D.; Akondy, R.; Moseley, N.; Laur, O.; Glidewell, J.; Monson, N.; Zhu, T.; Zhu, H.; et al. Case of yellow fever vaccine-associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. J. Infect. Dis. 2008, 198, 500–507. [Google Scholar] [CrossRef]
- Hallberg, P.; Smedje, H.; Eriksson, N.; Kohnke, H.; Daniilidou, M.; Ohman, I.; Yue, Q.Y.; Cavalli, M.; Wadelius, C.; Magnusson, P.K.E.; et al. Pandemrix-induced narcolepsy is associated with genes related to immunity and neuronal survival. EBioMedicine 2019, 40, 595–604. [Google Scholar] [CrossRef]
- Kaisho, T.; Akira, S. Toll-like receptors as adjuvant receptors. Biochim. Biophys. Acta 2002, 1589, 1–13. [Google Scholar] [CrossRef]
- Bayry, J.; Tchilian, E.Z.; Davies, M.N.; Forbes, E.K.; Draper, S.J.; Kaveri, S.V.; Hill, A.V.; Kazatchkine, M.D.; Beverley, P.C.; Flower, D.R.; et al. In silico identified CCR4 antagonists target regulatory T cells and exert adjuvant activity in vaccination. Proc. Natl. Acad. Sci. USA 2008, 105, 10221–10226. [Google Scholar] [CrossRef]
- Carreno, L.J.; Kharkwal, S.S.; Porcelli, S.A. Optimizing NKT cell ligands as vaccine adjuvants. Immunotherapy 2014, 6, 309–320. [Google Scholar] [CrossRef]
- Eisenbarth, S.C.; Colegio, O.R.; O’Connor, W.; Sutterwala, F.S.; Flavell, R.A. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 2008, 453, 1122–1126. [Google Scholar] [CrossRef]
- O’Hagan, D.T.; Ott, G.S.; De Gregorio, E.; Seubert, A. The mechanism of action of MF59-an innately attractive adjuvant formulation. Vaccine 2012, 30, 4341–4348. [Google Scholar] [CrossRef]
- Khurana, S.; Verma, N.; Yewdell, J.W.; Hilbert, A.K.; Castellino, F.; Lattanzi, M.; Del Giudice, G.; Rappuoli, R.; Golding, H. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci. Transl. Med. 2011, 3, 85ra48. [Google Scholar] [CrossRef]
- Galli, G.; Medini, D.; Borgogni, E.; Zedda, L.; Bardelli, M.; Malzone, C.; Nuti, S.; Tavarini, S.; Sammicheli, C.; Hilbert, A.K.; et al. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc. Natl. Acad. Sci. USA 2009, 106, 3877–3882. [Google Scholar] [CrossRef]
- Mosca, F.; Tritto, E.; Muzzi, A.; Monaci, E.; Bagnoli, F.; Iavarone, C.; O’Hagan, D.; Rappuoli, R.; De Gregorio, E. Molecular and cellular signatures of human vaccine adjuvants. Proc. Natl. Acad. Sci. USA 2008, 105, 10501–10506. [Google Scholar] [CrossRef]
- Caproni, E.; Tritto, E.; Cortese, M.; Muzzi, A.; Mosca, F.; Monaci, E.; Baudner, B.; Seubert, A.; De Gregorio, E. MF59 and Pam3CSK4 boost adaptive responses to influenza subunit vaccine through an IFN type I-independent mechanism of action. J. Immunol. 2012, 188, 3088–3098. [Google Scholar] [CrossRef]
- Garcon, N.; Vaughn, D.W.; Didierlaurent, A.M. Development and evaluation of AS03, an Adjuvant System containing alpha-tocopherol and squalene in an oil-in-water emulsion. Expert Rev. Vaccines 2012, 11, 349–366. [Google Scholar] [CrossRef]
- Morel, S.; Didierlaurent, A.; Bourguignon, P.; Delhaye, S.; Baras, B.; Jacob, V.; Planty, C.; Elouahabi, A.; Harvengt, P.; Carlsen, H.; et al. Adjuvant System AS03 containing alpha-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine 2011, 29, 2461–2473. [Google Scholar] [CrossRef]
- Didierlaurent, A.M.; Morel, S.; Lockman, L.; Giannini, S.L.; Bisteau, M.; Carlsen, H.; Kielland, A.; Vosters, O.; Vanderheyde, N.; Schiavetti, F.; et al. AS04, an aluminum salt-and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J. Immunol. 2009, 183, 6186–6197. [Google Scholar] [CrossRef]
- Anderson, J.; Olafsdottir, T.A.; Kratochvil, S.; McKay, P.F.; Ostensson, M.; Persson, J.; Shattock, R.J.; Harandi, A.M. Molecular Signatures of a TLR4 Agonist-Adjuvanted HIV-1 Vaccine Candidate in Humans. Front. Immunol. 2018, 9, 301. [Google Scholar] [CrossRef]
- Olafsdottir, T.A.; Lindqvist, M.; Nookaew, I.; Andersen, P.; Maertzdorf, J.; Persson, J.; Christensen, D.; Zhang, Y.; Anderson, J.; Khoomrung, S.; et al. Comparative Systems Analyses Reveal Molecular Signatures of Clinically tested Vaccine Adjuvants. Sci. Rep. 2016, 6, 39097. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, P.; Kim, Y.; Haste-Andersen, P.; Beaver, J.; Bourne, P.E.; Bui, H.H.; Buus, S.; Frankild, S.; Greenbaum, J.; et al. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008, 36, W513–W518. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Guan, P.; Flower, D.R. EpiJen: A server for multistep T cell epitope prediction. BMC Bioinform. 2006, 7, 131. [Google Scholar] [CrossRef]
- Zhang, L.; Udaka, K.; Mamitsuka, H.; Zhu, S. Toward more accurate pan-specific MHC-peptide binding prediction: A review of current methods and tools. Brief. Bioinform. 2012, 13, 350–364. [Google Scholar] [CrossRef]
- Singh, H.; Raghava, G.P. ProPred: Prediction of HLA-DR binding sites. Bioinformatics 2001, 17, 1236–1237. [Google Scholar] [CrossRef]
- El-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit 2008, 21, 243–255. [Google Scholar] [CrossRef]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.; et al. Meta-and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef]
- Joshi-Tope, G.; Gillespie, M.; Vastrik, I.; D’Eustachio, P.; Schmidt, E.; de Bono, B.; Jassal, B.; Gopinath, G.R.; Wu, G.R.; Matthews, L.; et al. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005, 33, D428–D432. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Sherman, B.T.; Huang da, W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Andorf, S.; Gomes, L.; Dunn, P.; Schaefer, H.; Pontius, J.; Berger, P.; Desborough, V.; Smith, T.; Campbell, J.; et al. ImmPort: Disseminating data to the public for the future of immunology. Immunol. Res. 2014, 58, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Brusic, V.; Gottardo, R.; Kleinstein, S.H.; Davis, M.M.; Hafler, D.A.; Quill, H.; Palucka, A.K.; Poland, G.A.; Pulendran, B.; Reinherz, E.L.; et al. Computational resources for high-dimensional immune analysis from the Human Immunology Project Consortium. Nat. Biotechnol. 2014, 32, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Zalocusky, K.A.; Kan, M.J.; Hu, Z.; Dunn, P.; Thomson, E.; Wiser, J.; Bhattacharya, S.; Butte, A.J. The 10,000 Immunomes Project: Building a Resource for Human Immunology. Cell Rep. 2018, 25, 1995. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
| Online Resource | Application & URL | References |
|---|---|---|
| Antigen Prediction | ||
| IEDB | http://www.iedb.org/ | Zhang et al., 2008 [44,45] |
| EpiJen | http://www.ddg-pharmfac.net/epijen/EpiJen/EpiJen.htm | Doytchinova et al., 2006 [46] |
| MULTIPRED2 | http://cvc.dfci.harvard.edu/multipred2/index.php | Zhang et al., 2011 [47] |
| Propred | http://crdd.osdd.net/raghava/propred/ | Singh et al., 2001 [48] |
| Bcepred | http://crdd.osdd.net/raghava/bcepred/ | Saha et al., 2004 [49] |
| Gene Set Enrichment and Network Analysis | ||
| Metascape | http://metascape.org/gp/index.html#/main/step1 | Tripathi et al., 2015 [50] |
| Reactome | https://reactome.org/ | Joshi-Tope et al., 2005 [51] |
| Cytoscape | https://cytoscape.org/ | Shannon et al., 2003 [52] |
| DAVID | https://david.ncifcrf.gov/ | Jiao et al., 2011 [53] |
| Data repositories and Analysis tools | ||
| ImmPort | https://www.immport.org/home | Bhattacharya et al., 2014 [54] |
| Immunespace | https://www.immunespace.org/ | Brusic et al., 2014 [55] |
| 10,000 Immunomes | http://10kimmunomes.ucsf.edu/ | Zalocusky et al., 2018 [56] |
| MSigDB | http://software.broadinstitute.org/gsea/msigdb | Liberzon et al., 2011 [25] |
| Gene Expression Omnibus | https://www.ncbi.nlm.nih.gov/geo/ | Edgar et al., 2002 [57] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, M.; Krammer, F.; García-Sastre, A.; Tripathi, S. Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era. Vaccines 2019, 7, 89. https://doi.org/10.3390/vaccines7030089
Sharma M, Krammer F, García-Sastre A, Tripathi S. Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era. Vaccines. 2019; 7(3):89. https://doi.org/10.3390/vaccines7030089
Chicago/Turabian StyleSharma, Mansi, Florian Krammer, Adolfo García-Sastre, and Shashank Tripathi. 2019. "Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era" Vaccines 7, no. 3: 89. https://doi.org/10.3390/vaccines7030089
APA StyleSharma, M., Krammer, F., García-Sastre, A., & Tripathi, S. (2019). Moving from Empirical to Rational Vaccine Design in the ‘Omics’ Era. Vaccines, 7(3), 89. https://doi.org/10.3390/vaccines7030089







